Abstract
Plasmodium falciparum parasites are responsible for the major global disease malaria, which results in >2 million deaths each year. With the rise of drug-resistant malarial parasites, novel drug targets and lead compounds are urgently required for the development of new therapeutic strategies. Here, we address this important problem by targeting the malarial neutral aminopeptidases that are involved in the terminal stages of hemoglobin digestion and essential for the provision of amino acids used for parasite growth and development within the erythrocyte. We characterize the structure and substrate specificity of one such aminopeptidase, PfA-M1, a validated drug target. The X-ray crystal structure of PfA-M1 alone and in complex with the generic inhibitor, bestatin, and a phosphinate dipeptide analogue with potent in vitro and in vivo antimalarial activity, hPheP[CH2]Phe, reveals features within the protease active site that are critical to its function as an aminopeptidase and can be exploited for drug development. These results set the groundwork for the development of antimalarial therapeutics that target the neutral aminopeptidases of the parasite.
Keywords: drug design, malaria, structural biology, protease
There are 300–500 million cases of clinical malaria annually, and 1.4–2.6 million deaths. Malaria is caused by apicomplexan parasites of the genus Plasmodium, with Plasmodium falciparum the most lethal of the 4 species that infect humans. Clinical manifestations begin when parasites enter erythrocytes, and most antimalaria drugs, such as chloroquine, exert their action by preventing the parasite development within these cells (1). As a result of the rapid spread of drug-resistant parasites, there is a constant need to identify and validate new antimalarial targets.
Intraerythrocytic parasites have limited capacity for de novo amino acid synthesis and rely on degradation of host hemoglobin (Hb) to maintain protein metabolism and synthesis, and an osmotically stable environment within the erythrocyte (1–4). Within the erythrocytes, malaria parasites consume as much as 75% of the cellular Hb (1). Hb is initially degraded by the concerted action of cysteine-, aspartyl-, and metallo-endoproteases, and a dipeptidase (cathepsin C) within a digestive vacuole (DV) to di- and tripeptide fragments (5, 6). These fragments are suggested to be exported to the parasite cytoplasm, where further hydrolysis to release free amino acids takes place [supporting information (SI) Fig. S1; see refs. 7 and 8].
The release of amino acids involves 2 metallo-exopeptidases: an alanyl aminopeptidase, PfA-M1 (9, 10), and a leucine aminopeptidase, PfA-M17 (7, 11, 12). We have demonstrated that the aminopeptidase inhibitor bestatin, an antibiotic and natural analogue of the dipeptide Phe-Leu derived from the fungus Streptomyces olivoretticul, prevents P. falciparum malaria growth in culture (13, 14). More recently, it was shown not only that synthetic phosphinate dipeptide analogues that inhibit metallo-aminopeptidases prevented the growth of wild-type and the chloroquine-resistant parasites in culture but also that one compound, hPheP[CH2]Phe (termed compound 4, Co4), reduced a murine infection of Plasmodium chabaudi chabaudi by 92% compared with controls (15, 16). Importantly, Co4 was found to cause no toxicity in these in vivo studies (16). Here, we functionally characterize PfA-M1 and validate it as a target for the inhibitory activity of bestatin and hPheP[CH2]Phe. We also present its 3D structure alone and complexed with both of these inhibitors, which sets the groundwork for the development of a previously undescribed class of antimalarial drugs.
Results and Discussion
Substrate Specificity of the P. falciparum Alanyl Aminopeptidase PfA-M1.
The M1 alanyl aminopeptidase gene (MAL13P1.56) (9), as annotated by PlasmoDB, is present in single copy and located on chromosome 13 of P. falciparum. The gene is 3,257 bp in length and encodes a protein, known as PfA-M1, of 1,085 aa and 126 kDa. The full-length amino acid sequence of PfA-M1 exhibits ≈70% sequence similarity with M1 aminopeptidase orthologues of the various rodent malaria species (Plasmodium berghei, P. chabaudi chabaudi, and Plasmodium yoelii); the sequences are most divergent at the large nonconserved N-terminal extension (≈194 aa), which contains 3 asparagine-rich low-complexity regions (LCRs) and a putative transmembrane domain (Fig. S2).
