Abstract
Medulloblastomas (MBs) are the most common brain tumors in children. Some are thought to originate from cerebellar granule neuron progenitors (GNPs) that fail to undergo normal cell cycle exit and differentiation. Because microRNAs regulate numerous aspects of cellular physiology and development, we reasoned that alterations in miRNA expression might contribute to MB. We tested this hypothesis using 2 spontaneous mouse MB models with specific initiating mutations, Ink4c−/−; Ptch1+/− and Ink4c−/−; p53−/−. We found that 26 miRNAs showed increased expression and 24 miRNAs showed decreased expression in proliferating mouse GNPs and MBs relative to mature mouse cerebellum, regardless of genotype. Among the 26 overexpressed miRNAs, 9 were encoded by the miR-17∼92 cluster family, a group of microRNAs implicated as oncogenes in several tumor types. Analysis of human MBs demonstrated that 3 miR-17∼92 cluster miRNAs (miR-92, miR-19a, and miR-20) were also overexpressed in human MBs with a constitutively activated Sonic Hedgehog (SHH) signaling pathway, but not in other forms of the disease. To test whether the miR-17∼92 cluster could promote MB formation, we enforced expression of these miRNAs in GNPs isolated from cerebella of postnatal (P) day P6 Ink4c−/−; Ptch1+/− mice. These, but not similarly engineered cells from Ink4c−/−; p53−/− mice, formed MBs in orthotopic transplants with complete penetrance. Interestingly, orthotopic mouse tumors ectopically expressing miR-17∼92 lost expression of the wild-type Ptch1 allele. Our findings suggest a functional collaboration between the miR-17∼92 cluster and the SHH signaling pathway in the development of MBs in mouse and man.
Keywords: cerebellum, microRNAs, oncomiR1, granule neuron progenitors
Medulloblastoma (MB), the most common pediatric malignant brain tumor, arises in the cerebellum with at least a subset originating from cerebellar granule neuron progenitor (GNP) cells that fail to properly migrate and differentiate (1, 2). Two inherited cancer syndromes induce MB in humans. Gorlin syndrome is characterized by skeletal abnormalities and large body size in combination with a high incidence of basal cell carcinoma and MB (3). Gorlin patients have mutations in PATCHED (PTCH), the receptor for Sonic Hedgehog (SHH) (4, 5, 6, 7). SHH is the major mitogen for GNP proliferation (8) and mutations in the SHH pathway induce MB in mice (9, 10). Turcots's syndrome is associated with colon cancer and malignant neuroepithelial brain tumors resulting from mutations of the adenomatous polyposis coli (APC) gene. APC is a member of the wingless (WNT) signaling pathway that regulates the proliferation and fate of neural progenitor cells (11, 12).
Both SHH/PTCH and WNT pathways have also been implicated in sporadic disease. Recent studies of a cohort of 46 St. Jude patients identified 5 distinct MB subgroups, on the basis of mRNA and genomic profiling. Two of these contained mutations that activate the SHH/PTCH (∼25%) and WNT (20%) signaling pathways, respectively (13).
The past several years have produced a strong appreciation that changes in noncoding RNAs can also impact tumorigenesis. MicroRNAs are endogenous triggers of the RNAi pathway. These 21–23 nucleotide (nt) RNAs are matured through a 2-step biogenesis mechanism from long, RNA polII transcripts (14). While some microRNAs are encoded from individual transcription units, others reside within introns of protein coding genes or exist as polycistrons that generate multiple microRNAs from a single primary transcript. MiRNAs reduce protein synthesis following recognition of the 3′ untranslated region (3′-UTR) of target genes via Watson–Crick base pairing interactions (15). Many miRNAs are conserved throughout evolution, suggesting that they perform fundamental biological functions during development.
Currently 678 human miRNAs and 472 mouse miRNAs have been confidently identified (miRBASE) (16). MicroRNAs have been linked to the initiation, progression, and metastasis of human malignancies, with some species displaying oncogenic and others tumor suppressive potential (17). MiRNAs are often expressed aberrantly in tumors as compared to normal tissues and are likely to contribute to tumorigenesis by dysregulating critical target genes (18).
