Summary
Mcm10 is an essential eukaryotic DNA replication protein required for assembly and progression of the replication fork. The highly conserved internal domain (Mcm10-ID) has been shown to physically interact with single-stranded (ss) DNA, DNA polymerase α, and PCNA. The crystal structure of Xenopus laevis Mcm10-ID presented here reveals a novel DNA binding architecture composed of an OB-fold followed in tandem by a variant and highly basic zinc finger. NMR chemical shift perturbation and mutational studies of DNA binding activity in vitro reveal how Mcm10 uses this unique surface to engage ssDNA. Corresponding mutations in Saccharomyces cerevisiae result in increased sensitivity to replication stress, demonstrating the functional importance of DNA binding by this region of Mcm10 to replication. In addition, mapping Mcm10 mutations known to disrupt PCNA, pol α, and DNA interactions onto the crystal structure provides insight into how Mcm10 may coordinate protein and DNA binding within the replisome.
Keywords: Mcm10, replication, OB-fold, zinc finger, DNA binding, ssDNA
Introduction
Eukaryotic DNA replication is carried out by large multiprotein machines that coordinate DNA unwinding and synthesis at the replication fork. During initiation, the replisome is assembled in stages at the G1/S transition through a series of protein complexes that recognize and denature origin DNA (Figure 1) (reviewed in Bell and Dutta, 2002; Garg and Burgers, 2005). First, origin recognition complex (ORC), Cdc6, Cdt1, and Mcm2-7 form a pre-replicative complex (pre-RC). Mcm10 loads onto origins after pre-RC assembly and is required for recruitment of Cdc45 (Wohlschlegel et al., 2002), which forms a helicase complex with Mcm2-7 and GINS to unwind DNA (Gambus et al., 2006; Moyer et al., 2006; Pacek et al., 2006). Phosphorylation of Mcm2-7 and several other replication factors by cyclin- and Dbf4-dependent kinases (CDK, DDK) stimulate origin unwinding, which is signaled by recruitment of RPA to the origin (Lei et al., 1997; Tanaka et al., 2007; Tanaka and Nasmyth, 1998; Zegerman and Diffley, 2007; Zou and Stillman, 2000). Mcm10, Cdc45, and RPA facilitate subsequent loading of DNA polymerase α-primase (pol α) (Ricke and Bielinsky, 2004; Walter and Newport, 2000). Finally, replicative DNA polymerases δ and ε and PCNA are recruited to form the intact replisome.
Figure 1. Initiation of eukaryotic DNA replication.
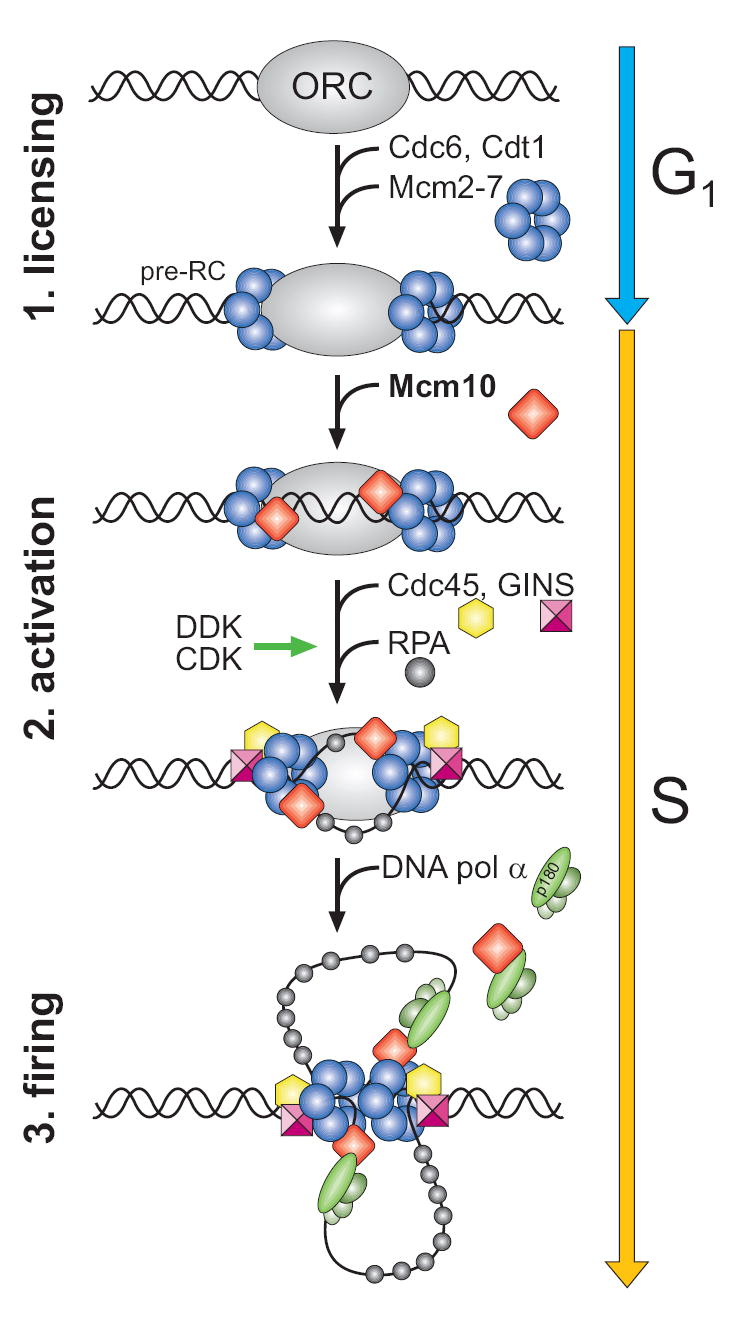
The schematic shows some of the key steps necessary for DNA unwinding and replication fork assembly at a eukaryotic origin of replication. Details are described in the text.
Mcm10 was identified from yeast genetic screens to be required for chromosomal DNA replication and plasmid maintenance (Merchant et al., 1997; Solomon et al., 1992), and is associated with chromatin throughout S-phase as a component of active replication complexes (Gambus et al., 2006; Pacek et al., 2006; Ricke and Bielinsky, 2004). A number of genetic and physical interactions have been observed between Mcm10 and proteins found in the pre-RC and at the replication fork (Christensen and Tye, 2003; Homesley et al., 2000; Izumi et al., 2000; Lee et al., 2003; Merchant et al., 1997; Zhu et al., 2007), indicating that Mcm10 is essential for both replisome assembly and fork progression. Interestingly, a diubiquitinated form of S. cerevisiae (sc)Mcm10 interacts with PCNA in budding yeast (Das-Bradoo et al., 2006). In addition, Mcm10 physically interacts with pol α (Chattopadhyay and Bielinsky, 2007; Ricke and Bielinsky, 2004, 2006), and affects the association between the polymerase and chromatin (Yang et al., 2005). In vitro, spMcm10 stimulates the polymerase activity of pol α (Fien et al., 2004) and has been shown to contain primase activity (Fien and Hurwitz, 2006), although Xenopus laevis Mcm10 (xMcm10) does not synthesize RNA primers under identical conditions (Robertson et al., 2008). The Mcm10-pol α interaction has led to the suggestion that Mcm10 helps to recruit the polymerase to the replisome and may regulate its activity.
