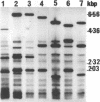
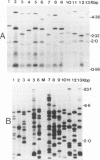
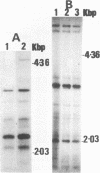
Abstract
Culture of Mycobacterium tuberculosis provides no information on the identity of a strain or the distribution of such a strain in the community. Strain identification of M. tuberculosis can help to address important epidemiological questions, e.g., the origin of an infection in a patient's household or community, whether reactivation of infection is endogenous or exogenous in origin, and the spread and early detection of organisms with acquired antibiotic resistance. To research this problem, strain identification must be reliable and accurate. Although genetic identification techniques already exist, it is valuable to have genetic identification techniques based on a number of genetic markers to improve the accurate identification of M. tuberculosis strains. We show that oligonucleotide (GTG)5 can be successfully applied to the identification of M. tuberculosis strains. This technique may be particularly useful in cases in which M. tuberculosis strains have few or no insertion elements (e.g., IS6110) or in identifying other strains of mycobacteria when informative probes are lacking.
Full text
PDF
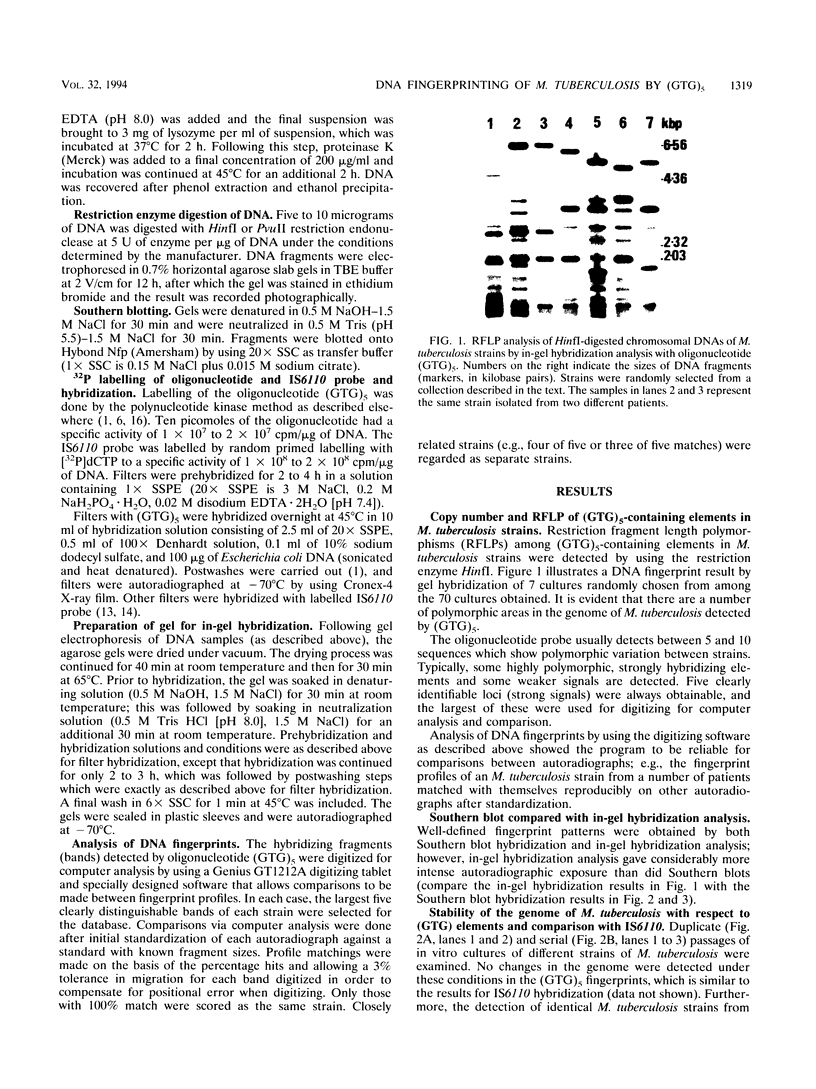
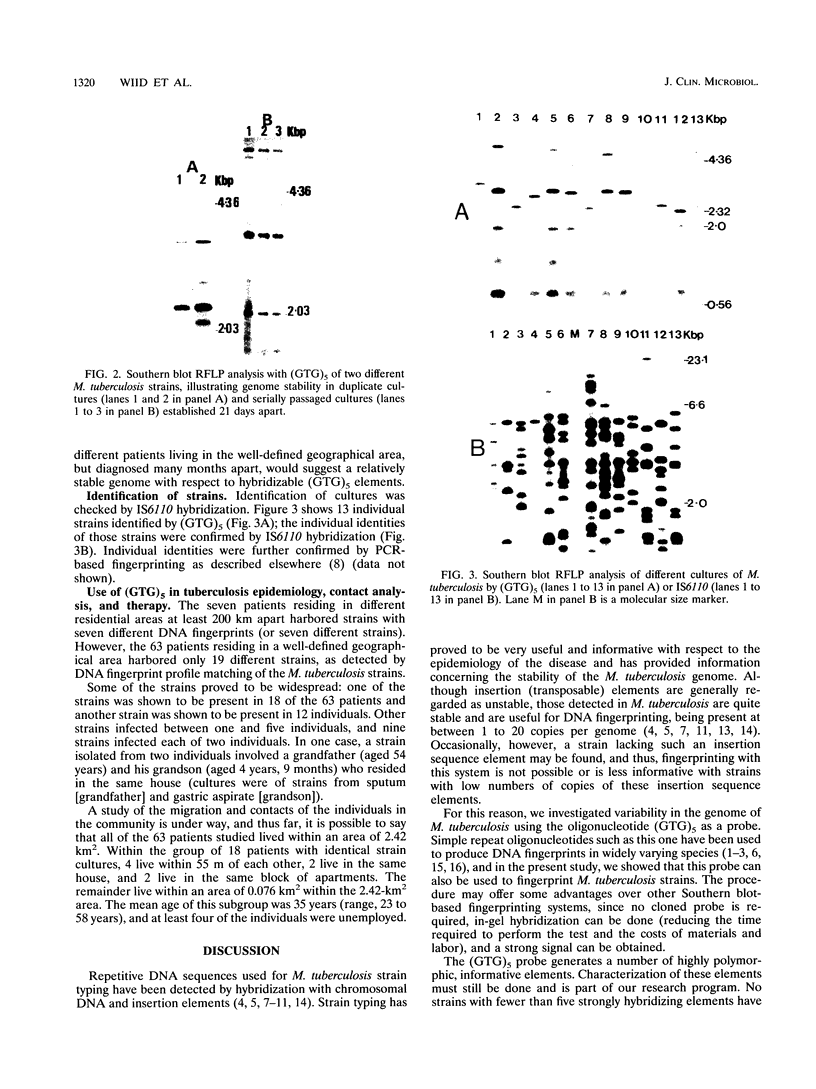

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ali S., Müller C. R., Epplen J. T. DNA finger printing by oligonucleotide probes specific for simple repeats. Hum Genet. 1986 Nov;74(3):239–243. doi: 10.1007/BF00282541. [DOI] [PubMed] [Google Scholar]
- Bettink S., Wullich B., Christmann A., Zwergel T., Zang K. D., Unteregger G. Genetic heterogeneity of prostatic carcinoma-derived cell lines as emphasized by DNA fingerprinting. Electrophoresis. 1992 Sep-Oct;13(9-10):644–646. doi: 10.1002/elps.11501301133. [DOI] [PubMed] [Google Scholar]
- Bierwerth S., Kahl G., Weigand F., Weising K. Oligonucleotide fingerprinting of plant and fungal genomes: a comparison of radioactive, colorigenic and chemiluminescent detection methods. Electrophoresis. 1992 Mar;13(3):115–122. doi: 10.1002/elps.1150130125. [DOI] [PubMed] [Google Scholar]
- Eisenach K. D., Crawford J. T., Bates J. H. Genetic relatedness among strains of the Mycobacterium tuberculosis complex. Analysis of restriction fragment heterogeneity using cloned DNA probes. Am Rev Respir Dis. 1986 Jun;133(6):1065–1068. doi: 10.1164/arrd.1986.133.6.1065. [DOI] [PubMed] [Google Scholar]