The miR-17∼92 cluster, also called Oncomir-1, was among the first miRNAs to be validated as showing oncogenic potential (19). The cluster has 2 paralogs, miR-106a∼363 and miR-106b∼25, that are located on different chromosomes and contain individual miRNAs that are highly similar to those encoded by the miR-17∼92 cluster. Despite their similarities, each of these miRNA clusters has distinct functions (20, 21). For example, miR-17∼92 alone is involved in B cell lymphomagenesis and in lung development (21).
We hypothesized that miRNAs might play an important role in normal cerebellar development and that dysregulation of miRNAs might contribute to MB formation. We found that miR-17∼92 is expressed in the developing mouse cerebellum and in proliferating GNPs but not in postmitotic differentiated neurons. The miR-17∼92 cluster is overexpressed in mouse and human MBs with an aberrantly activated SHH/PTCH signal pathway. Moreover, enforced expression of miR-17∼92 in P6 Ink4c−/−; Ptch1+/− GNPs induced MBs with complete penetrance after orthotopic transplant into the brains of recipient immunocompromised animals. These results provide the first evidence that the SHH/PTCH signaling pathway and miR-17∼92 functionally interact and contribute to MB development in mice and man.
Results
The miR-17∼92 Cluster Family Is Expressed During Mouse Cerebellar Development and in Mouse MBs.
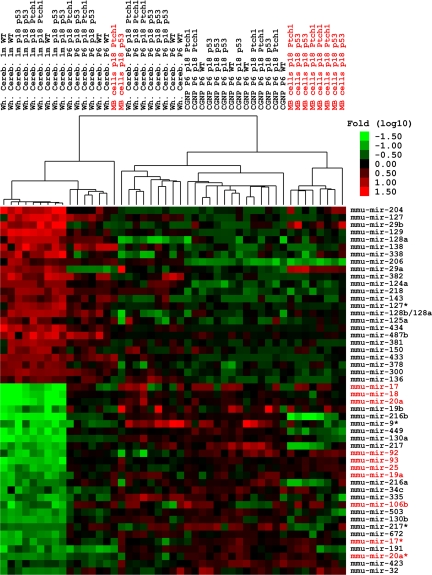
We performed unbiased Illumina deep sequencing and subsequent comparative expression analysis of small RNAs isolated from P6 and 1 month (M) wild-type, Ink4c−/−; p53−/− or Ink4c−/−; Ptch1+/− cerebella and GNPs purified from these tissues. These time points were chosen to coincide with the peak of proliferation of GNPs (P6) and with completion of cerebellar development (1M). miRNAs expression signatures of these tissues were compared with profiles generated from GNP-like tumor cells purified from MBs occurring spontaneously in Ink4c−/−; Ptch1+/− or Ink4c−/−; p53−/− mice (22) (Fig. 1).
Fig. 1.
MiRNA signature during cerebellar development and in mouse MB models. Illumina deep sequencing reveals 50 miRNAs differentially expressed in whole 1-month-old and P6 cerebella, and in purified P6 GNPs from cerebella of wild-type (WT), Ink4c−/−; Ptch1+/−, Ink4c−/−; p53−/− mice, or in MBs (red) from both genotypes. Tumor and P6 GNP samples, considered together, were contrasted against adult cerebella and miRNAs were filtered to require a 1% FDR and a 4-fold change between the groups.
Twenty-four miRNAs were expressed at higher levels in 1-month-old cerebella relative to P6 GNPs and MBs. These miRNAs might act as tumor suppressors and/or as regulators of genes that block differentiation. Among these was miR-124a, which increases during neuronal differentiation under the control of the repressor element-1 silencing transcription factor (REST) complex (23). Also showing greater abundance in postmitotic neurons was miR-138, a microRNA that was found to be expressed exclusively in neuronal tissues (24), and miR-128, that is highly expressed in the adult brain (25). Several miRNAs from 2 large clusters on mouse chromosome 12 were coordinately upregulated: miR-300,381,487b,382 and miR-433,127,434,136.