Using peptide antisera, Florent et al. (9) and Florent and coworkers (10), detected a 122-kDa PfA-M1 in membrane fractions of malaria parasites. However, this membrane form appeared to be further processed to smaller active soluble forms of 96 and 68 kDa. In our studies, expression of the full-length PfA-M1 was not successful, so a truncated form of the P. falciparum M1 aminopeptidase (residues 195-1085) correlating with the start of the M1 aminopeptidase of Escherichia coli PepN was prepared (17, 18). The construct lacked the 3 asparagine-rich LCRs and the putative transmembrane domain (Fig. S2) but was successfully expressed in E. coli and extracted as a soluble functional enzyme. The protein resolved as a major band at ≈100 kDa (predicted molecular mass 104.678 kDa) with a minor N-terminally truncated breakdown product of ≈55 kDa on reducing SDS/PAGE (Fig. S3).
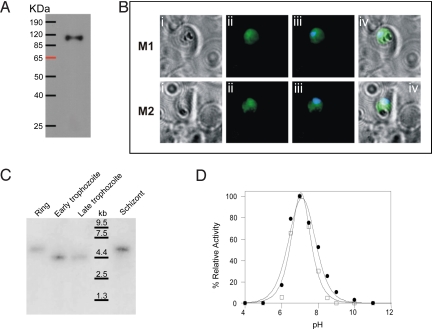
The purified recombinant PfA-M1 (rPfA-M1) exhibited a similar retention time on HPLC size analysis as the PfA-M1 in soluble extracts of P. falciparum parasites, both eluting between 80 and 100 kDa (Fig. S3). In support of the studies by Florent et al. (9), and Florent and coworkers (10), PfA-M1 antisera localized the enzyme to the parasite cytosol (data not shown). Also, P. falciparum D10 parasites transfected with a plasmid (pHTB-PfA-M1-cmycB) carrying the truncated PfA-M1 gene expressed and trafficked an ≈115-kDa product (Fig. 1A) to the parasite cytosol (Fig. 1B). These transgenic parasites exhibited a 2.8-fold higher level of PfA-M1 activity compared with D10 wild-type parasites, showing that the transgene product was functionally active within the parasite. Northern blotting experiments showed that the PfA-M1 is expressed at all stages in the development of the malaria parasites in the erythrocyte (Fig. 1C).
Fig. 1.
Analysis of PfA-M1 expression and activity. (A) Western blot of transgenic parasites expressing the product of the inserted transgene encoding PfA-M1. The blot was probed with a monoclonal anti-c-myc primary antibody followed by horseradish peroxidase-conjugated anti-mouse immunoglobulin antibodies and visualized by enhanced chemiluminescence. (B) Indirect immunofluorescence of transgenic parasites stained with monoclonal anti c-myc primary antibody followed by anti-mouse cy2. (i) Bright field; (ii) anti-c-myc antibody; (iii) anti-c-myc/nuclear stain merged; (iv) merge of i and iii. The data show that the PfA-M1 transgenic protein is localized to the parasite cytosol. (C) Northern blot analysis of stage-specific parasite RNA reveals that the endogenous PfA-M1 is expressed by parasites at all developmental stages within the erythrocyte. Developmental stages are indicated at the top: R, ring stage parasites; ET, early trophozoite parasites; LT, late trophozoite parasites; and S, schizont stage parasites. Size difference observed between trophozoite and ring/schizont is due to different transcription initiation sites. (D) The pH optima for activity of rPfA-M1 (circles) and native PfA-M1 in soluble extracts of parasites (squares) measured by using the fluorogenic peptides substrate H-Arg-NHMec.
The rPfA-M1 protein displayed a broad specificity for amino acids, demonstrated by its ability to cleave peptide bonds involving hydrophobic, basic, and aromatic amino acids, which is consistent with M1 aminopeptidases of E. coli PepN (17, 18) and Salmonella typhimurium PepN (19). The most efficiently cleaved P1 substrates were Leu (kcat/Km = 4,607 M−1·s−1), Ala (kcat/Km = 2,295 M−1·s−1), Arg (kcat/Km = 1,491 M−1·s−1), and Phe (kcat/Km = 924 M−1·s−1) (Table S1). However, proline was a poor substrate with kcat/Km a least 1,000-fold less than Leu (Table S1), consistent with the catalytic mechanism described by Ito et al. (20), who suggest that 2 conserved glutamate residues stabilize the reaction intermediate through the formation of hydrogen bonds with the terminal amino group, NH3. The presence of a proline in the P1 position may prevent such interactions forming due to the constraints imposed by the cyclized side chain. The rPfA-M1 protein did not cleave after the acidic amino acids Glu and Asp, which is consistent with our suggestion that another enzyme, a cytosolic M18 aspartyl aminopeptidase, is responsible for the specific removal of these residues from peptides (21).