In addition to its interactions with the replisome, Mcm10 binds both single (ss)- and double-stranded (ds) DNA (Fien et al., 2004; Robertson et al., 2008). The DNA binding function has been localized to a highly conserved, 200-residue internal domain (ID) and a C-terminal domain unique to higher eukaryotes (Robertson et al., 2008). In addition to a Cys3His-type zinc finger (Izumi et al., 2000), Mcm10-ID has been predicted to contain an oligonucleotide/oligosaccharide (OB)-fold (Ricke and Bielinsky, 2006), both of which are classic DNA binding motifs. Recently, human Mcm10 was reported to form ring-shaped hexameric assemblies that could encircle DNA (Okorokov et al., 2007). Despite these observations, the nature of Mcm10-DNA binding and its role in DNA replication remains unclear, in part due to a lack of high resolution structural information for the protein.
Remarkably, the mutations discovered from yeast genetic screens, as well as those identified to disrupt scMcm10 association with PCNA and pol α (Das-Bradoo et al., 2006; Ricke and Bielinsky, 2006), are all located within the Mcm10-ID (see Supporting Information). Collectively, these mutations demonstrate the importance of the ID in Mcm10 function and motivate structural analysis of this domain. Presented here is a structure-function analysis of the conserved internal domain of xMcm10. X-ray crystallography, NMR chemical shift perturbation, and site-directed mutational analysis of DNA binding reveal a unique ssDNA binding surface constructed from an arrangement of OB-fold and zinc finger motifs not yet observed in DNA processing proteins. Importantly, we show that mutation of DNA binding residues in scMcm10 increases the sensitivity of yeast that have been subjected to hydroxyurea-induced replication stress.
Results
We previously identified the domain architecture of xMcm10 (Robertson et al., 2008). Limited proteolytic digestion of the full-length protein produced N-terminal (NTD, aa 1-145), internal (ID, 230-417), and C-terminal (CTD, 596-860) domains that correspond to the sequence conservation among Mcm10 proteins (Figure 2A). The NTD encompasses an oligomerization function, while the ID and CTD both bind to ssDNA, dsDNA, and pol α (Robertson et al., 2008). Mcm10-ID is the only region of the 860-residue protein that shows significant homology across all species from vertebrates to yeast (Figure 2A), and is 73% and 24% identical to human and scMcm10, respectively. Extensive homology suggests that Mcm10-ID contains an essential function and prompted a rigorous structure-function analysis of this core domain.
Figure 2. Structure of the conserved central domain of Mcm10.
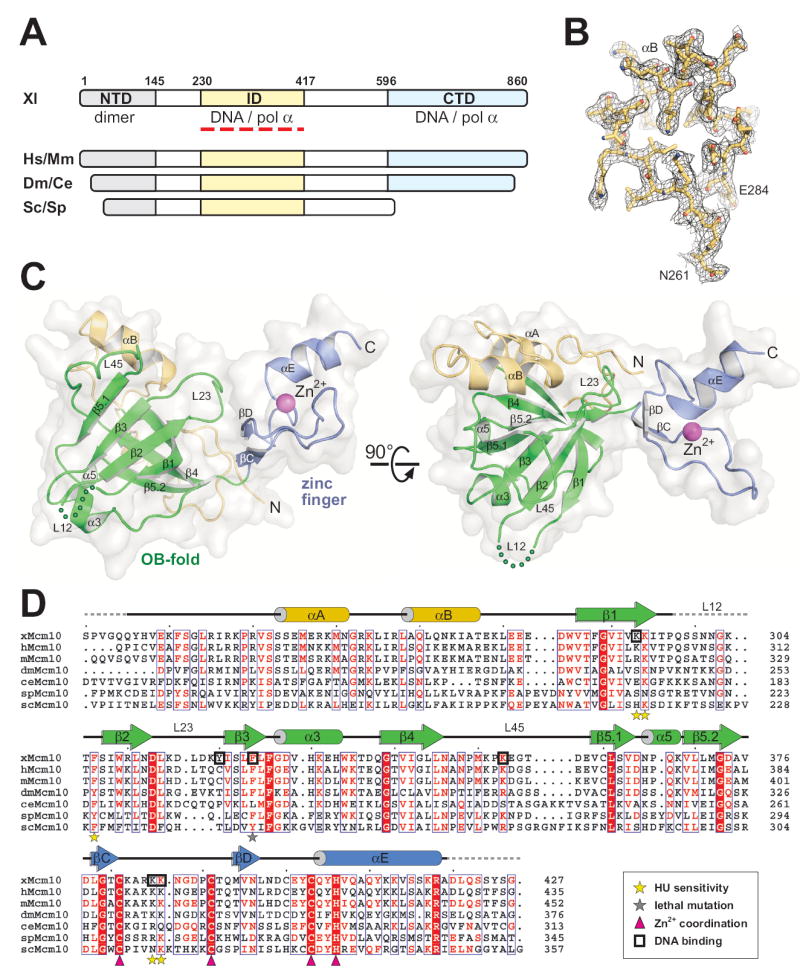
(A) Mcm10 domain architecture. The three structural domains identified from Xenopus laevis (Xl) Mcm10 are shown as colored bars and aligned with homologous regions of Mcm10 from Homo sapiens (Hs), Mus musculus (Mm), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp). (B) A representative section of the refined crystallographic model superimposed onto composite omit electron density contoured at 1σ. Stereoviews of electron density maps can be found as Supplementary Figure 1. (C) The crystal structure of Xenopus laevis Mcm10-ID (residues 230-427) is shown as a ribbon diagram with a white molecular surface. Two orthogonal views show the relative orientation of the OB-fold (green), zinc finger (blue ribbon, magenta Zn2+ sphere), and N-terminal α-helical/coil region (gold). (D) Sequence alignment of Mcm10-ID with schematic secondary structural elements colored as in panel C. xMcm10 residues identified by mutagenesis to interact with DNA in vitro are boxed. scMcm10 mutations that result in increased sensitivity of yeast to hydroxyurea are marked with yellow stars, and the grey star denotes a lethal mutation. Conserved Zn2+-coordinating residues are marked with magenta triangles.