In contrast, 26 miRNAs showed lower expression in 1-month-old cerebella relative to P6 GNPs and mouse MBs regardless of their genotypes. Among this group might be miRNAs that act as oncomiRs (Fig. 1). Remarkably, 9 members of this group were encoded by the miR-17∼92 cluster and its paralogs (20). We found high expression of the miR-17∼92 and miR-106b∼25 paralog clusters (Figs. 1, 2, and Fig. S1). In contrast, the miR-106a∼363 cluster encoded on mouse and human chromosome X was weakly expressed (Fig. 2 and Fig. S1). Although expression of individual miRNAs within the miR-106a∼363 cluster was 2 times higher in P6 GNPs and MBs compared to 1-month-old cerebella, overall levels remained very low compared to the paralogous clusters (Fig. 2 and Fig. S1). Because the pairs of miRNAs 19b-1 and 19b-2 and miRNAs 92–1 and 92–2 have identical sequences, we could not evaluate whether their expression came from the miR-17∼92 or the miR-106a∼363 cluster. Additionally, the miRNA signature obtained in P6 GNPs and GNP-like tumor cells versus differentiated neurons was similar in wild-type, Ink4c−/−; Ptch1+/−, and Ink4c−/−; p53−/− mice. These findings suggest that the miRNA signature that we obtained was characteristic of GNPs and that mutations in p53, Ptch1 and Ink4c have little influence on miRNA expression in this cell type under the conditions tested.
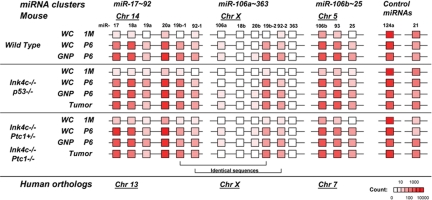
Fig. 2.
The miR-17∼92 cluster family is differentially expressed in proliferating GNPs and GNP-like tumor cells compared to postmitotic 1-month-old cerebella. Schematic expression profiles of the 3 miRNA clusters reveals increasing levels of the miR-17∼92 cluster family in proliferating GNPs and GNP-like tumor cells. Expression levels as sequencing read counts are indicated as color-scaled boxes. Each profile as a group was an average of multiple samples. Expression levels of miR-124a and miR-21, nonmembers of the miR-17∼92 cluster family, are shown as controls. wc, whole cerebellum; WT, wild type.
The Human MB Subgroup with an Activated SHH/PATCHED Signaling Pathway Overexpresses miRNAs from the miR-17∼92 Cluster.
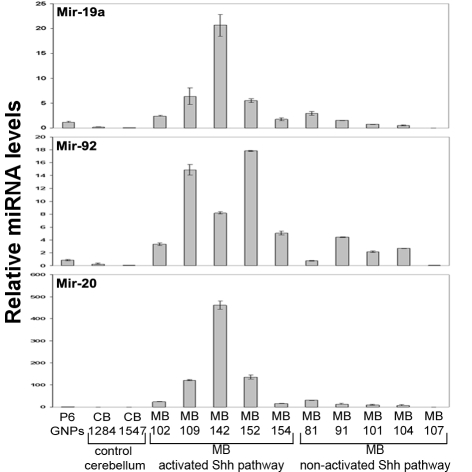
Mouse and human tumors that contain an activated SHH/PTCH pathway share similar global gene expression signatures (26, 13). We therefore tested whether human tumors with an activated SHH/PTCH pathway also upregulate the miR-17∼92 cluster. We performed quantitative real-time reverse transcriptase PCR (QRT-PCR) analysis of miRNAs in 5 normal control cerebella (generously provided by James Olson, Seattle, Washington) and 10 previously characterized human MBs (13) (Fig. 3). RNA isolated from P6 mouse GNPs was used for comparison. Among the MB samples, 5 tumors (MBs 102, 109, 142, 152, and 154) displayed a gene expression signature characteristic of an activated SHH/PTCH pathway; 3 of these (MBs 142, 152, and 154) contained inactivating mutations in PTCH. The 5 other human MBs did not display an activated SHH/PTCH signature, nor did they contain mutations in PTCH, SMOOTHENED (SMO), or SUFU (13). miR-19a (P = 0.003), miR-92 (P = 0.001), and miR-20a (P = 0.008) showed increased abundance specifically in MBs displaying the activated SHH/PTCH pathway signature over those showing a nonactivated SHH pathway (P = 0.026) (Fig. 3). These results implicate the miR-17∼92 cluster in the formation of human MBs that are driven by an aberrant SHH/PTCH pathway and suggest a functional relationship between Patched signaling and miR-17∼92.
Fig. 3.
