Abstract
Fanconi anemia (FA) is a recessive genome instability syndrome characterized by heightened cellular sensitivity to DNA damage, aplastic anemia and cancer susceptibility. Leukemias and squamous cell carcinomas (SCC) are the most predominant FA associated cancers, with the latter exhibiting markedly early disease onset and aggressiveness. While studies of hematopoietic cells derived from FA patients have provided much insight into bone marrow deficiencies and leukemogenesis, molecular transforming events in FA deficient keratinocytes, which are the cell type of origin for SCC, are poorly understood. We describe here the growth and molecular properties of FANCA-deficient versus FANCA-corrected, HPV E6/E7 immortalized keratinocytes in monolayer and organotypic epithelial raft culture. In response to DNA damage, FANCA-deficient patient-derived keratinocyte cultures displayed a G2/M phase arrest, senescence and apoptosis. Organotypic raft cultures exhibited DNA repair associated defects with more 53BP1 foci and TUNEL positive cells over their corrected counterparts. Interestingly, together with reduced rates of DNA damage, FA correction resulted in a marked decrease in epithelial thickness and the presence of fewer cell layers. The observed FANCA mediated suppression of hyperplasia correlated with the detection of fewer cells transiting through the cell cycle in the absence of gross differentiation abnormalities or apoptotic differences. Importantly, the knockdown of either FANCA or FANCD2 in HPV positive keratinocytes was sufficient for increasing epithelial hyperplasia. Our findings support a new role for FA pathways in the maintenance of differentiation-dependent cell cycle exit, with the implication that FA deficiencies may contribute to the high risk of FA patients for developing HPV-associated SCC.
Keywords: Fanconi anemia, human papillomavirus
Introduction
Fanconi anemia is a rare, recessive DNA instability syndrome and is usually diagnosed early in life based on congenital abnormalities, progressive aplastic anemia, cellular hypersensitivity to DNA damage, and a greatly increased risk of cancer. FA-associated anemia is often fatal and presents a median life expectancy of 20 years in the absence of a bone marrow transplant (Alter, 2003). Thirteen FA complementation groups were identified by cell fusion experiments (Wang, 2007) and more recently by retroviral complementation of specific FA genes (Chandra et al., 2005; Hanenberg et al., 2002). FA pathway functionality involves a high molecular weight nuclear core complex composed of FA proteins FANCA, B, C, G, E, F, G, L, M, FAAP24 and FAAP100 (Meetei et al., 2003a; Meetei et al., 2005; Niedernhofer et al., 2005). Complex activation in response to DNA damage or replication results in the monoubiquitination of FANCD2 (Garcia-Higuera et al., 2001; Timmers et al., 2001) and FANCI (Dorsman et al., 2007; Sims et al., 2007; Smogorzewska et al., 2007), and their colocalization with FANCD1/BRCA2 (Howlett et al., 2002), FANCN/PALB2 (Rahman et al., 2007; Reid et al., 2006) and FANCJ (BRIP1) (Bridge et al., 2005; Levitus et al., 2005) on sites of DNA repair. Complex activation then mediates DNA repair via poorly understood mechanisms that likely include the BRAFT complex, as FA proteins are components of the larger BRAFT DNA repair complex containing Bloom’s helicase (BLM), replication protein A (RPA) and topoisomerase IIIa (Meetei et al., 2003b). Other FA protein functions include roles in cellular survival and oxidative stress responses (Bagby & Alter, 2006; Pagano et al., 2005).
Molecular and biochemical analyses of FA patient cells have yielded important insights into the biology and importance of FA function in recent years. Research has focused mainly on the hematopoietic problems in FA patients, which can now be cured with a successful bone marrow transplant under optimal circumstances. However, FA patients who have undergone bone marrow transplants continue to be at high risk for developing solid tumors. Squamous cell carcinomas (SCCs) of the head and neck (HN), anogenital region and skin are the most commonly diagnosed solid tumors in FA patients (Kutler et al., 2003b), supporting important and independent FA functions in human keratinocytes. A potential link between HPV and FA (Kutler et al., 2003b; Kutler et al., 2003c) – albeit controversial (van Zeeburg et al., 2004; van Zeeburg et al., 2005) – is supported by a recent report showing accelerated chromosomal instability in HPV16 E7 oncogene expressing, FA deficient mouse embryo fibroblasts (Spardy et al., 2007). FA deficiency may accelerate tumorigenesis by promoting genomic instability that cooperates with HPV mediated inactivation of critical tumor suppressors. Examination of FA specific keratinocyte phenotypes in premalignant cells, particularly in the context of the HPV oncogenes, is critical for a deeper understanding of SCC development in FA patients.
Keratinocytes are the cell type of origin for SCC, but only one report has described some of the phenotypic characteristics of FA patient-derived epithelial cancer cell lines (van Zeeburg et al., 2005). Since keratinocyte differentiation as a major barrier to cancer has already been overcome in such tumors, the use of the above cell lines has not provided insights into the consequences of FA inactivation in the context of differentiation. Here we describe the culture, FA correction known as complementation, and characterization of HPV18 E6/E7 immortalized keratinocytes derived from a FANCA patient skin biopsy. This system explores the role of FA in an oncogenically challenged, premalignant state, and within three-dimensional epithelium. FANCA deficiency in E6/E7 positive monolayer cells did not result in significant changes in proliferation or cell death under normal culture conditions. However, in response to DNA damaging agents, phenotypes similar to those described for hematopoietic and stromal cells were evident. These included the lack of FANCD2 monoubiquitination, cell cycle arrest and increased rates of cell death. Interestingly, under differentiating conditions, FANCA complementation decreased HPV associated hyperplasia in organotypic epithelial rafts in correlation with a decrease in the proliferative index. Notable differences in the localization of differentiation markers were not observed. Conversely, FANCA or FANCD2 knockdown in HPV positive cells increased HPV associated hyperplasia. Taken together, our data demonstrate that HPV associated hyperplasia in FANCA mutant epithelium can be attenuated by the re-establishment of a functional FA pathway, and that the loss of FA pathways in HPV positive keratinocytes inhibits differentiation-associated cell cycle withdrawal.
Materials and Methods
Cell culture and vectors
Fresh skin biopsies were obtained through the Fanconi Anemia Comprehensive Care Center from patients undergoing clinically-indicated bone marrow aspiration, and keratinocytes were cultured on irradiated J2-3T3 feeder cells as described in (Nakahara et al., 2005). After colony outgrowth and replating, primary keratinocytes were transduced with the human papillomavirus type 18 E6 and E7 oncogenes by infection with the SF91-18E6/E7 retroviral vector, or transfected with episomal HPV16 DNA as previously described for primary normal keratinocytes (Hoskins et al., 2008). The SF91 viral backbone was a generous gift from the Baum laboratory (Schambach et al., 2000). For generation of the retroviral vector, the HPV18 E6/E7 open reading from was cloned into pGEM Easy (Promega, Madison, WI), excised using Not I and subcloned into Not I digested pSF91 vector. Proper insert orientation was verified by PCR and the resulting vector was sequenced. E6/E7 and HPV immortalized cells were carried for 12 passages to ensure relative culture stability, and then complemented with either FANCA expressing retrovirus or with empty vector backbone. These vectors were previously described (Chandra et al., 2005), and were provided by the CCHMC Viral Vector Core and Translational Trials Development and Support Lab. A pure population of GFP positive cells was obtained by flow cytometric sorting in the CCHMC Flow Core Facility.