The unique structure of the conserved domain of Mcm10
The crystal structure of Mcm10-ID from Xenopus laevis was determined to 2.3 Å resolution (Figures 2B and S1). Experimental phases were obtained from a multiwavelength anomalous dispersion (MAD) experiment using a single gold derivative crystal. The atomic model, which consists of three Mcm10-ID molecules in the asymmetric unit, was built into 3.0 Å Au-MAD electron density and refined against 2.3 Å native diffraction data (Table 1) to a crystallographic residual of 20.2% (Rfree = 24.7%). The accuracy of the structure is reflected in part by 91.5% and 8.3% of the 515 total residues residing within the favored and allowed regions of the Ramachandran plot, respectively.
Table 1.
Data collection, phasing and refinement statistics for xMcm10-ID
| Native
|
K2Au(CN)2 |
|||
|---|---|---|---|---|
| Data collectiona | ||||
| Space group | P21 | P21 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 54.6, 94.4, 69.8 | 54.5, 94.4, 69.8 | ||
| α, β, γ (°) | 90, 112.8, 90 | 90, 112.6, 90 | ||
| Peak | Inflection | Remote | ||
| Wavelength (Å) | 1.000 | 1.0388 | 1.0370 | 1.0311 |
| Resolution (Å) | 50-2.3 (2.38-2.3) | 50-2.8 (2.9-2.8) | 50-2.8 (2.9-2.8) | 50-2.8 (2.9-2.8) |
| Rsym or Rmerge | 0.074 (0.425) | 0.034 (0.098) | 0.034 (0.131) | 0.038 (0.129) |
| I / σI | 27.7 (2.3) | 25.7 (7.9) | 19.3 (4.4) | 24.0 (6.5) |
| Completeness (%) | 99.6 (97.0) | 99.6 (100.0) | 79.2 (80.7) | 99.7 (99.8) |
| Redundancy | 6.7 (3.9) | 3.7 (3.6) | 2.3 (2.1) | 3.7 (3.6) |
| Refinement | ||||
| Resolution (Å) | 50-2.3 | |||
| No. reflections | 27,448 | |||
| Rwork / Rfree | 0.202 / 0.247 | |||
| No. atoms | ||||
| Protein | 4128 | |||
| Ligand/ion | 3 | |||
| Water | 149 | |||
| B-factors (Å2) | ||||
| Protein | 56.3 | |||
| Ligand/ion | 47.2 | |||
| Water | 54.3 | |||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.014 | |||
| Bond angles (°) | 1.572 | |||
Values in parentheses refer to the highest-resolution shell.
Mcm10-ID forms a globular domain consisting of an OB-fold (β1-β5.2) flanked by an α-helical/random coil region (αA-αB) at the amino-terminus and a zinc finger motif (βC-αE) at the carboxy-terminal end (Figure 2C). The α-helical region is packed against the back side of the OB-fold to form an essentially flat molecular surface. The zinc finger protrudes to one side of the structure and makes extensive electrostatic and van der Waals contacts with the OB-fold L23 loop and the α-helical/coil region (Figure 2C). The three crystallographically independent Mcm10-ID molecules in the asymmetric unit superimpose with an r.m.s. deviation of 0.7 Å for all atoms with the zinc finger in the same relative position with respect to the OB-fold in each protomer. Thus, there is no evidence to suggest free movement between the OB-fold and zinc finger, despite the cluster of invariant glycine residues 339, 373, and 379 at the OB-fold/zinc finger junction (Figure 2D). Moreover, each protomer has a unique crystal packing environment, and thus we rule out any crystal lattice effect on the structure. In summary, Mcm10-ID forms a single structural domain with the OB-fold and zinc finger motifs in an orientation that makes them both accessible to DNA.
The two DNA binding motifs in Mcm10 form a unique molecular surface based on several key structural features. Firstly, the spatial arrangement of OB-fold and zinc finger in Mcm10-ID is different from other DNA processing proteins that contain both structural motifs (Figure 3A). Whereas zinc motifs of RPA70, T4 gp32, NAD+-dependent DNA ligase, and archaeal MCM helicase reside within or adjacent to the L12 loop and have been suggested to play a structural, rather than ligand-binding role (Bochkarev et al., 1997; Fletcher et al., 2003; Lee et al., 2000; Shamoo et al., 1995), the zinc finger of Mcm10 is on the opposite (L23) face of the OB-fold. Secondly, Mcm10’s variant Cys3His-type zinc finger contains an extended loop between βC and βD that is oriented perpendicular to the α-helix as a result of Zn2+ coordination at the N-terminal end of the helix (Figure 3B). Consequently, the zinc loop is positioned immediately adjacent to the putative DNA binding cleft of the OB-fold (Figure 2C).
Figure 3. Comparison of OB-fold and zinc finger motifs in DNA binding proteins.
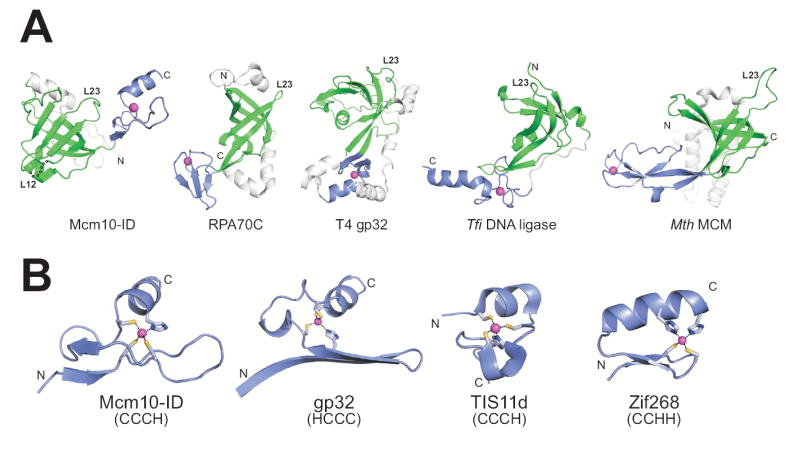
(A) Structures of five proteins that contain an OB-fold (green) and zinc motif (blue) in the same domain. The structures are oriented with respect to the OB-fold β1 strand, with the L23 loops labeled to illustrate the relative positions of the zinc-binding motifs. (B) Variant zinc fingers from Mcm10-ID (CX9CX11CX2H), T7 gp32 (HX12CX9CX2C), and TS11d (CX8CX5CX3H), and the archetypical zinc finger from Zif268 (CX2CX12HX3H) are oriented with the long axis of the beta sheet running horizontally across the plane of the page. These structures suggest that the zinc finger fold depends more on the number of intervening residues between cysteines and histidines than on the order of the coordinating residues themselves. PDB IDs: Mcm10, 3ebe (this work); RPA70C, 1jmc (Bochkarev et al., 1997); gp32, 1gpc (Shamoo et al., 1995); DNA ligase, 1v9p (Lee et al., 2000); MCM, 1ltl (Fletcher et al., 2003); TIS11d, 1rgo, (Hudson et al., 2004); Zif268, 1zaa, (Pavletich and Pabo, 1991).