Human MBs with a SHH/PATCHED gene signature express individual miRs from the miR-17∼92 cluster. Quantitative RT-PCR analysis of selected microRNAs from the miR-17∼92 cluster using RNA extracted from human MBs, purified GNPs from P6 mice, and normal human cerebella tissue. MB tumor patient samples were previously molecularly characterized (13).
Enforced Expression of miR-17∼92 in GNPs from Ink4c−/−; Ptch1+/− Mice Accelerates MB in Vivo.
To determine whether expression of the miR-17∼92 cluster can lead to oncogenic transformation of primary P6 GNPs, we enforced the expression of the entire cluster in primary GNPs purified from the cerebella of P6 Ink4c−/−; p53−/− and Ink4c−/−; Ptch1+/− mice. These mice normally spontaneously develop MBs within 5 months of life. GNPs were infected with mouse stem cell virus (MSCV)-based retroviruses encoding the miRNA cluster along with green fluorescent protein under the control of the SV40 promoter (MLS). Cells were injected immediately into the cortices of immunocompromised naïve recipient mice without prior cell culture (27). All (9/9 injections) populations of Ink4c−/−; Ptch1+/− GNPs, but none (0/4 injections) from Ink4c−/−; p53−/− mice transduced with human hsa-miR-17∼92 cluster formed tumors, with a latency of 1.5 to 5 months (Table 1).
Table 1.
Enforced expression of the miR-17∼92 cluster in GNPs from Ink4c−/−; Ptch1+/− but not Ink4c−/−; p53−/− mice induces medulloblastoma
| Plasmids | Ink4c−/−; Ptch1−/− | Ink4c−/−; p53−/− |
|---|---|---|
| Control (MSCV or MLS empty vector) | 5/15 (2.5–4 months) | 0/10 |
| miR-17∼92 | 9/9 (1.5–5 months) | 0/4 |
| miR-106a∼92 | 0/5 | 0/4 |
| miR-21 | 0/3 | 0/6 |
| N-Myc | 6/7 (1–4.5 months) | 11/17 (1.5–6 months) |
| c-Myc | 8/8 (1.5–4 months) | 16/21 (1–2.5 months) |
| Cyclin D1 | 3/6 (3–6 months) | 2/5 (3.5 months) |
| Cyclin D2 | 5/6 (2–4.5 months) | 1/5 (6.5 months) |
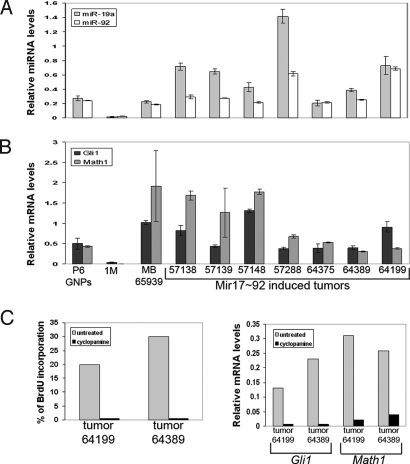
Promotion of MB development was specific to the miR-17∼92 cluster. Enforced expression of the miR-106a∼92 cluster or oncomiR miR-21 (28) failed to induce MBs. (Table 1). When infected with retroviruses carrying N-Myc, c-Myc, or cyclins D1 or D2, GNPs from Ink4c−/−; p53−/− mice formed MBs with the same efficiency as those from Ink4c−/−, Ptch1+/− animals, suggesting that they were competent for transformation (Table 1). Histopathologic analysis of excised tumors characterized them as MBs with a characteristic biphasic pattern found in human MBs (Fig. S2). The majority of tumor cells were GFP positive, suggesting that they were retrovirally infected. We confirmed by QRT-PCR that these tumor cells expressed the miR-17∼92 cluster at elevated levels and found that miR-19a and miR-92 were expressed in engineered tumors (Fig. 4A). QRT-PCR analysis also revealed that all miR-17∼92 engineered tumors lost expression of the wild-type Ptch1 allele, demonstrating that Patched1 is a bona fide tumor suppressor in engineered and spontaneously derived MBs (Fig. S3). Finally miR-17∼92-induced tumors expressed high levels of Math1 and Gli1 mRNAs (Fig. 4B). Together, these findings demonstrated that the Shh/Patched signaling pathway was constitutively activated in these engineered MBs.
Fig. 4.