FANCA and FANCD2-specific RNA interference
HPV positive normal immortalized keratinocytes (NIKs) were a gift from Paul Lambert, University of Wisconsin-Madison, and were carried as previously described (Nakahara et al., 2005). Irradiated J2-3T3 cells were used as feeder cells. Non-targeting, FANCA and FANCD2 specific shRNA-expressing and puromycin resistant lentiviral vectors were obtained through the Sigma™ MISSION®shRNA program (Sigma-Aldrich, St. Louis, MO). Relevant product numbers were SHC 002V for the nontargeting control vector (NT), TRCN0000082841 for the FANCD2sh4 targeting vector, and TRCN0000118982 and TRCN0000118985 for the FANCA targeting vectors. Keratinocytes were infected with either lentiviral nontargeting control vector or with the FANCD2 shRNA expression vectors, and selected in 1ug/ml puromycin for four days. For FANCA knockdown, five vectors were pre-screened for functionality in HeLa cells overexpressing FANCA, and the two most potent vectors were selected, combined and used for the HPV-NIKs transduction. Successful FANCA knockdown was confirmed functionally by western blot detection of the mono- versus nonubiquitinated FANCD2 forms following cell exposure to HU for 16 hrs.
Organotypic epithelial raft culture
Organotypic rafts were generated as described in (Nakahara et al., 2005). Briefly, a total of 1×106 human keratinocytes was plated on a collagen matrix with embedded feeder fibroblasts. Exposure to the liquid-air interface resulted in the generation of stratified epithelium with differentiation properties that reflect its natural human counterpart. The tissue was fixed in 4% paraformaldehyde 12 days after lifting, embedded in paraffin, sectioned and morphologically examined by hematoxylin/eosin staining.
Immunofluorescence
Monolayer cultures: Keratinocytes were plated onto polylysine-coated coverslips and allowed to grow at 37°C and 5% CO2. When the cells reached 80% confluency, they were either left untreated or treated with 0.5 mM hydroxyurea for 16 hours to induce DNA damage. Keratinocytes were then extracted for 35 minutes on ice in supplemented CSK buffer (10 mM PIPES, 100 mm NaCl, 300 mM sucrose, 1 mM MgCL2, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 0.2% Triton X-100, 1X protease inhibitor cocktail), washed with PBS, and fixed in 100% methanol for five minutes on ice. Immunofluorescence staining was performed by incubating keratinocytes with anti-FANCD2 (Novus, Littleton, CO) or anti-phosphorylated histone H2AX antibody (Upstate, Charlottesville, VA) for one hour at 37°C, 5% CO2 and then either in polyclonal rhodamine– or monoclonal FITC–conjugated antibodies (Jackson Immunoresearch, West Grove, PA) for 30 minutes at 37°C, 5% CO2. Vectashield mounting medium with DAPI (Vector laboratories, Burlingame, CA) was used for nuclear staining. A Zeiss (Thornwood, NY) Axioplan 2 Imaging microscope and Axiovision Release 4.6 software were utilized to take images. Keratinocytes containing at least five FANCD2 or γH2AX foci were counted as positive and the data was expressed as percent positive relative to the total cell number. Organotypic rafts. Tissue sections were deparaffinized with xylene and rehydrated. Following antigen retrieval in 10mM sodium citrate, sections were blocked with 5% donkey serum (Jackson Immunoresearch) in PBS for 30 min, and then incubated with mouse monoclonal K10 and rabbit polyclonal K14 (both from Covance Research Products, Richmond, CA), mouse monoclonal E-cadherin, or mouse monoclonal Ki67 (both from BD Biosciences, San Jose, CA) antiserum in IF antibody dilution buffer (3% BSA, 0.5% Triton-X100, 0.05% Tween-20, 0.04% sodium azide in PBS) overnight at 4°C. Detection was with the above secondary antibodies (Jackson Immunoresearch) in antibody dilution buffer. BrdU detection was similar using an anti-BrdU antibody (ZBU-30) from Invitrogen, Carlsbad, CA. TUNEL staining was performed using the In situ Cell Death Detection Kit, TMR Red (Roche Applied Science, Indianapolis, IN) as per manufacturer instructions. The sections were mounted in Vectashield mounting media containing DAPI and analyzed using the microscope and software described in A. In order to analyze the organotypic raft cultures for the presence of 53BP1 and FANCD2 foci, we used a polyclonal antibody against 53BP1 (Novus Biologicals, Littleton, CO) or FANCD2 (Genetex, San Antonio, TX) followed by a Fluorescein isothiocyanate- or Rhodamine Red-conjugated secondary antibody (Jackson Immunoresearch).
Apoptosis Analysis by Flow Cytometry
Cells were split at a density of 4×105 cells per 60mm dish on feeder fibroblasts, and either left untreated, or treated the following day with either 10 ng/ml or 50 ng/ml mitomycin C (Sigma-Aldrich, St. Louis, MO). On day 5, cells were trypsinized and 1×106 cells from each treatment were washed in 1× PBS and fixed for 20 minutes. Subsequent washes were performed using BD Perm/Wash Buffer (BD Biosciences, San Jose, CA). Cells were incubated for 1 hour in the dark with 20ul biotinylated anti-active caspase-3 rabbit monoclonal antibody diluted in 100ul BD Perm/Wash buffer, followed by a 30 minute incubation in 500ul BD Perm/Wash containing 5ng of streptavidin-APC (BD Pharmingen, San Jose, CA). Cells were acquired on a BD FACSCalibur instrument (BD Biosciences, San Jose, CA) and the results were analyzed using the FCS3 Express software (De Novo, Los Angeles, CA).
Cell Cycle Analysis by Flow Cytometry
Cells were split as for the apoptosis assay, but either left untreated or treated on the following day with 0.25ug/ml melphalan (Sigma-Aldrich, St. Louis, MO). After 48 hours, cells were trypsinized and 5×105 cells were washed in 1× PBS and fixed in 100ul BD Cytofix/Cytoperm (BD Biosciences, San Jose, CA). The protocol was followed for the APC BrdU Flow Kit (BD Pharmingen, San Jose, CA). Cell cycle profiles were detected using 7AAD, with samples acquired on a BD FACSCalibur instrument (BD Biosciences, San Jose, CA) and the results were analyzed using the FCS3 Express software (De Novo, Los Angeles, CA).
Western blot analysis
The cells were washed with PBS and whole cell lysates were harvested with RIPA buffer (1% Triton, 1% DOC, 0.1% SDS, 0.16M NaCl, 10mM Tris pH 7.4, and 5mM EDTA) supplemented with a protease inhibitor cocktail (Pharmingen, San Diego, CA). Protein concentrations were determined using Bradford Reagent (BioRad, Hercules, CA). Equal amounts of protein were boiled in SDS sample buffer and resolved by SDS-PAGE. Proteins were transferred to a PVDF membrane (BioRad, Hercules, CA) and probed with mouse monoclonal FANCA (5G9, a generous gift from Maureen Hoatlin, OHSU), rabbit polyclonal FANCD2 (Novus Biologicals, Littleton, CO), or an actin specific monoclonal antibody, a generous gift from James Lessard. On the next day, the membrane was washed with TNET (10mM Tris, 2.5mM EDTA, 50mM NaCl, and 0.1% Tween 20) and secondary anti-mouse or anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham, Piscataway, NJ) were added for 30 minutes. Membranes were then exposed to enhanced chemilluminescence reagents (Perkin Elmer, Boston,MA) and the protein bands were detected by autoradiography.