A novel DNA binding platform
In order to expand our understanding of nucleic acid binding by Mcm10-ID, ssDNA binding was investigated by monitoring perturbations in NMR chemical shifts as DNA was titrated into 15N-enriched Mcm10-ID (Figure 4A). Sequence-specific resonance assignments (Figure S2A and Table S1) were obtained using standard multi-dimensional triple resonance experiments performed on 13C,15N-enriched Mcm10-ID. The addition of ssDNA to Mcm10-ID resulted in a shift of a significant number but not all of the peaks in the 2D 15N-1H HSQC spectrum (Figure S2B), which is consistent with a combination of effects from DNA binding and small conformational changes in the protein. Over the course of the titration, the peaks shifted continuously (fast exchange) with changes saturated at a 1:3 protein:DNA ratio (data not shown). These observations are consistent with the 3 μM binding affinity measured by fluorescence anisotropy (Robertson et al., 2008). In order to determine the DNA binding site of Mcm10-ID, the residues exhibiting the most significant NMR chemical shift perturbations were mapped onto the crystal structure (Figure 4B). The largest shifts were observed for residues lining the β-barrel of the OB-fold and, surprisingly, the extended loop of the zinc finger. Very few perturbations were observed on the opposite face of the protein, demonstrating that DNA primarily contacts the common OB-fold/zinc loop surface.
Figure 4. Mapping the Mcm10 DNA binding site.
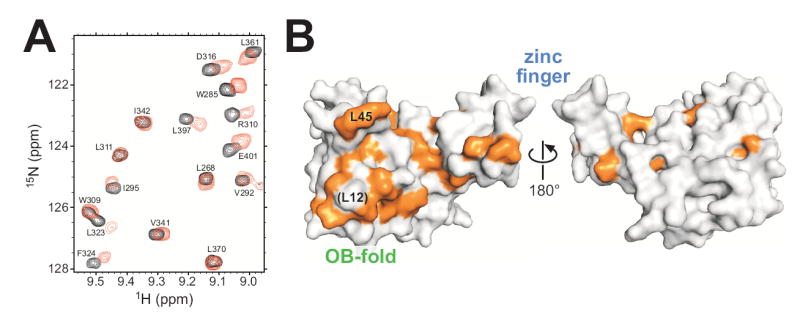
(A) NMR chemical shift perturbations in response to ssDNA binding to Mcm10. An overlay of a region of 15N-1H HSQC spectra of 15N-enriched Mcm10 in the absence (black) and presence (red) of ssDNA (1:0.4 protein:DNA ratio) is shown. Peak assignments are labeled. (B) Surface representation of Mcm10-ID with all assigned residues showing significant chemical shift perturbation (>0.057 ppm) colored orange. Two orientations rotated 180° with respect to one another show that perturbations occur almost exclusively on one surface of the protein.
A comparative structure search using the DALI server (Holm and Sander, 1993) revealed that the OB-fold in Mcm10-ID is most similar to those of the high affinity ssDNA binding domains from the 70-kD subunit of human RPA (RPA70AB) (Figure 5A,B). RPA70AB is composed of tandem OB-folds oriented with their ssDNA binding surfaces side-by-side, which allows 8 nucleotides of ssDNA to traverse both binding pockets in a linear fashion (Figure 5A). Superposition of the Mcm10-ID and RPA70A OB-folds places the zinc finger in the same location as the second RPA70B OB-fold, suggesting that 8-10 nucleotides are needed to span the OB-fold/zinc loop surface. This correlates well with the length dependence of DNA binding to Mcm10-ID determined by fluorescence anisotropy (Figure S3), which showed that 10-nucleotide oligomers were the shortest DNA that supported high affinity binding for ssDNA. Thus, the OB-fold and extended zinc loop in Mcm10-ID forms a molecular surface analogous to the DNA binding platform of the two RPA70AB OB-folds.
Figure 5. The DNA binding surface of Mcm10.