The efficient cleavage of the amino acid Arg is of particular interest, because this amino acid is not cleaved by the other major malaria aminopeptidase, PfA-M17 (12). This observation facilitated the development of a specific assay for measuring PfA-M1 in malaria extracts. Thus, by using Arg as a substrate, we showed that rPfA-M1 and native PfA-M1 activity within soluble extracts of malaria parasites exhibited optimal activity at pH 7.0, with <20% activity below pH 6.0, which is consistent with a cytosolic function for the enzyme (Fig. 1D) (12). Also, both rPfA-M1 and native PfA-M1 activity depends on a divalent metal ion as metal chelators (1 mM EDTA or o-phenanthroline) were found to inhibit their activity. Collectively, the above data demonstrate that despite lacking the N-terminal domain, rPfA-M1 exhibits physicobiochemical characteristics identical to those of the native soluble malaria PfA-M1 enzyme.
X-Ray Crystal Structure of rPfA-M1 Confirms Bacterial Aminopeptidase Fold.
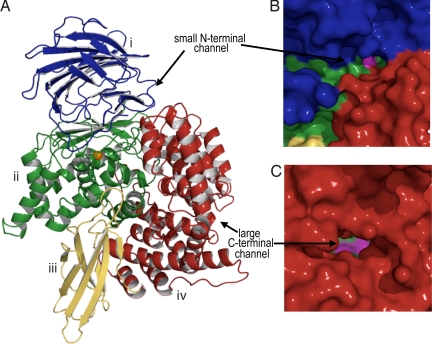
Outside malaria, PfA-M1 shows closest identity (≈35%) to the bacterial M1 aminopeptidases, which do not contain the extended N-terminal domain. We determined the X-ray crystal structure rPfA-M1 to 2.1 Å (Table S2). The structure confirmed that rPfA-M1 adopts the bacterial aminopeptidase N-fold (20, 22, 23), and comprises 26 α-helices and 7 β-sheets divided into 4 domains (Fig. 2A). The catalytic domain II (residues 392–649) adopts a thermolysin-like fold and contains the active site, incorporating the zinc-binding motif H496EYFHX17KE519 and the well-conserved G490AMEN motif involved in substrate recognition (20, 22, 23). The catalytic zinc ion is coordinated by Nε2 atoms of His496 and His500, the carboxyl Oε atom of Glu519, and a water molecule that acts as the nucleophile that attacks the carbonyl carbon of the substrate (20). This water molecule forms a slightly longer metallo-bond with the zinc ion and is also coordinated by Glu497 and Glu463. It is not clear what the catalytic base of the reaction is, but the structure suggests that it may be the Glu497 residue.
Fig. 2.
The structure of rPfA-M1. (A) Diagram of unbound rPfA-M1 colored by domain: I (blue), II (green), III (yellow), and IV (red). (B and C) Molecular surface of PfA-M1 (colored as in A) showing small (B) and large (C) openings to active site (active site cavity shown in magenta).
Inspection of the molecular surface of PfA-M1 reveals 2 openings to the active site cavity. The first opening (N-terminal channel) comprises a shallow 8-Å-long groove at the junction of domains I and IV (Fig. 2B). The second and larger opening (C-terminal channel) is formed by the C-terminal domain IV, which comprises 8 pairs of α-helices arranged in 2 layers to form a cone-shaped superhelical structure. This domain interacts with the catalytic domain II and contains an ≈30-Å-long channel leading toward the active site (Fig. 2C). At the entrance, a helix (α14) with a 90° bend confines the pore size to ≈13-Å diameter.
The rPfA-M1 Has a Buried Active Site That Does Not Require Conformational Change to Bind Substrate.
Bestatin is an antibiotic originally isolated from filtrates of the fungus S. olivoretticul, but is now available in synthetic form (24). Its structure N-[(2S,3R)-3-amino-2-hydroxy-4-phenylbutanoyl]-l-leucine is an analogue of the dipeptide Phe-Leu. Thus, bestatin is a competitive inhibitor of many metallo-aminopeptidases (24). Accordingly, kinetic studies performed with rPfA-M1 revealed that bestatin is an effective inhibitor of rPfA-M1, with an inhibitory constant Ki = 478.2 nM.