Molecular characterization of “engineered” MBs in mice after enforced expression of the miR-17∼92 cluster in GNPs from P6 Ink4c−/−; Ptch1+/− mice. Enforced expression of miR-17∼92 in P6 GNPs from Ink4c−/−; Ptch1+/− mice induced MBs in recipient mice. QRT-PCR analysis was performed in (A) and (B) using RNA extracted from purified GNPs from P6 mouse cerebella, 1-month (1M) whole mouse cerebellum, and GNP-like tumor cells from MBs engineered and spontaneously arising in Ink4c−/−; Ptch1+/− mice (MB 65939) (the numbers indicate the recipient mice). (A) miR-17∼92 expression in induced tumors was confirmed by analysis of selected individual microRNAs from the cluster (miR-19a, miR-92). (B) Induced tumors express high levels of Math1 and Gli1 mRNA. (C) GNP-like tumor cells overexpressing the miR-17∼92 cluster remain sensitive to cyclopamine: tumor cells from 2 independently derived MBs (tumor nos. 64199 and 64389) treated or not with cyclopamine were measured for proliferation by BrdU incorporation (Left panel) and for expression of Gli1 and Math1 RNA expression by QRT-PCR (Right panel).
GNP-like tumor cells are sensitive to cyclopamine, an inhibitor of SHH pathway signaling (29). We found that tumor cells from miR-17∼92 expressing MBs were also sensitive to cyclopamine, which decreased their proliferation to the same extent as it affected P6 GNPs or tumor cells from spontaneously derived MBs (Fig. 4C, Left panel). In addition, as expected, Math1 and Gli1 expression were concomitantly downregulated following treatment (Fig. 4C, Right panel).
Finally, we addressed whether any of the reported miR-17∼92 targets were downregulated in MBs. When we compared the protein levels of PTEN, p27Kip1, p130, and E2F-1 from purified tumor cells from miR-17∼92-induced MBs with those in purified GNPs from P6 cerebella, 1-month-old cerebella, and purified tumor cells from spontaneous tumors, we were unable to convincingly show that these reported miR-17∼92 targets were affected by its overexpression (data not shown).
Discussion
We have characterized the miRNA expression pattern during mouse cerebellar development and in MBs from 2 mouse models. A comparison of proliferating GNPs from P6 wild-type and tumor-prone mice and tumor cells to postmitotic differentiated neurons revealed 26 miRNAs that showed increased expression and 24 miRNAs that showed decreased expression. Among the 26 overexpressed miRNAs, 9 were encoded by the miR-17∼92 cluster or its paralogs. As we observed for mouse MBs, the subset of human MBs with a SHH signature also overexpress miRNAs from the miR-17∼92 cluster. Ectopic overexpression of this cluster, but not miR-21, another oncomiR, in proliferating GNPs from P6 Ink4c−/−, Ptch1+/− animals causes mice to develop MBs with complete penetrance.
The miR-17∼92 Cluster Is Overexpressed Specifically in Mouse and Human MBs with a SHH Signature.
All current mouse MB models harbor a Shh/Patched signature regardless of the mutations used to predispose it to tumor development. This suggests that the miRNA signature obtained from tumors in our mouse models might be similar across all models, if alterations in the SHH pathway or those mutations that cooperate with such lesions exert dominant effects on miRNA expression. Because only human MBs with a SHH signature expressed individual miRNAs from the miR-17∼92 cluster, other subgroups of human MBs with unknown mutations or mutations in the WNT pathway may express a distinct subset of miRNAs that contribute to tumor development in that genetic setting. Thus future prospective analysis of miRNAs on a larger cohort of human MBs with different gene signatures might provide a more refined analysis and might provide not only prognostic value but also clues to the full range of miRNAs that impact human MB.
Functional Interrelationship Between Patched Signaling and the miR-17∼92 Cluster.
Purified GNPs from P6 Ink4c−/−; Ptch1+/− mice infected with a GFP-expression control retroviral vector gave rise to rare MBs when orthotopically implanted into recipient animals. These tumors occur with low penetrance. This strongly argues for an impact of stochastic secondary mutations that transform a subset of transplanted cells irrespective of whether they had or had not integrated the viral vector.