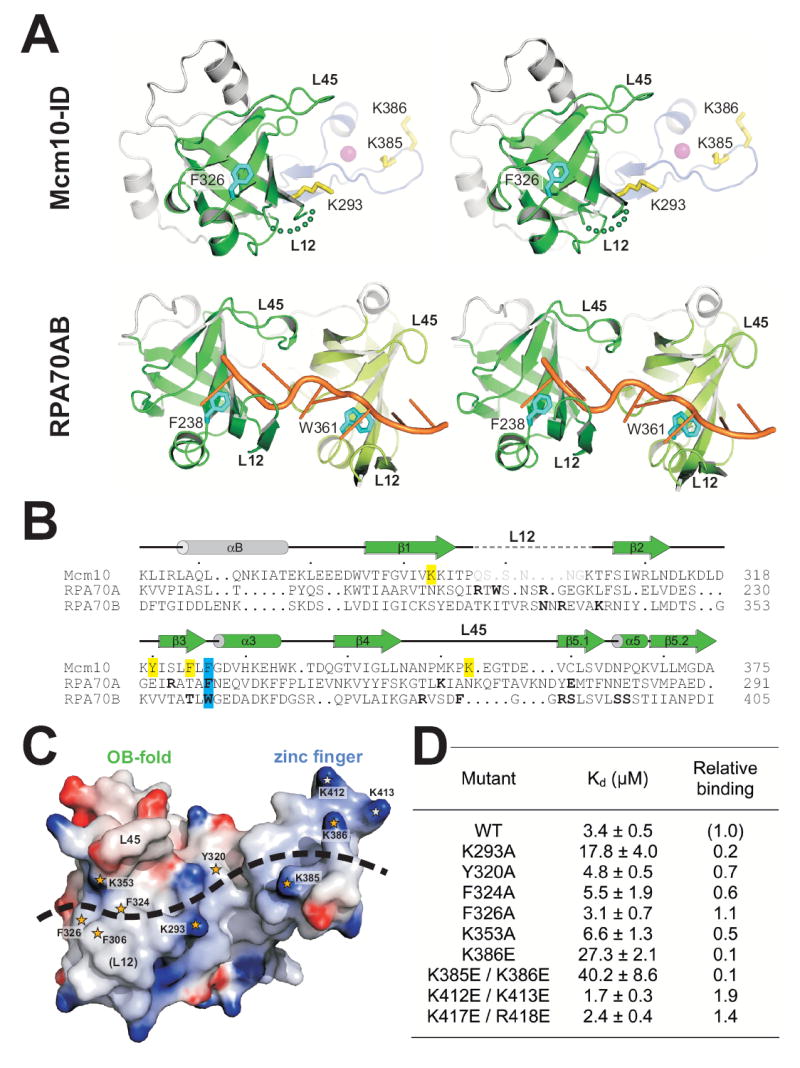
(A) Stereodiagram of crystal structures of Mcm10-ID and an RPA70AB/ssDNA complex. OB-folds are colored green, zinc finger blue with magenta Zn2+, and ssDNA orange. Residues important for DNA binding to Mcm10 and RPA are rendered as yellow and cyan sticks, respectively. (B) Structure-based sequence alignment of OB-folds from Mcm10, RPA70A, and RPA70B. Residues shown by mutagenesis to affect in vitro DNA binding in Mcm10-ID are highlighted yellow, and conserved aromatic residues contacting ssDNA in RPA are highlighted cyan. DNA-binding residues identified from the RPA crystal structure are in bold-face, and disordered residues in the Mcm10-ID crystal structure are grey. (C) Mcm10-ID engages ssDNA at both the OB-fold and zinc loop surface. Mcm10-ID is shown as an electrostatic potential surface (blue, positive; red, negative). Residues implicated in DNA binding are highlighted with orange stars, and the positions of L12 and L45 loops known to contact ssDNA in other OB-folds are labeled. (D) ssDNA binding activity of Mcm10-ID mutants. The dissociation constants (Kd) for wild-type and mutant Mcm10-ID proteins for 25mer ssDNA were determined in vitro using fluorescence anisotropy.
In order to validate that Mcm10 utilizes the entire OB-fold/zinc loop surface to engage DNA, mutational studies of Mcm10’s DNA binding activities were performed. The assessment of mutants was based on DNA binding affinities for 25mer ssDNA measured by the fluorescence anisotropy assay. A significant difference between Mcm10’s putative DNA binding surface and that of RPA70AB is the cluster of basic residues (Lys293, Lys385, Lys386) on the zinc finger loop and in the cleft formed between the two motifs (Figure 5). Mutations in this electropositive region had a marked effect on Mcm10 binding to DNA. Most strikingly, a Lys385Glu/Lys386Glu double mutant on the extended zinc loop exhibited a 10-fold reduction in ssDNA binding affinity (Figure 5C,D). Lysine → glutamate substitutions along the zinc finger helix (Lys412Glu/Lys413Glu and Lys417Glu/Arg418Glu), on the other hand, had a modest stimulatory effect on DNA binding. At the cleft between the OB-fold and zinc finger, Lys293Ala reduced the affinity for ssDNA 5-fold with respect to wild-type Mcm10-ID, and Tyr320Ala had a marginal but significant effect (Figure 5D).
On the OB-fold side, a cluster of three phenylalanine side chains (Phe306, Phe324, Phe326) and Lys353 are poised to interact with ssDNA in our model. Indeed, Phe324Ala on strand β3 and Lys353 in the L45 loop had a modest effect on DNA binding (Figure 5D). Substitution of any residue within the L12 loop, including Phe306, resulted in insoluble protein, which precluded analysis of L12 in our DNA binding assay. In RPA70AB, both OB-folds clamp the ssDNA between loops L12 and L45, and aromatic residues Phe238 (RPA70A) and Trp361 (RPA70B) at the C-terminus of β3 form DNA base stacking interactions (Bochkarev et al., 1997). Phe326 in xMcm10 is invariant among Mcm10 orthologs and superimposes with RPA Phe238 and Trp361 (Figure 5A,B). Surprisingly, substitution of Phe326 with alanine did not affect DNA binding to Mcm10-ID (Figure 5D). However, a single Phe238Ala mutation in RPA70A was also reported as not having a measurable effect on ssDNA binding, despite the observation of a direct contact to ssDNA in the crystal structure (Walther et al., 1999). Thus, it is not possible to draw specific conclusions from the mutational data alone. The data do, however, reflect the redundancy in protein-DNA contacts along the extended binding site in Mcm10-ID. Taken together, the NMR and mutational data indicate that DNA spans the hydrophobic cleft of the OB-fold and the extended, positively-charged loop of the variant zinc finger.
Functional relevance of DNA binding to Mcm10
In order to establish that the residues affecting DNA binding of xMcm10 in vitro have a role during DNA replication in vivo, we introduced the corresponding mutations into the endogenous MCM10 locus of S. cerevisiae and tested for sensitivity to hydroxyurea (HU). Mid-log phase cultures were incubated in 0.2 M HU for 60, 120 or 180 min before they were diluted and plated in the absence of HU to determine the rate of recovery. Figure 6A shows that all mutants were expressed at levels similar to wild-type Mcm10. Under our test conditions, wild-type cells doubled in number, and mutations that showed no effect on DNA binding in vitro (Asn313Ala/Lys314Ala) behaved in the same manner. Those mutants exhibiting a modest decrease in DNA binding (His215Ala/Lys216Ala, corresponding to xMcm10 Lys293Ala) displayed a 2-fold decline in survival (Figure 6B). Viability was more strongly compromised in Phe230Ala/Phe231Ala mutants. Either of these two phenylalanines correspond to Phe306 in xMcm10, which is implicated in DNA binding by our ssDNA docking model, but we were unable to test this directly because the protein was insoluble. Most strikingly, when Asn313 and Lys314 (corresponding to Lys385/386 in xMcm10) were changed to glutamic acid instead of alanine, survival was drastically reduced by more than 7-fold to about 30% even after a very short exposure to HU (Figure 6B). Taken together, these results extend our in vitro DNA binding studies and suggest that the corresponding residues in scMcm10 have an important role in DNA replication. Because neither Phe230/231 (located in the OB-fold) nor Asn313/Lys314 (located in the zinc finger loop) lie within the binding sites for pol α or PCNA (Das-Bradoo et al., 2006; Ricke and Bielinsky, 2006), it is likely that the HU sensitivity we detected is directly due to a defect of scMcm10 in DNA binding.
Figure 6. Mutations in the OB-fold and zinc finger domain of scMcm10 affect cell viability in hydroxyurea.