Senescence assay
Staining for perinuclear SA-βgal activity was performed as previously described (Dimri et al., 1995).
Results
FA patient derived, HPV18 E6/E7 immortalized keratinocytes exhibit DNA damage phenotypes
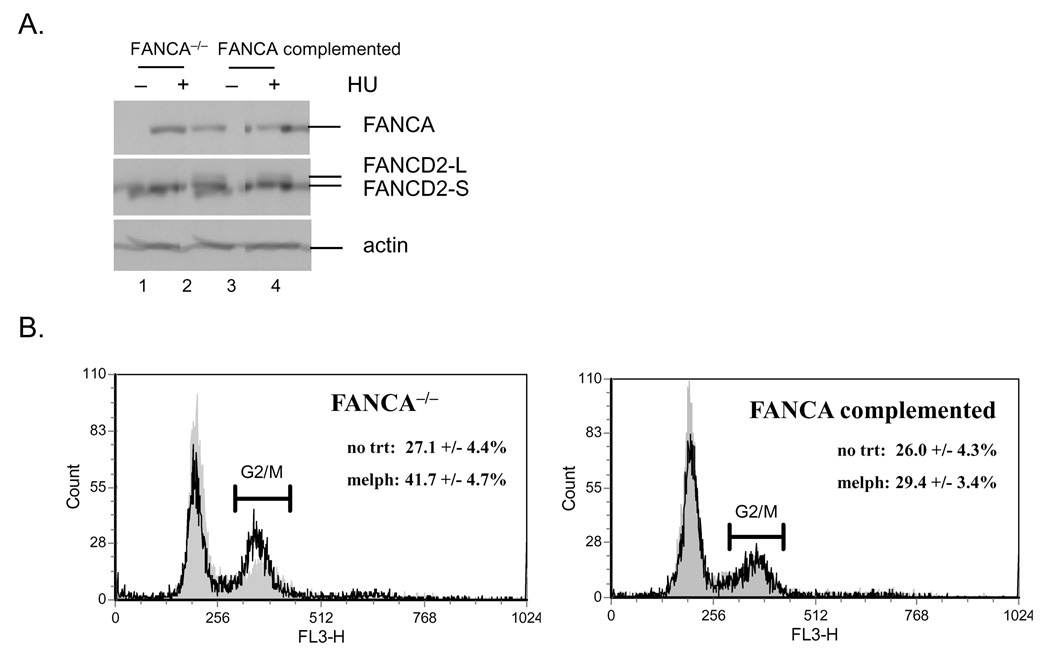
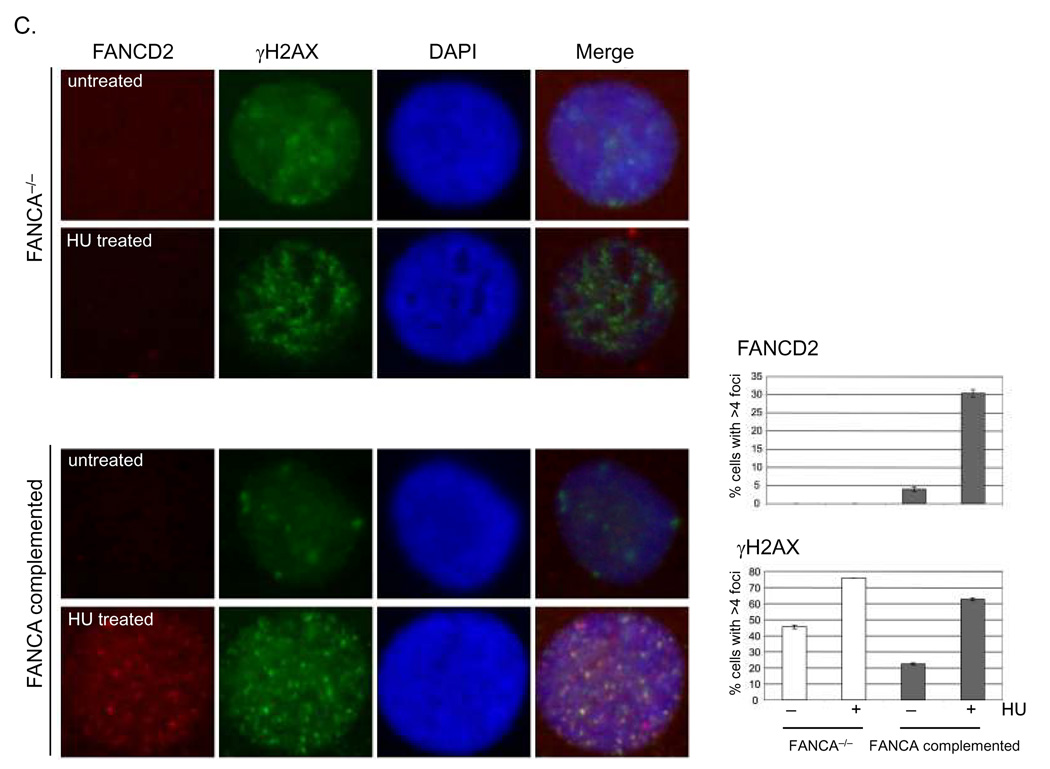
To determine the consequences of FA deficiency on epithelial growth and differentiation, we isolated and cultured keratinocytes from a skin biopsy of a FANCA deficient patient. The cells were immortalized by retroviral transduction with an HPV18 E6/E7 expression vector, and then either complemented with a FANCA expressing retroviral vector or transduced with a control vector expressing only EGFP. A pure population of GFP positive cells was obtained following flow cytometric sorting, and phenotypic differences were analyzed in the presence and absence of DNA damaging agents. FANCA expression in the complemented cells was confirmed at the level of protein expression (Fig. 1A, lanes 3 and 4). Defects similar to those reported for hematopoietic cells and fibroblasts (Tamary & Alter, 2007) were observed following treatment with the DNA damaging agents hydroxyurea (HU) and melphalan (Fig. 1). These included defective monoubiquitination of FANCD2 (Fig. 1A) and reduced FANCD2 nuclear foci (Fig. 1C) following hydroxyurea (HU) treatment, as well as the accumulation of cells with a G2/M content following melphalan treatment (Fig. 1B). Control staining was performed with γH2AX specific antibodies (1C). Importantly, partial phenotypic complementation could be achieved for each phenotype through the reintroduction of the FANCA cDNA.
Figure 1. Complementation and characterization of FANCA deficient keratinocyte cell lines.
Primary keratinocytes from a FANCA deficient patient were immortalized with an HPV18 E6/E7 expressing retroviral vector and then complemented either with empty MIEG or FANCA expressing retrovirus. A. Western blot analysis was performed after treatment with 0.5 mM HU. Monoubiquitinated FANCD2: FANCD2-L. Nonubiquitinated FANCD2: FANCD2-S. B. Cell cycle profiles were obtained following treatment with 0.25 ug/ml melphalan for 48 hrs using 7-AAD incorporation and flow cytometry. Only one representative profile is shown. The numbers represent an average of three independent experiments as percentage of cells with G2/M content together with the standard deviation. C. Nuclear FANCD2 and γH2AX foci were detected by immunofluorescence as described in (Montes de Oca et al., 2005) and quantitated.