In stark contrast, similar GNPs engineered to ectopically express miR-17∼92 by retroviral transfer developed into MBs following transplantation with complete penetrance in 9/9 recipient mice. Interestingly, enforced expression of the miR-106∼25b paralog, which maps to chromosome X, never gave rise to MBs. This finding is consistent with the fact that this paralog is expressed at low or negligible levels in GNPs and MBs and suggests that lower expression is not determined by the endogenous promoter of this cluster alone but also by posttranscriptional mechanisms (30). miR-21 has been shown to be overexpressed in many human tumors; however, it was not expressed at all during cerebellum development or in tumors, suggesting specificity in the nature of prooncogenic microRNAs that can contribute to tumor development in the cerebellum. Consistent with the implications of expression profiling, ectopic expression of miR-21 in GNPs from the cerebella of P6 Ink4c−/−; Ptch1+/− mice did not induce tumors.
Ink4c; p53 doubly deficient mice sometimes spontaneously develop MBs. In this genetic context, overexpression of miR-17∼92 in GNPs from the cerebella of P6 mice with this genotype did not induce tumors above this background level upon transplantation of engineered cells into the cortices of recipient mice. Enforced expression of miR-17∼92 did efficiently cooperate with Patched loss to induce tumors with complete penetrance, arguing strongly for a functional link between the Patched pathway and this microRNA cluster.
Because we were unable to convincingly show that any of the previously reported targets, including PTEN, were affected at the protein level by ectopic expression of miR-17∼92, it is not at present possible to propose a specific molecular mechanism by which miR-17∼92 affects the phenotype of GNPs. Our data support the idea that loss of Patched is necessary for miR-17∼92 to exert its tumorigenic effect. This could be the result of a direct or indirect interaction between a gene that is activated by SHH signaling, including N-Myc, Gli1, Math1, or other genes, and miR-17∼92. One could envision several models by which cooperation could occur. For example, (i) miR-17∼92 could repress target(s) that otherwise buffer or counteract the oncogenic signals emanating from the SHH pathway or (ii) miR-17∼92 could amplify the effects of SHH signaling, perhaps by inhibiting a transcriptional repressor of a subset of SHH target genes. In this case, these genes would be activated only when both SHH signaling is on and miR-17∼92 is inhibiting the repressor.
Patched might also impact the transcription of the miR-17∼92 cluster directly, with loss of Patched through Gli1 or other Shh mRNA targets upregulating miRNA expression. While this could explain the specific overexpression of the cluster in tumors with activated SHH signaling, it is difficult to rationalize this hypothesis with the fact that activated SHH signaling is required to uncover the oncogenic effects of miR-17∼92 in GNPs. miR-17∼92 might repress genes that induce cell cycle arrest and/or differentiation or in turn be regulated by such growth arrest and differentiation pathways. In accord with this idea, we previously found that BMP-2, -4, and -7 oppose Shh/Ptch-induced proliferation of GNPs and MBs, by inducing their differentiation and the downregulation of genes in this pathway in mouse and human MBs, but only in tumors with a SHH signature (31). Thus, BMPs might repress miR-17∼92 levels and thus prevent promotion of MB development.
Whether miR-17∼92 is required for the initiation, progression, and/or maintenance of the disease will require experiments that address the consequences of lost miR-17∼92 function. Unfortunately, primary GNPs cannot be efficiently transfected or electroporated, preventing effective suppression of miR-17∼92 by antagomirs. Experiments using miR-17∼92-floxed mice in which miR-17∼92 can be conditionally deleted by the Cre recombinase (21) will enable us to test whether miR-17∼92 is essential for GNP proliferation and tumor development when Ptch1 is mutated.
miR-17∼92 Is a Myc Target.
Myc is one of the most potent oncogenic agent in human cancers. Interestingly, N-Myc is expressed at high levels in all MBs from our mouse models, suggesting that N-Myc could activate the miR-17∼92 cluster in cerebellar development and MBs. We found that enforced expression of N-Myc or c-Myc in primary purified GNPs from cerebella of P6 Ink4c−/−; Ptch1+/− mice induces MBs after orthotopic transplant in the cortices of recipient animals (Table 1). The miR-17∼92 cluster is a direct c-Myc target (32) and collaborates with c-Myc in B cell lymphoma development (19). In this context, miR-17∼92 may provide cells with a proliferative advantage by preventing cell cycle exit or differentiation and thus collaborate with oncogenic signals to transform GNPs, but would not be oncogenic on its own.