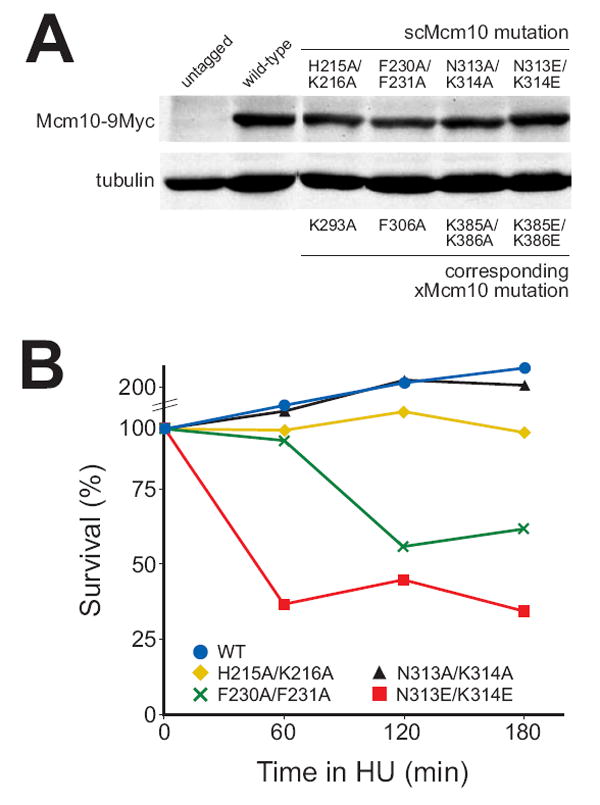
(A) Total protein extracts prepared from mid-logarithmic phase cycling cells were analyzed by western blot with anti-Myc and anti-α-tubulin antibodies. (B) Survival of wild type, mcm10-H215A/K216A, mcm10-F230A/F231A, mcm10-N313A/K314A, and mcm10-N313E/K314E cells after treatment with 0.2 M hydroxyurea for 60, 120 or 180 min is shown in one representative experiment.
Discussion
Mcm10-ID is the highly conserved region of the protein that binds both DNA and pol α. The crystal structure of Xenopus Mcm10-ID reveals a unique arrangement of OB-fold and zinc finger motifs. NMR chemical shift perturbation and mutational analysis show that DNA spans both the hydrophobic β barrel of the OB-fold and the electrostatic, extended loop of the zinc finger. This model is further substantiated by the finding that substitutions of conserved key residues within the OB-fold and zinc finger in scMcm10 decrease cell viability in the face of replication stress.
At this time we are unable to reconcile the high-resolution structure of the core domain of Mcm10 with the proposed hexameric structure of the full-length human protein (Okorokov et al., 2007). Efforts to dock our high-resolution crystal structure into the large volume within the hexameric EM reconstruction did not result in a clear solution. Moreover, the novel OB-fold/zinc finger configuration in Mcm10-ID bears no structural or functional resemblance to the OB-fold/zinc finger domain in the ring-shaped archaeal MCM helicase (Figure 3A) (Fletcher et al., 2003). In addition, burying the ID within a hexameric assembly would likely occlude one or more sites of DNA or protein interactions described below. Nevertheless, the Mcm10 structures serve as an important launching point for further work to investigate the function of this essential eukaryotic replication factor.
The unique DNA binding platform observed here raises interesting questions regarding Mcm10 association with chromatin. Given that Mcm10 possesses two DNA binding domains and can bind to both ss- and dsDNA (Robertson et al., 2008), it is likely that the protein is anchored to DNA throughout replisome assembly. Interestingly, NMR titration of Mcm10-ID with dsDNA resulted in chemical shift perturbations of largely the same residues as for ssDNA, strongly implying that the binding sites for ssDNA and dsDNA are similar. The binding of both ssDNA and dsDNA to the same site is unexpected given the existence of classic ssDNA and dsDNA binding motifs. Further analysis is required to understand the molecular basis for the complex nature of the DNA binding to Mcm10-ID and the results of such studies will be reported in due course. Nevertheless, the lack of specificity for a particular DNA structure and a common ss/ds-DNA binding site within the ID raises the possibility that Mcm10 acts as a molecular scaffold to stabilize the replisome on DNA. Indeed, the effect of DNA binding residues on HU sensitivity in yeast suggests that Mcm10’s DNA binding function is critical for fork integrity during DNA synthesis.
The structure of Mcm10-ID enables the location of residues that effect DNA replication and cell viability (Supplementary Figure S5). The cdc23-1E2 Cys239Tyr (Grallert and Nurse, 1997) and cdc23-M30 Leu287Pro (Liang and Forsburg, 2001) mutations map to xMcm10 Leu323 and Leu369, respectively, which point into the core of the OB-fold β-barrel and are thus likely to disrupt the protein fold. Similarly, cdc23-M36 Asp232Gly (Nasmyth and Nurse, 1981) relates to an invariant aspartate (xMcm10 Asp313) on the interior of the L23 loop, and thus likely alters the conformation of L23. This loop might mediate Mcm10-protein interactions given its surface exposed location immediately outside of the DNA binding region. Similarly, cdc23-M36 Val265Ile and mcm10-1 Pro269Leu mutations (Maine et al., 1984; Nasmyth and Nurse, 1981) map to solvent exposed positions on L45 and thus likely mediate intermolecular interactions. These residues are adjacent to the putative pol α binding surface, and extensive NMR chemical shift perturbation was observed in the L45 loop upon addition of DNA (Figure 4B).
The relative positions of residues that mediate protein-protein and protein-DNA interactions provide further insight into Mcm10’s role at the replication fork. Of Mcm10’s protein binding partners, PNCA and pol α are the most extensively characterized. The putative PCNA interacting protein (PIP) box predicted from the scMcm10 sequence (Ricke and Bielinsky, 2006) coincides with the OB-fold β3 strand (Supplementary Figure S5). scMcm10 Tyr245 was found to be important for an interaction between diubiquinated scMcm10 and PCNA (Das-Bradoo et al., 2006). This residue (xMcm10 Phe324) is located on the concave, DNA-binding face of β3 (Figure 5B,C) and had a modest effect on DNA binding. We therefore speculate that it might contribute to DNA binding in unmodified Mcm10, but alters its interaction with DNA upon Mcm10 ubiquitination, which has been suggested to trigger a conformational change (Das-Bradoo et al., 2006). This conformational change likely affects the β3 strand within the OB-fold because extensive mutational analysis suggests that Mcm10 interacts directly with PCNA via its PIP box motif (Das-Bradoo et al., 2006). Further work to elucidate the site of ubiquitination and its structural consequences will be required to understand the mechanistic basis for how Mcm10 modulates its interactions with DNA and PCNA.