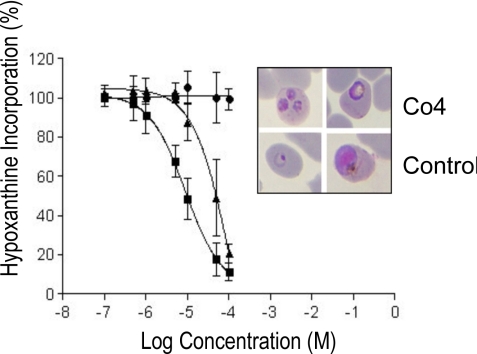
More recently, a new class of metallo-aminopeptidase inhibitors, phosphinate dipeptide analogues, were designed by using a computer-aided approach by Grembecka et al. (15); 1 of these hPheP[CH2]Phe, Co4, exhibited more potent inhibition of rPfA-M1 (Ki = 79 nM) than bestatin. Both bestatin and Co4 had similar killing activity against malaria parasites in culture; IC50 bestatin range 8–14 μM and Co4 is 24–62 μM (Fig. 3). However, bestatin only showed low-level (33%) antimalarial activity in murine (P. c. chabaudi) models, whereas Co4 was much more potent and reduced infection by 92% compared with controls (16).
Fig. 3.
The inhibitory effect of the aminopeptidase inhibitor bestatin (squares), and Co4 (triangles) on P. falciparum clone 3D7 growth in culture compared with parasites grown in the absence of inhibitor (circles). Data are presented as mean ± SD of 3 independent, triplicate experiments. Parasites grown in the presence of bestatin or Co4 for 24 h exhibit cellular damage and stunted development compared with control parasites grown in the absence of drug (Inset).
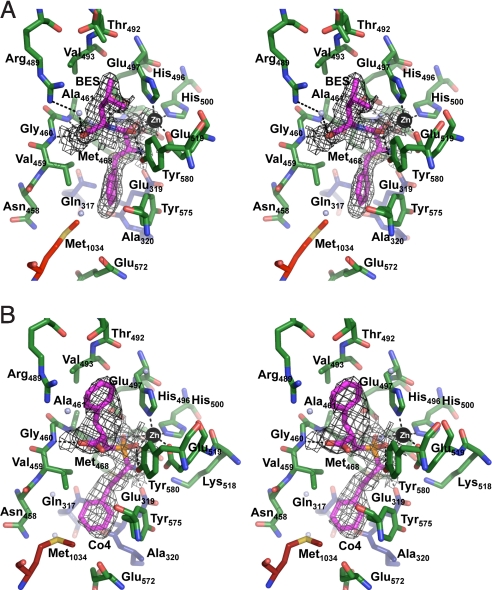
To elucidate the mechanism by which bestatin and Co4 bound to the active site of rPfA-M1, we determined the X-ray crystal structure of rPfA-M1 complexed to each inhibitor to 1.65 and 2.0 Å, respectively (Table S2). Given that the active site of the enzyme is buried in the middle of the protein, we were particularly interested in understanding whether conformational change is required for substrate/inhibitor entry. Interestingly, comparison of the unbound and ligand-bound structures revealed that the enzyme does not undergo any global conformational rearrangements on binding either inhibitor [rmsd of 0.14 (bestatin) and 0.16 Å (Co4) >890 Cα atoms]. The omit electron density of ligands within the active site was well defined (Fig. 4), and the dipeptide analogues slot neatly into the large catalytic cavity without causing any major alterations to active site residues.
Fig. 4.
Stereo diagram of inhibitors binding to active site of rPfA-M1. (A) 1.65-Å rPfA-M1-BES and (B) 2.0-Å rPfA-M1-Co4 active site showing inhibitors bound in the active site. Inhibitors (BES/Co4) are colored in magenta. Carbon atoms of residues are colored by domain: I (blue), II (green), IV (red), and Zn is shown as black sphere. Water molecules are shown as blue spheres. Hydrogen bonds are indicated (dashed lines). Electron density is a composite omit map contoured at 1.0 σ calculated by using a model containing only PfA-M1 atoms (no zinc, ligand, or water was included in the calculation).