FANCA deficient keratinocytes are hypersensitive to mitomycin C induced cell death
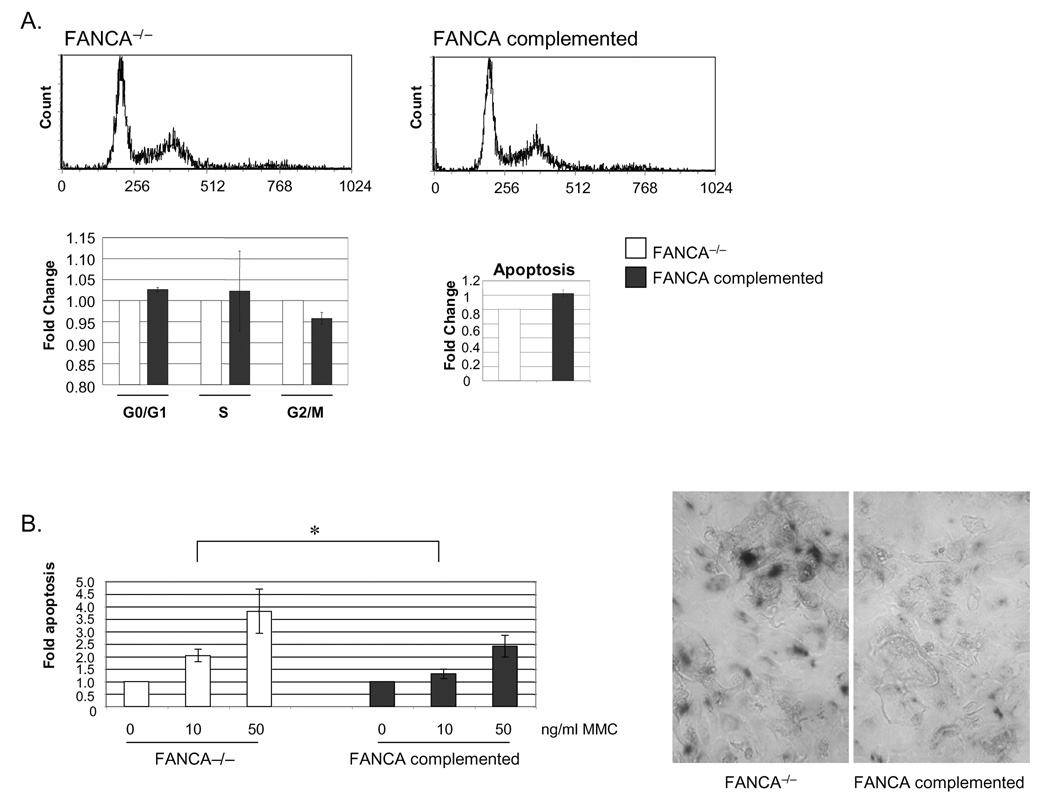
Analyses of complemented versus noncomplemented cells in the absence of DNA damaging agents did not reveal elevated rates of cell cycle arrest or apoptosis in the FA deficient cells (Fig. 2A). We next determined whether these same cells were hypersensitive to DNA damaging agents similar to FA hematopoietic and stromal cells. Keratinocyte populations were treated with increasing doses of mitomycin C (MMC), and subjected to flow cytometry based measurements of apoptosis using activated caspase 3 antibodies. As observed with immortalized, FA mutant hematopoietic cells and fibroblasts, FA deficient keratinocytes were more susceptible to apoptosis following MMC treatment (Fig. 2B). In addition, cell treatment with 50 ng/ml MMC and staining for the detection of senescence associated beta gal activity (SA-βGal) after 10 days revealed heightened susceptibility to senescence (Fig. 2B, right panels). These data support a scenario whereby the steady state growth of immortalized, FA deficient keratinocytes is not significantly impacted under standard in vitro conditions, but is compromised through apoptosis and senescence in response to exogenous DNA damage induction.
Figure 2. FANCA deficient human keratinocytes are susceptible to mitomycin C induced cell death and senescence.
A. FANCA deficient and complemented cells were subjected to cell cycle and apoptosis measurements under standard culture conditions as described in the materials and methods. B. Cells were exposed to increasing doses of mitomycin C and subjected to apoptosis analyses. The asterisk indicates a P value of less than 0.05. Cells treated with 50 ng/ml MMC were stained for SA-bGal activity 10 days post exposure and imaged.
DNA damage in FANCA deficient epithelium
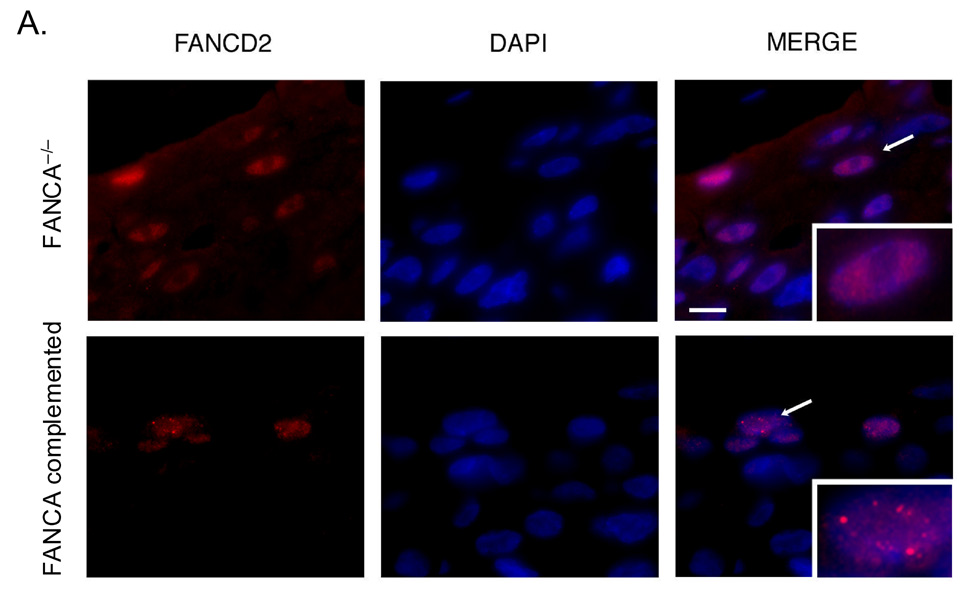
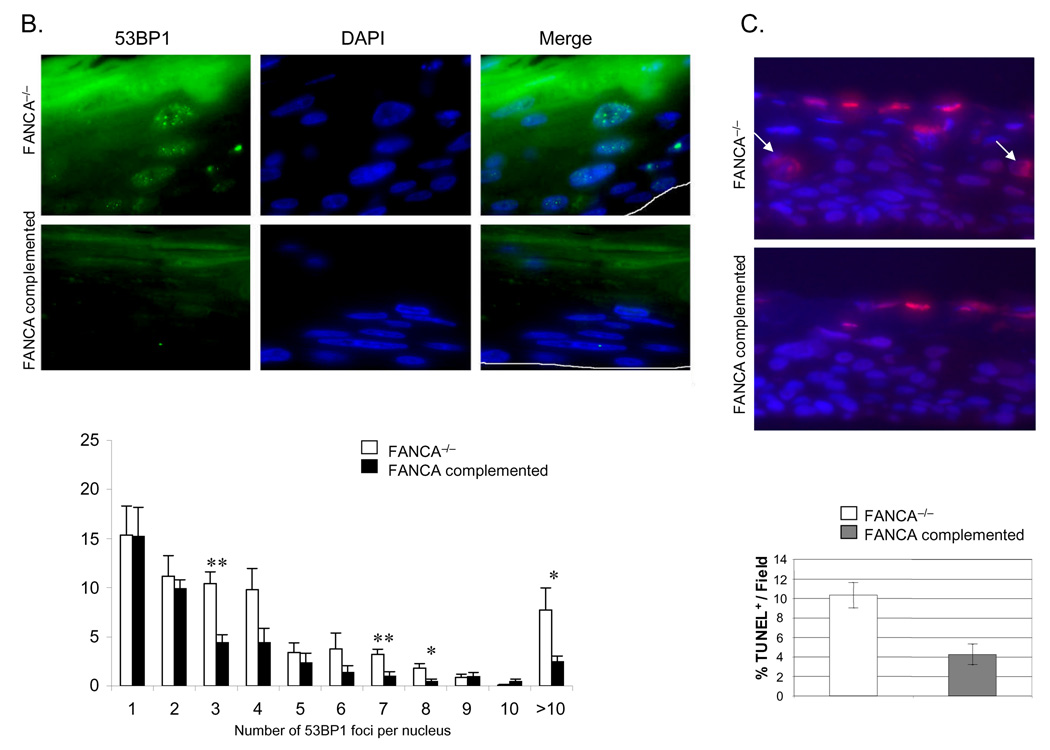
In order to determine whether the absence of a functional FA pathway is associated with increased accumulation of DNA damage in HPV positive human epithelium, we utilized the FA deficient and complemented cultures for generation of organotypic rafts. Detection of FANCD2 foci in FANCA expressing, but not deficient rafts confirmed successful complementation in this system (Fig. 3A). We next quantitated nuclear 53BP1 foci in FANCA deficient versus complemented rafts (Fig. 3B). 