Conclusions
Although the exact mechanism by which Patched and miR-17∼92 collaborate during cerebellar development and in MB formation is still unclear, future studies on miR-17∼92 regulation and the targets that it regulates are warranted, both by expression studies in mouse and human MBs and by functional studies in mice. Our findings indicate that antigomirs to the miR-17∼92 cluster might provide a potential new therapeutic strategy for patients with MBs harboring a constitutively activated SHH/PATCHED signaling pathway.
Methods
MB Patients' Samples.
Human MB samples were previously molecularly characterized (13).
Animal Husbandry and Mouse Tumor Samples.
Breeding and genotyping of mice were done as reported previously (22). Mice were housed in an accredited facility of the Association for Assessment of Laboratory Animal Care in accordance with National Institutes of Health guidelines. The Animal Care and Use Committee of St. Jude Children's Research Hospital approved all procedures. Mice exhibiting signs of illness (abnormal head movements, cranial expansion, reduced activity, or ataxia) were killed. Tumors were removed and a portion was used for purifying tumor cells. In parallel, a piece of tumor was fixed in 10% neutral-buffered formalin, embedded in paraffin, and subjected to histopathology and immunohistochemistry analysis.
Isolation of Small RNAs and miRNAs, Sequencing, and Data Analysis.
Total RNA from GNPs, GNP-like tumor cells, and tissues was extracted using Trizol with an extra chloroform wash and precipitated with isopropanol. Five micrograms (μg) of total RNA were used to amplify and sequence small RNAs as described previously (33), with the introduction of a barcoded multiplexed strategy. RNA adapters ending with a 6-nt barcode followed by a 6-nt constant region were used in the 5′-ligation step. Resulting miRNA libraries were PCR amplified with primers containing Illumina adapter sequences, such that the miRNA would be read in reverse complement followed by the barcode sequence, mixed into pools of 10 libraries with different barcodes, and sequenced on Illumina sequencers. Lack of barcode bias was confirmed by amplifying and sequencing a HeLa sample with the 10 different barcodes. The reads were deconvoluted by barcode, collapsed to unique sequences, aligned to the genome, and annotated. Read counts for miRNA sequences were cubic-spline normalized then log transformed. The R-project software (http://www.R-project.org), including Bioconductor were used for data analysis. For identification of up- and downregulated miRNAs in tumors and P6 GNPs versus adult cerebella, the Bioconductor package linear modeling for microarray (limma) was used to build a model of tumor and P6 GNP samples contrasted against adult cerebella. P-values from limma were transformed to Q-values for estimation of the false discovery rate (FDR). The set of miRNAs corresponding to a 1% FDR were filtered to require a 4-fold change between the groups. The remaining 50 miRNAs were considered significantly different in the adult cerebella relative to the tumor and P6 GNP samples. For sequences, see Methods, SI Text (Fig. S4). For expression profiling, the normalized read counts for miRNAs encoded by miR-17∼92 cluster family were collected, while undetected sequences were scaled to the minimum count of the data set.
GNP and GNP-Like MB Tumor Cell Purification, Retroviral Infection, Orthotopic Injections, and Tumor Cell Cultures.
Purification of GNPs from mouse cerebella and MBs were done as described (22). Infection of GNPs and orthotopic injections were carried out as described using 2 × 106 cells (27). To assess the infection efficiency, 3 × 105 infected cells were cultured for 48 h, then fixed and immunostained with anti-GFP. In each experiment, the infection efficiency was at least 30%. MicroRNAs were cloned into a MSCV-SV40 promoter-green fluorescent protein (GFP) vector (19). Tumor cells were purified, cultured, and treated with cyclopamine, as previously described (31).
Quantitative Real-Time PCR.
Quantitative real-time reverse transcriptase PCR (QRT-PCR) for Ptch1, Math1, and Gli1 on RNA extracted from purified GNP-like tumor cells was done as described (22). QRT-PCRs for miR-19a, miR-92, and miR-20 were performed using Applied Biosystems TaqMan MicroRNA assays according to manufacture's recommendations (ABI, Foster City, CA). Briefly, cDNAs were synthesized from total RNA using miRNA-specific primers according to the TaqMan MicroRNA reverse transcription protocol. Reverse transcription was performed using the following program: 30 min at 16 °C, 30 min at 42 °C, 5 min at 85 °C, and then held at 4 °C. Q-PCR on the cDNAs was performed using a 7900HT sequence detection system and the TaqMan Universal MasterMix reagents. Cycling conditions were 10 min at 95 °C, followed by 40-cycle amplification for 15 sec at 95 °C and 1 min at 60 °C. The internal RNU6B RNA levels were used for normalization. Each measurement was performed in triplicate. Data were analyzed with SDS version 2.0 software (ABI).