In E. coli aminopeptidase N protein Met454, positioned immediately preceding the GAMEN exopeptidase motif, is postulated to function as a cushion to accept substrates (20), altering the size of the active site pocket. This residue is conserved in bacterial aminopeptidases; however, the equivalent position (469) in eukaryote M1 aminopeptidases is commonly a smaller valine or alanine residue. We noted no movement of the rPfA-M1 Val469 residue to accommodate either the bestatin or Co4 molecule. Indeed, only 2 residues showed significant movement after ligand binding, the Glu526 side chain that moves away from the active site, removing what would otherwise form a close contact with P1′ position of either inhibitor (Leu moiety of bestatin or the Phe ring of Co4). However, in the unbound form of rPfA-M1, the Met1034 residue adopts 2 alternative conformations in the ligand-bound structures, this side chain adopts a single conformation and forms packing interactions with the inhibitor moieties (P1 position).
The lack of any large scale conformational changes in the bound structure suggests that it is unlikely a conformational change is required for active site access. Indeed, initial attempts to obtain data from a ligand-bound complex involved soaking the inhibitor into rPfA-M1 crystals. This procedure caused no deterioration of crystal quality (however, it did result in variable occupancy of the active site, hence cocrystallization was used), also indicating that it is unlikely that large-scale rearrangements of the protein molecules are required for substrate/inhibitor binding. Therefore, we suggest that the active site of this monomeric aminopeptidase is buried, substrate access is achieved by means of the C-terminal domain vortex, and that control of substrate hydrolysis can be achieved, and depends on, the size of this channel.
Both inhibitors primarily interact with the enzyme through a pentahedral coordination with the catalytic zinc ion (Fig. 4). Aside from bonds with His496, His500, and Glu519, the hydroxyl group (O2) and the carbonyl group (O3) of bestatin and the central phosphoryl O atoms of Co4 are coordinated to the zinc ion (Fig. S4). Pentahedral zinc coordination, rather than the tetrahedral geometry observed in the unbound structure, is required for the transition state of the enzyme that exists after the nucleophilic attack at the carbonyl carbon of the substrate. The bestatin carbonyl carbon (O3) and the Co4 central phosphoryl O atoms (O3 and O4; Fig. S4) also form hydrogen bonds with the side chain of Tyr580, stabilizing this reaction intermediate. A cis-peptide (Glu319-Ala320) allows the side chain of Glu319 to extend into the active site, where it forms a hydrogen bond with the N2 atom of bestatin and the amino group (NH2) of Co4 (Fig. 4). The GAMEN recognition motif residues also contribute hydrogen bonds to ligand binding with the side chain of Glu463 and main-chain amide of Gly460, with both inhibitors (Fig. 4). Bestatin also forms a hydrogen bond with the main-chain amide of Ala461 (Table S3). When bound, Co4 resides in an extended conformation (≈11 Å from Phe-ring centroids) and has a total contact area of 19.9Å2 (compared with bestatin total buried surface area of 17.7 Å2). The 2 Phe rings of Co4 form favorable hydrophobic interactions; the Phe ring at P1′ position (C1-C6, Fig. S4) packs against side-chains of residues Arg489, Thr492, and Val493, whereas the Phe ring at position P1 (C10-C16, Fig. S4) forms hydrophobic contacts with side chains of Gln317, Val459, Met462, Tyr575, and Met1034. In bestatin, the Leu moiety is flanked by Val459 and Thr492; 2 water molecules also coordinate the O1 atom (bestatin) with the Nη atoms of Arg489. The Phe-like amino acid is cushioned by hydrophobic interactions with Gln317, Met462, Tyr575, and Met1034. In total, bestatin makes 8 hydrogen bonds with the active site, whereas Co4 forms only 6 hydrogen bonds (Table S3). However, Co4 is further stabilized by 10 residues contributing to its hydrophobic packing within in the active site core in contrast to bestatin, which has only 7 such interactions with only 1 residue (Thr492), contributing to the stability of the leucyl moiety in the P1′ position.