53BP1 is a component of the DNA damage response and sensor of DNA double-strand breaks (Huyen et al., 2004). Numbers of cells with detectable nuclear 53BP1 foci as well as numbers of foci per nucleus were substantially increased in FANCA deficient compared to complemented rafts. The observed overactivated 53BP1 response is likely a consequence of intracellular accumulation of DNA damage in FA deficient rafts. To further support this notion, we quantitated double stranded DNA ends on FANCA deficient and complemented rafts by TUNEL staining. In total, over 1100 nuclei were counted for each raft. Excluding enucleating cells in the cornified layer, we detected increased numbers of TUNEL positive cells in the FANCA deficient rafts (Fig. 3C). We conclude that the repair of baseline and/or E6/E7-induced DNA damage in keratinocytes involves FA pathways.
Figure 3. Increased rates of DNA damage in FANCA deficient epithelium.
Cell lines from Figure 1 were utilized for the generation of organotypic raft cultures as described in the materials and methods. Rafts were fixed, embedded in paraffin and sectioned. A and B. FANCD2 and 53BP1 staining was performed as follows: tissue sections were baked overnight, deparaffinized in xylene and dehydrated in 100% ethanol. After rehydration in a graded ethanol series, slides were washed in distilled dH2O, microwave treated in 0.01 M sodium citrate (pH 6.0) for 10 min (53BP1) or 30 min (FANCD2), and rinsed in dH2O. For FANCD2 staining, sections were digested using pepsin. All sections were then washed in PBS and blocked in 10% normal donkey serum for 30 min at room temperature. Slides were incubated with either a polyclonal antibody against 53BP1 or FANCD2 followed by a Fluorescein isothiocyanate- or Rhodamine Red-conjugated secondary antibody. Nuclei were stained with DAPI. C. TUNEL staining was performed as per the Roche In situ Cell Death Detection Kit, TMR Red instructions. TUNEL positive nuclei below the parakeratotic compartment are indicated by arrows. Nuclei were counterstained with DAPI.
HPV E6/E7 associated hyperplasia is attenuated by FANCA complementation and FANCA loss in HPV positive cells stimulates hyperplasia
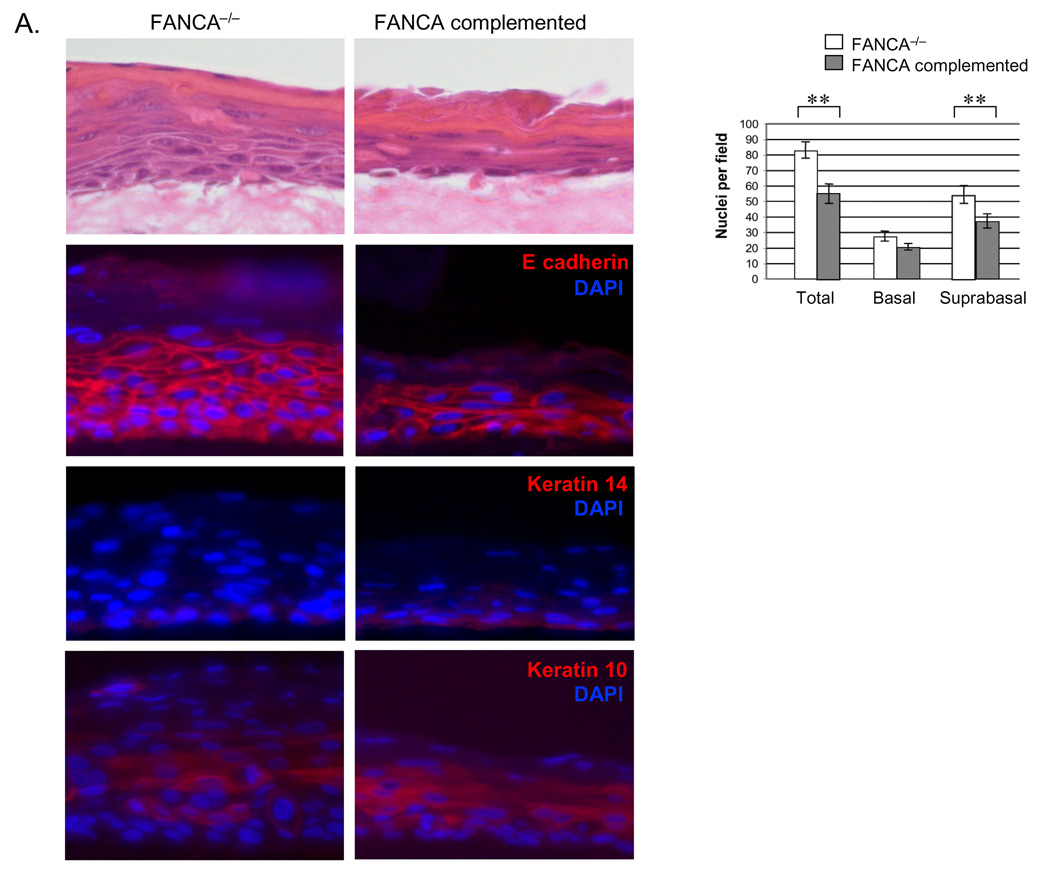
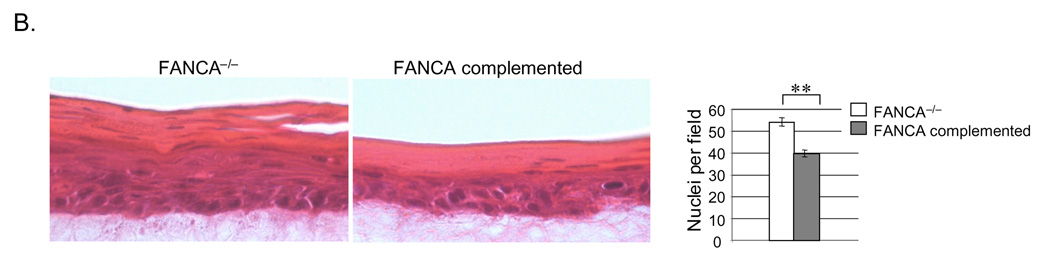
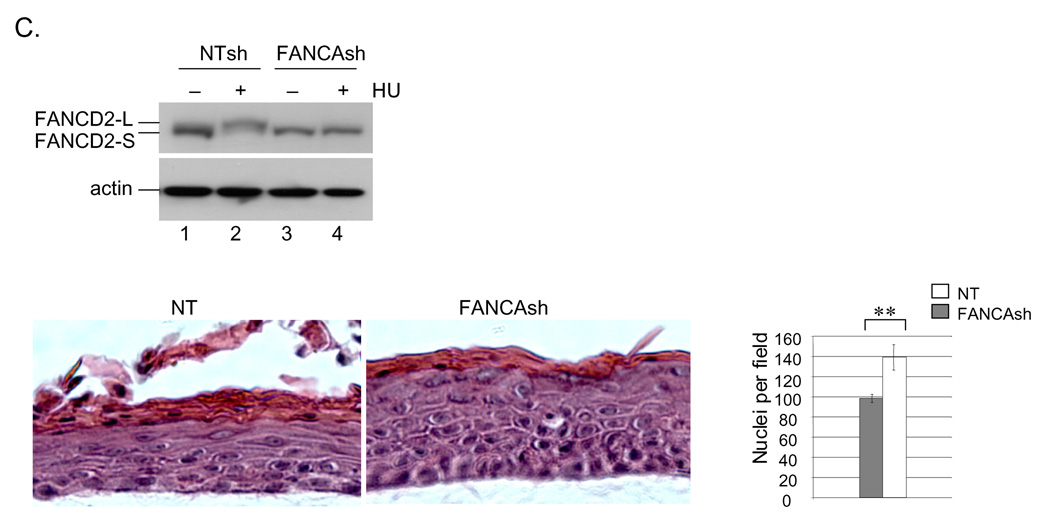