- Eisenach K. D., Crawford J. T., Bates J. H. Repetitive DNA sequences as probes for Mycobacterium tuberculosis. J Clin Microbiol. 1988 Nov;26(11):2240–2245. doi: 10.1128/jcm.26.11.2240-2245.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epplen J. T., Ammer H., Epplen C., Kammerbauer C., Mitreiter R., Roewer L., Schwaiger W., Steimle V., Zischler H., Albert E. Oligonucleotide fingerprinting using simple repeat motifs: a convenient, ubiquitously applicable method to detect hypervariability for multiple purposes. EXS. 1991;58:50–69. doi: 10.1007/978-3-0348-7312-3_4. [DOI] [PubMed] [Google Scholar]
- Hermans P. W., van Soolingen D., Dale J. W., Schuitema A. R., McAdam R. A., Catty D., van Embden J. D. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990 Sep;28(9):2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palittapongarnpim P., Chomyc S., Fanning A., Kunimoto D. DNA fingerprinting of Mycobacterium tuberculosis isolates by ligation-mediated polymerase chain reaction. Nucleic Acids Res. 1993 Feb 11;21(3):761–762. doi: 10.1093/nar/21.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi P. P., Talwar G. P., Khandekar P. S. Repetitive DNA sequence of Mycobacterium tuberculosis: analysis of differential hybridization pattern with other mycobacteria. Int J Lepr Other Mycobact Dis. 1988 Dec;56(4):592–598. [PubMed] [Google Scholar]
- Shoemaker S. A., Fisher J. H., Jones W. D., Jr, Scoggin C. H. Restriction fragment analysis of chromosomal DNA defines different strains of Mycobacterium tuberculosis. Am Rev Respir Dis. 1986 Aug;134(2):210–213. doi: 10.1164/arrd.1986.134.2.210. [DOI] [PubMed] [Google Scholar]
- Thierry D., Cave M. D., Eisenach K. D., Crawford J. T., Bates J. H., Gicquel B., Guesdon J. L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990 Jan 11;18(1):188–188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D., Hermans P. W., de Haas P. E., van Embden J. D. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol. 1992 Jul;30(7):1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D., de Haas P. E., Hermans P. W., Groenen P. M., van Embden J. D. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993 Aug;31(8):1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]