Bestatin is a natural analogue of the dipeptide Phe-Leu, but is a much weaker inhibitor of PfA-M1 than Co4. When analyzing the active site contacts of the 2 ligands, a possible distinction between potency to Co4 of bestatin may be due to a lack of contacts made by the leucyl side chain of bestatin, leaving a open hydrophobic cleft that is filled when Co4 is bound. Interestingly, the phosphinate dipeptide hPheP[CH2]Gly (Co2) is a significantly poorer inhibitor than Co4 [having a 13-fold greater Ki of 1,030.0 nM than Co4 (hPheP[CH2]Phe)] (16). We suggest that this reduced potency is probably also due to the lack of contacts at the P1′ position within this large active-site cavity. Because of its high Ki, compound hPheP[CH2]Gly does not exhibit antimalarial activity in vitro. However, these data clearly highlight the impact such ligand-binding analysis may have for structure-based drug design in conjunction with biological analysis.
The active site of PfA-M1 is buried deep in the structure, providing a solution to the problem of controlling appropriate substrate entry, as well as preventing hydrolysis of unwanted substrates. By comparison with the available bacterial aminopeptidases structures (20, 22, 23), PfA-M1 can, thus, be considered to adopt a “closed” conformation. Interestingly, structural studies on the homologous tricorn-interacting factor 3, F3, reveals this protease undergoes a substantial conformational changes on substrate entry, where domain IV swings away from domain II, exposing the active site. However, for the aminopeptidase N structures available, including PfA-M1, there is no movement of the C-terminal domain noted, even in the presence of inhibitors. We argue that in PfA-M1, the large C-terminal channel functions to permit substrate entry whereby Hb-derived peptides access the buried active site leaving the smaller sized opening for exit of released amino acids. A maximal pore size of 10 Å has been proposed to serve as the entry point in other multimeric proteases structures (22, 25); here, we suggest that for such an opening is sufficient to allow access to the buried active site of this monomeric protease.
Given our detailed knowledge of the active site of the PfA-M1 enzyme, high-throughput screening of chemical libraries followed by medicinal chemistry would allow the development of additional lead antimalarial compounds. The static nature of this family of proteases allows the opportunity for a 2-fold approach to rational drug design; discovery of lead compounds that bind to and irreversibly block the active site, and/or compounds that block egress of digestion products by occluding the N-terminal channel, both essentially trapping the molecule in an inactive state.
Conclusions
There is a paucity of new antimalarial drugs entering the development pipeline. Our report of the unique active site structure of PfA-M1, in complex with the antimalarial inhibitors bestatin and Co4, provides the groundwork for the de novo discovery of a previously unrecognized class of antimalarials by using high-throughput chemical screening and medicinal chemistry. Co4 also inhibits the second important neutral aminopeptidase of malaria, PfA-M17 (12, 26), creating the possibility of developing a 2-target or combination therapy that would be more resilient to the emergence of drug-resistant malaria parasites.
Materials and Methods
Parasite Preparation.
P. falciparum clone D10 was cultured as described (27). For experiments investigating the stage specific expression of PfA-M1, parasites were synchronized by using 2 rounds of sorbitol treatment (28), and stage specific parasites harvested at ring, trophozoite, and schizont stage. Details of parasites, and methods of immunoblotting, Northern blotting, and transfection can be found in SI Methods.
Enzymatic Analysis.
Aminopeptidase activity was determined by measuring the release of the fluorogenic leaving group, 7-amino-4-methyl-coumarin (NHMec) from the fluorogenic peptide substrates (SI Methods).
In Vitro Sensitivity of P. falciparum Malaria Parasites to Aminopeptidase Inhibitors.
The in vitro sensitivity of each parasite population to bestatin, Co4 and Co2 was determined by using [3H]-hypoxanthine incorporation (for further details see SI Methods) (29). IC50 values were determined by linear interpolation of inhibition curves (SI Methods).
Crystallization, X-Ray Data Collection, Structure Determination, and Refinement.
A truncated form of PfA-M1 (residues 195-1085, rPfA-M1) was purified from E. coli (SI Methods); rPfA-M1 at 5 mg/mL in 50 mM Hepes pH 8.5; 300 mM NaCl 5% (vol/vol) glycerol was crystallized in 22% (vol/vol) polyethylene glycol 8000, 10% (vol/vol) glycerol, 0.1 M Tris (pH 8.5), and 0.2 M MgCl2. Crystals of the ligand bound rPfA-M1 complexes were obtained by cocrystallization under similar conditions in the presence of 1 mM ligand. The diffraction data for the unbound, bestatin-bound, and Co4-bound protease were collected to 2.1-, 1.65-, and 2.0-Å resolution, respectively (Table S2). Crystallographic analysis was performed by using CCP4i (30–33). The structure was determined by using the program PHASER (34) (using 2GTQ.pdb as a search probe; see ref. 23). Refinement was performed by using REFMAC (35). All model building was done by using COOT (for further crystallization details, see SI Methods) (36).