In addition to evidence of DNA damage suppression in Fig. 3, the FANCA complemented raft exhibited decreased hyperplasia upon morphological comparison (Fig. 4). Hematoxylin and eosin staining demonstrated increased raft thickness and the presence of additional spinous cell layers in the FANCA deficient compared to the complemented rafts. Similar phenotypes were noted in two additional, independently derived epithelial rafts obtained from the same cell populations (data not shown). Hyperplasia was reflected by increased numbers of nuclei in the suprabasal compartment. Closer morphological examination revealed approximately equal numbers of dysplastic and dyskeratotic cells, likely related to E6/E7 expression, in both rafts (data not shown). Epithelial differentiation and maturation properties were also similar as verified by comparable patterns of expression of the basal cell marker K14 and the suprabasal cell marker K10 by immunofluorescence (Fig. 4A, bottom two panels). Detection of the adherens junction component E-cadherin revealed more E-cadherin positive cell layers, likely a reflection of increased raft thickness, in the FANCA deficient epithelium. However, we did not note obvious E-cadherin mis-localization and/or expression at a single cell level, indicating similar cell-cell contact properties in the FANCA deficient versus complemented epithelium. A marked reduction in hyperplasia was also observed upon FANCA complementation of HPV16 episome-immortalized FA patient keratinocytes (Fig. 4B). Conversely, we detected an increase in hyperplasia following lentiviral knockdown of FANCA in HPV positive normal human keratinocytes (NIKs) and subsequent raft generation (Fig. 4C), thus ruling out the possibility that clonal selection might have been associated with the observed differences between FANCA deficient versus sufficient patient-derived keratinocytes. We cannot fully exclude the possibility that increased hyperplasia is a consequence of subtle cellular adaptation to FA loss that was undetectable in 2D culture. However, if proliferative gains and epithelial hyperplasia are adaptive in nature, our data would support that they remain reversible through the re-instatement of functional FA pathways by complementation.
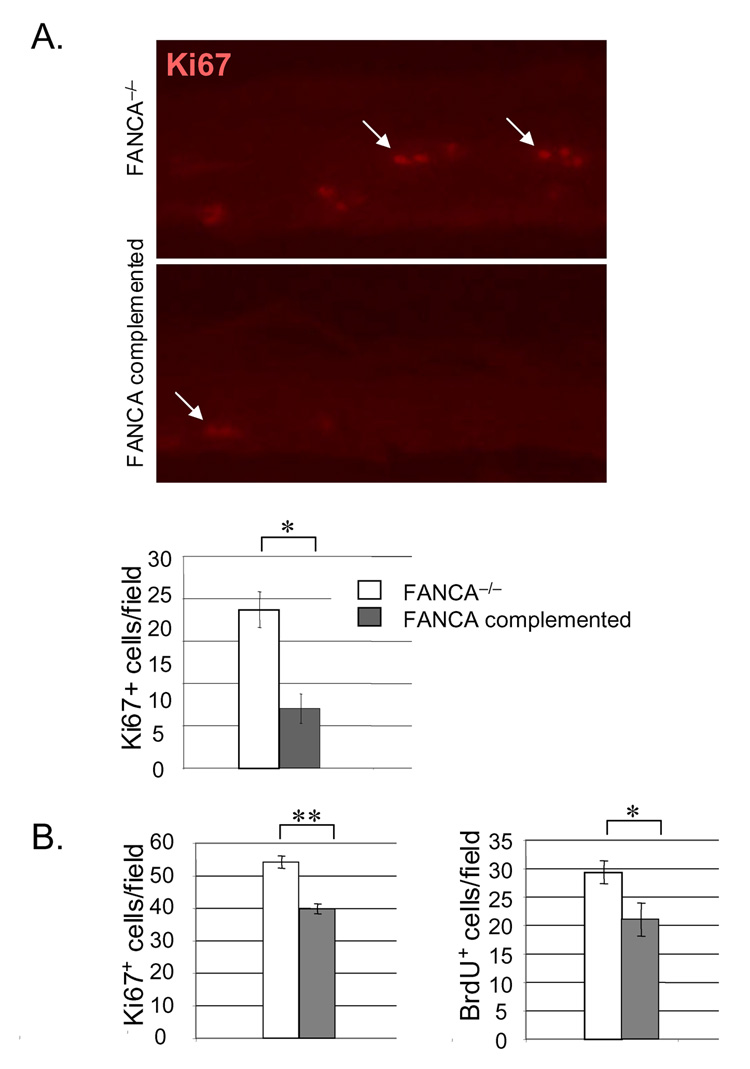
Figure 4. Suppression of HPV associated epithelial hyperplasia by FANCA complementation.