In S. cerevisiae, the primary interaction site for pol α is confined to a hydrophobic stretch, the heat-shock protein (Hsp)10-like motif (Ricke and Bielinsky, 2006), which lies adjacent to Mcm10’s PIP box (Das-Bradoo et al., 2006) within Mcm10-ID. scMcm10 Asn268 (xMcm10 Asn346) is important for Mcm10 stabilization of pol α and maps to the C-terminal end of the β4 strand (Supplementary Figure S5). We have previously shown that Mcm10-ID binds to the N-terminus of the catalytic p180 subunit of pol α in vitro (Robertson et al., 2008). The fact that Asn346 is surface exposed is consistent with a role for this residue in binding pol α. In addition, Asn346 is clearly located outside of the DNA binding interface. This raises the possibility that Mcm10-ID can bind DNA and pol α simultaneously and is consistent with the proposal that Mcm10 acts to recruit pol α to the origin (Ricke and Bielinsky, 2004, 2006). Furthermore, evidence is mounting to suggest that Mcm10 likely associates with pol α during elongation (Chattopadhyay and Bielinsky, 2007; Pacek et al., 2006; Ricke and Bielinsky, 2004, 2006; Yang et al., 2005). Structures of Mcm10 in complex with its protein binding partners will be critical to understand the physical basis for function of this modular, multi-functional protein.
Experimental Procedures
Mcm10 Purification
Xenopus laevis Mcm10-ID (amino acids 230-427) was expressed and purified as previously reported (Robertson et al., 2008). Briefly, Mcm10-ID was overexpressed as a N-terminal thioredoxin-His6 fusion protein from a modified pET-32a expression vector (Novagen) in E. coli BL21(DE3) cells at 16° C. For NMR experiments, protein was uniformly enriched with 13C and 15N by propagating cells in M9 minimal medium supplemented with 2 mg/ml 13C6-glucose and/or 1 mg/ml 15NH4Cl (Cambridge Isotope Laboratories). Cells were harvested in 50 mM Tris pH 7.5, 500 mM NaCl, and 10% glycerol, and lysed under pressure (25,000 psi) using an Avestin EmulsiFlex C3 homogenizer. Mcm10-ID was isolated using Ni-NTA affinity chromatography (Qiagen). Following cleavage of the affinity tag, the protein was further purified by ssDNA-cellulose (Sigma) and gel filtration chromatography using a Superdex™ 200 preparative column (GE Healthcare) equilibrated in 20 mM Tris pH 7.5, 150 mM NaCl, and 5% glycerol.
X-ray Crystallography
Mcm10-ID crystals were grown by hanging drop vapor diffusion from 100 mM Tris pH 8.0, 100 mM KSCN, and 40% PEG 4000, and were flash frozen in mother liquor containing 10% glycerol prior to data collection. X-ray diffraction data were collected at beamline 22-ID at the Advanced Photon Source (Argonne, IL) and processed with HKL2000 (Otwinowski and Minor, 1997). Mcm10-ID crystallized in space group P21 with three molecules in the asymmetric unit.
Experimental X-ray phases were obtained from a multiwavelength anomalous dispersion (MAD) experiment using a single crystal that was soaked for 5 h at 25°C in mother liquor supplemented with 1 mM KAu(CN)2. Diffraction data (Table 1) were collected at 110 K at energies corresponding to the peak (1.0388 Å), inflection (1.0370 Å) and high-energy remote (1.0311 Å) settings for the gold LIII absorption edge. The positions of 10 gold atoms in the asymmetric unit were located by automated Patterson searching using SHELXD (Uson and Sheldrick, 1999), and initial phases to 3 Å were refined by solvent flattening using the SOLOMON option within autoSHARP (Vonrhein et al., 2006). The model containing all three proteins in the asymmetric unit was built manually into the experimentally phased electron density using XtalView/Xfit (McRee, 1999). Electron density for residues 230-234 (N-terminus), 420-427 (C- terminus), and 298-304 (loop L12) were unobserved.
The model was refined against the native X-ray data (50-2.3 Å) with a maximum likelihood target for experimental phases as implemented in REFMAC 5.2 (Murshudov et al., 1997). Improvements to the model were made by manual inspection of σA-weighted 2mFo-DFc and mFo-DFc electron density maps, and they were judged successful by a decrease in Rfree during refinement. Translation/libration/screw-rotation (TLS) refinement in REFMAC was used to model anisotropic motion of each protein domain (three in total). Individual anisotropic B-factors were derived from the refined TLS parameters and held fixed during subsequent rounds of refinement, which resulted in a decrease in both R and Rfree and a noticeable improvement in the electron density maps (Supplementary Figure S1).
Analysis of the final structure using PROCHECK (Laskowski et al., 1993) showed 91.5% and 8.3% of the total of 515 residues to be within the favored and allowed regions of the Ramachandran plot, respectively. Only one residue, located at the extreme N-terminus, resided in the disallowed region. The coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession number 3EBE.
NMR Spectroscopy
Spectra were acquired at 25 °C on Bruker DRX 500, 600 and 800 NMR spectrometers equipped with cryoprobes. Backbone resonance assignments were made using 3D triple resonance experiments acquired on the DRX 600: HNCA, HNCACB, CBCA(CO)NH, (H)C(CO)NH-TOCSY, and HNCO. Chemical shift perturbation data were collected by titrating unlabelled 15mer oligonucleotide d(GGCGCATTGTCGCAA) into 250 μM 15N-Mcm10-ID in 20 mM Tris-d11 (pH 7.0), 75 mM NaCl and 5% D2O. Gradient enhanced 15N-1H HSQC NMR spectra were recorded at protein/DNA ratios of 1:0, 1:0.5, 1:1, 1:3, and 1:5. The observation of chemical shift perturbations in the fast exchange limit (on the NMR time scale) enabled the peaks in the free protein and DNA complexes to be correlated. All spectra were processed and analyzed using Topspin v1.3 (Bruker, Billerica, MA) and Sparky v3.1 (University of California, San Francisco, CA).