Supplementary Material
Acknowledgments.
J.C.W. is an Australian Research Council (ARC) Federation Fellow; A.M.B. is a National Health and Medical Research Council (NHMRC) Senior Research Fellow; and C.J.P. is a NHMRC Training (Peter Doherty) Fellow. J.P.D., K.R.T., and D.G. are supported by the ARC Discovery Project Grant DP0666128; K.R.T. and D.G. by the NHMRC Program Grant 290208 and by generous donation from Mark Nicholson, Alice Hill, and the Tudor Foundation; and T.S-A. by a University of Queensland Postdoctoral Fellowship and a Ramaciotti Development grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3EBG, 3EBH, and 3EBI).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807398106/DCSupplemental.
References
- 1.Rosenthal PJ. Antimalarial drug discovery: Old and new approaches. J Exp Biol. 2003;206:3735–3744. doi: 10.1242/jeb.00589. [DOI] [PubMed] [Google Scholar]
- 2.Divo AA, Geary TG, Jensen JB. Oxygen- and time-dependent effects of antibiotics and selected mitochondrial inhibitors on Plasmodium falciparum in culture. Antimicrob Agents Chemother. 1985;27:21–27. doi: 10.1128/aac.27.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lew VL, Macdonald L, Ginsburg H, Krugliak M, Tiffert T. Excess hemoglobin digestion by malaria parasites: A strategy to prevent premature host cell lysis. Blood Cells Mol Dis. 2004;32:353–359. doi: 10.1016/j.bcmd.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc Natl Acad Sci USA. 2006;103:8840–8845. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemba M, Gluzman I, Goldberg DE. A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. J Biol Chem. 2004;279:43000–43007. doi: 10.1074/jbc.M408123200. [DOI] [PubMed] [Google Scholar]
- 6.Rosenthal PJ. Hydrolysis of erythrocyte proteins by proteases of malaria parasites. Curr Opin Hematol. 2002;9:140–145. doi: 10.1097/00062752-200203000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Curley GP, et al. Aminopeptidases from Plasmodium falciparum, Plasmodium chabaudi chabaudi and Plasmodium berghei. J Eukaryot Microbiol. 1994;41:119–123. doi: 10.1111/j.1550-7408.1994.tb01483.x. [DOI] [PubMed] [Google Scholar]
- 8.Kolakovich KA, Gluzman IY, Duffin KL, Goldberg DE. Generation of hemoglobin peptides in the acidic digestive vacuole of Plasmodium falciparum implicates peptide transport in amino acid production. Mol Biochem Parasitol. 1997;87:123–135. doi: 10.1016/s0166-6851(97)00062-5. [DOI] [PubMed] [Google Scholar]
- 9.Florent I, et al. A Plasmodium falciparum aminopeptidase gene belonging to the M1 family of zinc-metallopeptidases is expressed in erythrocytic stages. Mol Biochem Parasitol. 1998;97:149–160. doi: 10.1016/s0166-6851(98)00143-1. [DOI] [PubMed] [Google Scholar]
- 10.Allary M, Schrevel J, Florent I. Properties, stage-dependent expression and localization of Plasmodium falciparum M1 family zinc-aminopeptidase. Parasitology. 2002;125:1–10. doi: 10.1017/s0031182002001828. [DOI] [PubMed] [Google Scholar]
- 11.Gavigan CS, Dalton JP, Bell A. The role of aminopeptidases in hemoglobin degradation in Plasmodium falciparum-infected erythrocytes. Mol Biochem Parasitol. 2001;117:37–48. doi: 10.1016/s0166-6851(01)00327-9. [DOI] [PubMed] [Google Scholar]
- 12.Stack CM, et al. Characterization of the Plasmodium falciparum M17 leucyl aminopeptidase. A protease involved in amino acid regulation with potential for antimalarial drug development. J Biol Chem. 2007;282:2069–2080. doi: 10.1074/jbc.M609251200. [DOI] [PubMed] [Google Scholar]
- 13.Gavigan CS, Machado SG, Dalton JP, Bell A. Analysis of antimalarial synergy between bestatin and endoprotease inhibitors using statistical response-surface modelling. Antimicrob Agents Chemother. 2001;45:3175–3181. doi: 10.1128/AAC.45.11.3175-3181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nankya-Kitaka MF, Curley GP, Gavigan CS, Bell A, Dalton JP. Plasmodium chabaudi chabaudi and P. falciparum: Inhibition of aminopeptidase and parasite growth by bestatin and nitrobestatin. Parasitol Res. 1998;84:552–558. doi: 10.1007/s004360050447. [DOI] [PubMed] [Google Scholar]