A. Organotypic raft sections were stained with hematoxylin and eosin for morphological evaluation (top panel). Hyperplasia as observed in the FANCA deficient rafts was additionally quantitated by nuclear counts. The data are represented in the graph on the right. Immunofluorescence detection of E-cadherin, K14 and K10 are shown in the second, third and fourth panel from the top, respectively. Two asterisks indicate a p value of less than 0.01. B. Keratinocytes were immortalized by transfection with HPV16 genomic DNA as described in (Hoskins et al., 2008). Subsequent complementation, sorting and organotypic raft generation for morphological evaluation was carried out as described for A. The data are quantitated by nuclear counts, Fig. 5B depicts quantitation of Ki67 and BrdU positivity. C. HPV-NIKs were infected with nontargeting control or FANCA knockdown vectors, selected in puromycin and used for organotypic raft generation. Western blot analysis demonstrates cellular inability to respond to DNA damage by the monoubiquitination of FANCD2 (compare lanes 4 and 2). Morphological evaluation was carried out following H&E staining as in B.
Reduced rates of proliferation in FANCA complemented epithelium
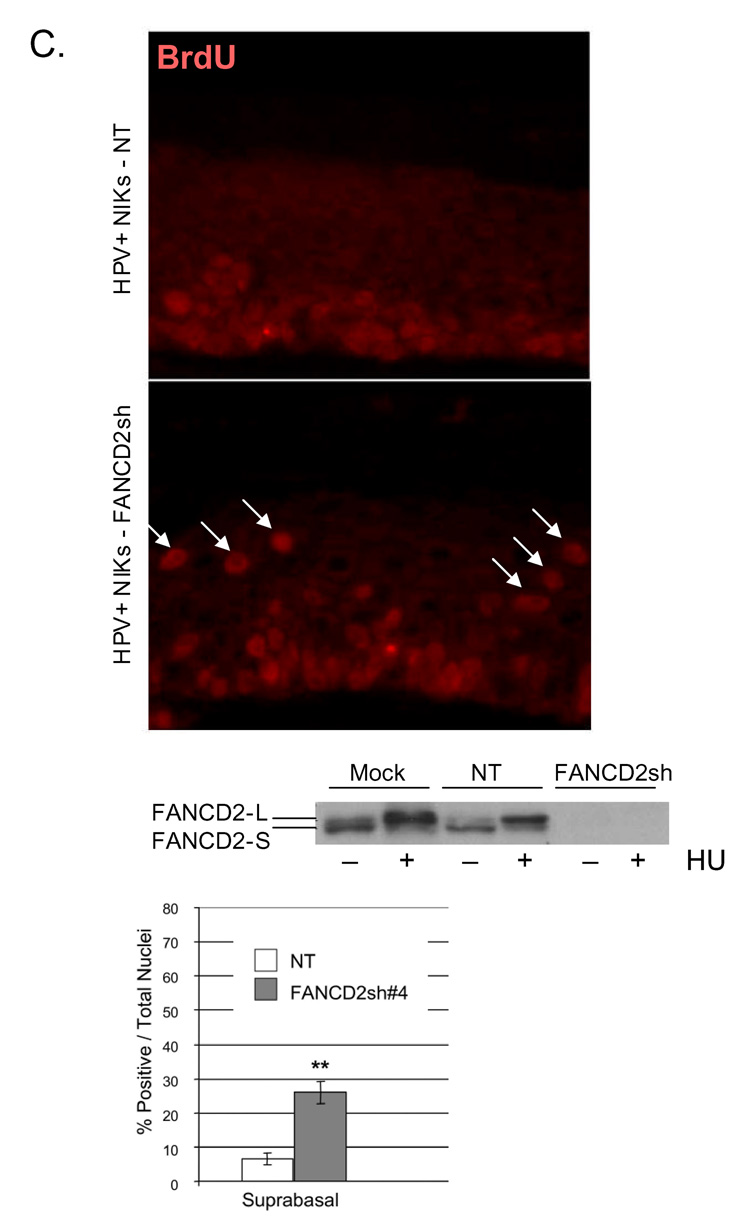
We next determined directly whether reduced proliferation might be responsible for the suppression of hyperplasia by FANCA (Fig. 5A). Detection of the proliferation marker Ki67 in the HPV18 E6/E7 positive patient derived rafts from Fig. 4A revealed a decreased proliferative index in the FANCA complemented compared to the deficient rafts (Fig. 5A). Quantitation of Ki67 and BrdU positive cells in the HPV16 immortalized rafts from Fig. 4B revealed similar decreases in the number of cells transiting through the cell cycle following complementation. Immunofluorescence detection of activated caspase 3 did not reveal an increased apoptotic index in the FANCA complemented rafts (data not shown). We concluded that increased proliferative rates were responsible for the hyperplastic differences in the FANCA-deficient compared to the complemented rafts. In order to further confirm this notion and to determine whether FA complex formation rather than a FANCA-specific effect was responsible, HPV positive NIKs were utilized for the lentiviral knockdown of FANCD2 (Fig. 5B). Cells were infected as described in the materials and methods, selected in puromycin and carried on feeders without obvious differences in cellular growth. Organotypic rafts were generated, and 10uM BrdU was added 48 hrs prior to raft harvest. BrdU detection by immunofluorescence revealed increased proliferation in the FANCD2 knockdown rafts. Taken together, our data support the notion that FA pathway deficiency weakens control of differentiation-associated cell cycle exit and that increased hyperplasia in FA deficient epithelium is reversible by FA correction.
Figure 5. FANCA suppression of hyperplasia correlates with decreased rates of cellular proliferation.
A. Detection of the Ki67 proliferation marker in the HPV18 E6/E7 positive organotypic raft sections is shown, together with quantitation of Ki67 positive cells. B. The HPV16 genome positive rafts were incubated with 10uM BrdU 48 hrs prior to tissue fixation, and quantitation of both BrdU and Ki67 positive cells is depicted. One or two asterisks represent a P value of less than 0.05 or 0.01, respectively. C. HPV genome positive NIKs, either transduced with a nontargeting (NT) vector or with a FANCD2 knockdown vector (FANCD2sh) were verified by western blot analysis using equal amounts of protein, and utilized for the generation of organotypic rafts. Rafts were incubated with 10uM BrdU 48 hrs prior to harvesting. Sections were utilized for the detection of BrdU positive cells within the rafts. BrdU positive cells were strikingly overrepresented in the suprabasal, K14 negative, compartment and are denoted with white arrows. K14 staining is not shown. The asterisks indicate statistical significance with a p value of less than 0.01.
Discussion
Our data support the notion that FA deficient human keratinocytes – like hematopoietic and stromal cells – respond to DNA damage by exaggerated cell death. However, clinical observations suggest keratinocyte specific differences, perhaps through differential adaptation, might also exist. Specifically, extreme radiosensitivity of FA mutant oropharyngeal epithelium is not shared by fibroblast cultures derived from the same patient (Marcou et al., 2001). Such cell type differences may influence the timing of disease manifestation in distinct organs: anemia and leukemia are often diagnosed early, whereas SCC development is generally not observed until later (Alter, 2003). Significant insight into the role of FA proteins in hematopoietic cells and fibroblasts has emerged in the past years, but no information is available on the role of FA pathways in differentiating keratinocytes. In light of likely keratinocyte specific FA phenotypes and their importance to SCC development, we chose to examine effects of FANCA loss on epithelial cells using patient-derived, FANCA deficient keratinocytes.