Mutagenesis and in vitro DNA Binding Assays
xMcm10 mutants were prepared using a Quik-Change Kit (Stratagene) and purified similarly to wild-type protein, except that the ssDNA-cellulose affinity step was replaced with an SP-sepharose (GE Healthcare) ion exchange step. DNA binding to Mcm10 mutants was measured by following an increase in fluorescence anisotropy as protein was added to a fluorescently-labeled oligonucleotide d(ATGGTAGGCAACCATGTAGTAGTCA) containing a 6-carboxyfluorescein moiety at the 3’-end. For DNA length dependence measurements, 5-, 10-, and 15mer oligonucleotides were derived from the 5’-end of the 25mer sequence above. Protein was added over the concentration range of 0.1-50 μM to a solution containing 25 nM DNA in 20 mM Tris pH 7.5, 100mM NaCl, and 5% glycerol. Polarized fluorescence intensities using excitation and emission wavelengths of 495 and 515 nm, respectively, were measured for 30 s (1/s) and averaged. Anisotropy (r) was calculated using the equation r = (Ipar-Iperp)/(Ipar+2Iperp), where Ipar and Iperp are the observed fluorescence intensities recorded through polarizers oriented parallel and perpendicular to the direction of vertically polarized light. Dissociation constants (Kd) were derived by fitting a simple two-state binding model to data from three experiments using Kaleidagraph 3.5 (Synergy Software, PA).
Hydroxyurea Survival Assay
scMcm10 mutant yeast strains were constructed as previously described (Das-Bradoo et al., 2006) and survival in HU was assayed according to an earlier report (Allen et al., 1994). Briefly, pRS406-MCM10-9MYC (ABy491), pRS406-MCM10-9MYC-H215A/K216A (ABy492), pRS406-MCM10-9MYC-F230A/F231A (ABy496), pRS406-MCM10-9MYC-N313E/K314E (ABy503), and pRS406-MCM10-9MYC-N313A/K314A (ABy525) were integrated at the endogenous MCM10 locus of ABy014 (W303). Total protein extracts were prepared from mid-logarithmic phase cycling yeast cultures (OD600 = 0.6) as described previously (Ricke and Bielinsky, 2006). Proteins were transferred to a nitrocellulose membrane and probed by western blot with anti-Myc (9E11, LabVision Neomarkers) for Myc-tagged scMcm10 and anti-α-tubulin (MMS-407R, Covance). For the hydroxyurea survival assay, cells were grown to mid-logarithmic phase (OD600 = 0.6). All mutants tested had doubling times comparable to wild type (data not shown). A 100 μl aliquot of cells was removed from each culture before adding 0.2 M hydroxyurea. 100 μl aliquots were removed at timed intervals, diluted, and colony-forming units were scored for viability on YPD plates as described previously (Allen et al., 1994). Percentage survival was determined relative to cells that were not exposed to hydroxyurea at the beginning of the experiment.
Supplementary Material
Acknowledgments
The authors are indebted to Audrey Metz for her assistance in crystal preparation, and to Johannes Walter for stimulating discussions. We thank the SER-CAT staff at the Advanced Photon Source (Argonne, IL), and Hassane Mchaourab and Albert Beth for access to fluorimeters. This work was funded by the National Institutes of Health (R01 GM080570 to B.F.E.; R01 GM065484 to W.J.C.; R01 GM074917 to A.-K.B.). Additional support for facilities came from the Vanderbilt Center in Molecular Toxicology (P30 ES000267) and the Vanderbilt Discovery Grant Program. E.M.W. was supported by the Molecular Biophysics Training Grant (T32 GM08320).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Bielinsky AK. Human Mcm10 Regulates the Catalytic Subunit of DNA Polymerase-{alpha} and Prevents DNA Damage during Replication. Mol Biol Cell. 2007;18:4085–4095. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen TW, Tye BK. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol Biol Cell. 2003;14:2206–2215. doi: 10.1091/mbc.E02-11-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo S, Ricke RM, Bielinsky AK. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol Cell Biol. 2006;26:4806–4817. doi: 10.1128/MCB.02062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fien K, Cho YS, Lee JK, Raychaudhuri S, Tappin I, Hurwitz J. Primer utilization by DNA polymerase alpha-primase is influenced by its interaction with Mcm10p. J Biol Chem. 2004;279:16144–16153. doi: 10.1074/jbc.M400142200. [DOI] [PubMed] [Google Scholar]
- Fien K, Hurwitz J. Fission yeast Mcm10p contains primase activity. J Biol Chem. 2006;281:22248–22260. doi: 10.1074/jbc.M512997200. [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Critical reviews in biochemistry and molecular biology. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. An approach to identify functional homologues and suppressors of genes in fission yeast. Curr Genet. 1997;32:27–31. doi: 10.1007/s002940050244. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000;14:913–926. [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- Izumi M, Yanagi K, Mizuno T, Yokoi M, Kawasaki Y, Moon KY, Hurwitz J, Yatagai F, Hanaoka F. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 2000;28:4769–4777. doi: 10.1093/nar/28.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Lee JK, Seo YS, Hurwitz J. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc Natl Acad Sci U S A. 2003;100:2334–2339. doi: 10.1073/pnas.0237384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim HK, Kwon ST, Suh SW. Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications. Embo J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DT, Forsburg SL. Characterization of Schizosaccharomyces pombe mcm7(+) and cdc23(+) (MCM10) and interactions with replication checkpoints. Genetics. 2001;159:471–486. doi: 10.1093/genetics/159.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Merchant AM, Kawasaki Y, Chen Y, Lei M, Tye BK. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3261–3271. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, A A, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Okorokov AL, Waugh A, Hodgkinson J, Murthy A, Hong HK, Leo E, Sherman MB, Stoeber K, Orlova EV, Williams GH. Hexameric ring structure of human MCM10 DNA replication factor. EMBO reports. 2007;8:925–930. doi: 10.1038/sj.embor.7401064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J Biol Chem. 2006;281:18414–18425. doi: 10.1074/jbc.M513551200. [DOI] [PubMed] [Google Scholar]
- Robertson PD, Warren EM, Zhang H, Friedman DB, Lary JW, Cole JL, Tutter AV, Walter JC, Fanning E, Eichman BF. Domain Architecture and Biochemical Characterization of Vertebrate Mcm10. J Biol Chem. 2008;283:3338–3348. doi: 10.1074/jbc.M706267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- Solomon NA, Wright MB, Chang S, Buckley AM, Dumas LB, Gaber RF. Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast. 1992;8:273–289. doi: 10.1002/yea.320080405. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. Embo J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr Opin Struct Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods in molecular biology (Clifton, N J) 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walther AP, Gomes XV, Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry. 1999;38:3963–3973. doi: 10.1021/bi982370u. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- Yang X, Gregan J, Lindner K, Young H, Kearsey SE. Nuclear distribution and chromatin association of DNA polymerase alpha-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast. BMC Mol Biol. 2005;6:13. doi: 10.1186/1471-2199-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