- 15.Grembecka J, Mucha A, Cierpicki T, Kafarski P. The most potent organophosphorus inhibitors of leucine aminopeptidase. Structure-based design, chemistry, and activity. J Med Chem. 2003;46:2641–2655. doi: 10.1021/jm030795v. [DOI] [PubMed] [Google Scholar]
- 16.Skinner-Adams TS, et al. Identification of phosphinate dipeptide analog inhibitors directed against the Plasmodium falciparum M17 leucine aminopeptidase as lead antimalarial compounds. J Med Chem. 2007;50:6024–6031. doi: 10.1021/jm070733v. [DOI] [PubMed] [Google Scholar]
- 17.Chandu D, Nandi D. PepN is the major aminopeptidase in Escherichia coli: Insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology. 2003;149:3437–3447. doi: 10.1099/mic.0.26518-0. [DOI] [PubMed] [Google Scholar]
- 18.Golich FC, Han M, Crowder MW. Over-expression, purification, and characterization of aminopeptidase N from Escherichia coli. Protein Expr Purif. 2006;47:634–639. doi: 10.1016/j.pep.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Nandi D. Characterization and role of peptidase N from Salmonella enterica serovar Typhimurium. Biochem Biophys Res Commun. 2007;353:706–712. doi: 10.1016/j.bbrc.2006.12.073. [DOI] [PubMed] [Google Scholar]
- 20.Ito K, et al. Crystal structure of aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli and conformational change of methionine 260 involved in substrate recognition. J Biol Chem. 2006;281:33664–33676. doi: 10.1074/jbc.M605203200. [DOI] [PubMed] [Google Scholar]
- 21.Teuscher F, et al. The M18 aspartyl aminopeptidase of the human malaria parasite Plasmodium falciparum. J Biol Chem. 2007;282:30817–30826. doi: 10.1074/jbc.M704938200. [DOI] [PubMed] [Google Scholar]
- 22.Addlagatta A, Gay L, Matthews BW. Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site. Proc Natl Acad Sci USA. 2006;103:13339–13344. doi: 10.1073/pnas.0606167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nocek B, Mulligan R, Bargassa M, Collart F, Joachimiak A. Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis. Proteins. 2007;273:273–279. doi: 10.1002/prot.21276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scornik OA, Botbol V. Bestatin as an experimental tool in mammals. Curr Drug Metab. 2001;2:67–85. doi: 10.2174/1389200013338748. [DOI] [PubMed] [Google Scholar]
- 25.Russo S, Baumann U. Crystal structure of a dodecameric tetrahedral-shaped aminopeptidase. J Biol Chem. 2004;279:51275–51281. doi: 10.1074/jbc.M409455200. [DOI] [PubMed] [Google Scholar]
- 26.Gardiner DL, Trenholme KR, Skinner-Adams TS, Stack CM, Dalton JP. Overexpression of leucyl aminopeptidase in Plasmodium falciparum parasites. Target for the antimalarial activity of bestatin. J Biol Chem. 2006;281:1741–1745. doi: 10.1074/jbc.M508955200. [DOI] [PubMed] [Google Scholar]
- 27.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 28.Lambros C, Vanderburg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 29.Geary TG, Delaney EJ, Klotz IM, Jensen JB. Inhibition of the growth of Plasmodium falciparum in vitro by covalent modification of hemoglobin. Mol Biochem Parasitol. 1983;9:59–72. doi: 10.1016/0166-6851(83)90057-9. [DOI] [PubMed] [Google Scholar]
- 30.Leslie AGW. Joint CCP4 + ESF-EAMCB Newslett Prot Crystallogr. Warrington, UK: Daresbury Laboratory; 1992. p. 26. [Google Scholar]
- 31.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 32.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 33.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Davis IW, et al. Molprobity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.