Supplementary Material
Acknowledgments.
We thank all members of the Roussel/Sherr laboratory and Charles J. Sherr for helpful comments; Jerold E. Rehg and Dorothy Bush for histopathology and immunohistochemistry analysis; Robert Jenson for mouse genotyping; John Killmar for helping with cranial surgery; Twala Hogg for technical assistance; James Olson (University of Washington, Children's Hospital, Seattle, Washington) for normal human cerebella specimens; Reuven Agami (The Netherlands Cancer Institute) for miRNA libraries; and Scott Lowe, Mike Hemann, and Ross Dickins (Cold Spring Harbor Laboratory, New York) for the MLS vector expressing miR-17∼92. This work was funded by grants from National Cancer Institue PO1CA-096832 (M.F.R. and R.J.G.), CA-129541 (R.J.G.), K99 Pathway to Independence Award (L.H.), Core Grant P30CA-21765 (M.F.R. and R.J.G.), the Children's Brain Tumor Foundation (M.F.R. and T.U.), the Emily Dorfman Foundation for Children through the American Brain Tumor Association (T.U.), the Collaborative Ependymoma Research Network (CERN) (R.J.G.), the American Cancer Society Postdoctoral Fellowship PF-07–058-01-GMC (F.V.K.), and the American Lebanese–Syrian Associated Charities of St. Jude Children's Research Hospital. G.H. is an investigator at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing data reported in this paper has been deposited in the NCBI CEO database, accession number GSE14470.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809579106/DCSupplemental.
References
- 1.Oliver TG, et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medullolastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 2.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 3.Herzberg JJ, Wiskemann A. Basal cell nevus with hereditary malformation and medulloblastoma. Dermatologica. 1963;126:106–123. [PubMed] [Google Scholar]
- 4.Evangelista M, Tian H, de Sauvage FJ. The hedgehog signaling pathway in cancer. Clin Cancer Res. 2006;12:5924–5928. doi: 10.1158/1078-0432.CCR-06-1736. [DOI] [PubMed] [Google Scholar]
- 5.Hahn H, et al. A mammalian patched homolog is expressed in target tissues of sonic hedgehog and maps to a region associated with developmental anomalies. J Biol Chem. 1996;271:12125–12128. doi: 10.1074/jbc.271.21.12125. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RL, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 7.Smyth I, et al. Isolation and characterization of human patched 2 (PTCH2), a putative tumour suppressor gene in basal cell carcinoma and medulloblastoma on chromosome 1p32. Hum Mol Genet. 1999;8:291–297. doi: 10.1093/hmg/8.2.291. [DOI] [PubMed] [Google Scholar]
- 8.Hatten ME. Expansion of CNS precursor pools: a new role for Sonic Hedgehog. Neuron. 1999;22:2–3. doi: 10.1016/s0896-6273(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 9.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 10.Hallahan AR, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton SR, et al. The molecular basis of Turcot's syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 12.Schüller U, Rowitch DH. Beta-catenin function is required for cerebellar morphogenesis. Brain Res. 2007;1140:161–169. doi: 10.1016/j.brainres.2006.05.105. [DOI] [PubMed] [Google Scholar]
- 13.Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 19.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uziel T, et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev. 2005;19:2656–2667. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smirnova L, et al. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y, et al. A molecular fingerprint for medulloblastoma. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- 27.Zindy F, et al. Genetic alterations in mouse medulloblastomas and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res. 2007;67:2676–2684. doi: 10.1158/0008-5472.CAN-06-3418. [DOI] [PubMed] [Google Scholar]
- 28.Nicoloso MS, Calin GA. MicroRNA involvement in brain tumors: from bench to bedside. Brain Pathol. 2008;18:122–129. doi: 10.1111/j.1750-3639.2007.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman DM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 30.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 33.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.