Primary cells from an FANCA deficient patient biopsy were immortalized with the HPV18 E6/E7 oncogenes, which are well characterized for their ability to inhibit cellular tumor suppressor pathways and to cause chromosomal instability (Duensing & Munger, 2004). Immortalized cells were subsequently complemented with FANCA by retroviral gene transfer. This culture system probed the effect of FANCA deficiency in the context of oncogenic challenge, but not in the primary state. The difficulty of culturing primary cutaneous keratinocytes from fresh specimen, both FA and normal, has so far precluded such analyses. Extending the short cellular life span of post-natal skin keratinocytes through better culture conditions will be required before meaningful growth comparisons between cells from FA patients versus normal donors can be initiated. Whether similar FA-dependent phenotypes will be observed in primary patient derived cells or in cells immortalized with other viral and cellular oncogenes remains to be determined in the future. However, E6/E7 immortalization is particularly relevant for these studies since a link has been suggested between FA and HPV associated disease. Specifically, in one study, 84% of FA associated SCCs were reported to contain genomic DNA from the high-risk human papillomaviruses compared to only 36% of SCCs in the general population (Kutler et al., 2003c). A link between FA and HPV is further supported by a recent report on accelerated chromosomal instability in FA deficient, HPV16 E7 oncogene expressing cells (Spardy et al., 2007). Importantly, HPV DNA was not detectable in a cohort of Dutch FA patients with head and neck SCCs (van Zeeburg et al., 2004; van Zeeburg et al., 2005). Reconciling these findings will require more extensive epidemiological and molecular studies of FA SCC samples in a manner that considers geographical and technical differences between the above studies.
Our studies uncover several specific defects in FANCA deficient keratinocytes which are similar to those described for FA hematopoietic and stromal cells. These relate to the known roles of FA genes in the maintenance of genome stability. Under standard monolayer culture conditions, FA status in the keratinocyte populations did not significantly affect steady state cell growth (Fig. 2A). In contrast, typical hallmarks of FA deficient hematopoietic and stromal cells were evident under conditions of exogenous DNA damage. These included a lack of FANCD2 monoubiquitination and focus formation, increased G2/M cell cycle arrest in response to melphalan, and apoptosis and senescence induction in response to mitomycin C. These phenotypes have already been utilized for the clinical diagnosis of FA mutations in various settings (Taniguchi & D'Andrea, 2006), and add to the reported finding that FA cancer cell lines exhibit sensitivity to cisplatin (van Zeeburg et al., 2005). Hallmarks of DNA damage further extended to FA deficient three-dimensional epithelium as evidenced by 53BP1 and TUNEL positivity (Fig. 3). Taken together, like immortalized hematopoietic and stromal cells, keratinocytes require a functional FA pathway under conditions of genotoxic stress.
The HPV oncogenes are well known to mediate unscheduled re-entry of differentiated cells into S phase and resulting hyperplasia, but their presence does not inhibit full execution of the cellular differentiation program. Uncoupling cell cycle exit from differentiation provides optimal replication conditions for the virus (Longworth & Laimins, 2004). HPV18 E6/E7-associated hyperplasia in FA deficient epithelium was observed as expected, and importantly, could be significantly attenuated by the re-introduction of FANCA expression (Fig. 4A). Similarly, HPV16-associated cellular hyperplasia was attenuated by complementation (Fig. 4B). Together, these data suggest that FA complementation may be a useful approach for the treatment of high risk HPV positive lesions, and may apply to lesions that maintain episomal HPV DNA. The observed differences between FA mutant and complemented epithelia correlated with decreased proliferative rates in a small subpopulation of cells within the suprabasal compartment. Conversely, FANCD2 knockdown in an HPV genome positive immortalized keratinocyte cell line increased the proliferative index (Fig. 5), thus supporting the hypothesis that FA deficiency stimulates hyperplasia in differentiated epithelium. Increased numbers of proliferating cells in FANCA and FANCD2 knockdown rafts would appear to implicate core complex- rather than FANCA-specific functions. Importantly, proliferative differences that are described in Fig. 5 are only observed in a 3D context, and represent a small proportion of the raft compared to the vast excess of differentiated cells. It is therefore not surprising that the underlying biochemical mechanisms are not yet known. However, since some of the known cellular targets of E7, including the Rb pocket proteins and p21, represent critical links between cellular differentiation and cell cycle control, these molecules might be considered possible candidate mediators for FA-dependent cell cycle control.
Relatively subtle abnormalities within FA deficient epithelium as observed would explain the clinical lack of obvious skin defects, yet a high risk of SCC, in FA patients. The fact that FA complementation can inhibit E6/E7 driven hyperplasia suggests that intact FA functions can serve as a barrier to E6/E7– stimulated cell cycle abnormalities. It will be important to determine whether somatic FA mutations may be selected for during HPV SCC development in the general population. The loss of such functions in the FA patient population may explain the up to thousand fold increased risk for developing SCC (Kutler et al., 2003b), and we suggest that the pursuit of FA gene correction for the treatment of FA SCC might be a worthwhile clinical endeavor in the future. Taken together, our findings point to a novel role for the FA pathway in suppressing viral oncogene induced cellular abnormalities.
SCC in FA patients presents at a particularly early patient age, generally with aggressive disease course and poor prognosis (Kutler et al., 2003a): surgical treatment remains the mainstay, but relapse-free, two-year survival rates are below 50%. Short term toxicity following the radiotherapy of FA patients – both as a conditioning regimen and as a treatment for SCC – has been noted in bystander epithelium (Alter, 2002). Our in vitro studies confirm at a molecular level that FA deficient cells and epithelium exhibit hypersensitivity to DNA damage induction (Fig. 2 and 3). Subsequent formation of secondary cancers within the same area of FA patients has overshadowed the initial success of the above patient treatments (Bremer et al., 2003; Guardiola et al., 2004), and was perhaps due to the selection of surviving clones in an environment of genomic instability. We suggest that the hyperplastic phenotype in FA deficient epithelium (Fig. 4) might represent a clinically relevant precursor to SCC. Continued studies of this model will explore the underlying molecular pathways involved in SCC progression, and will provide a foundation for the preclinical testing of new therapies. FA is a rare genetic disease; however, somatic inactivation of the FA pathway has been widely observed in sporadic cancers (Mathew, 2006). In this way, the above model system is also expected to advance our understanding of FA deficient SCC in the general population.
Acknowledgements
We thank Paul Andreassen, Ruhikanta Meetei, Qishen Pang, James Mulloy, Harmut Geiger, Yi Zheng and Stella Davies for helpful discussions and critical comments on the manuscript. We thank members of the Fanconi Anemia Comprehensive Care Center (FACCC), particularly Lars Wagner, Richard Harris and Robin Mueller for human tissue samples, as well as Susan Radtke for regulatory support. We thank the patients of the FACCC for samples. We are grateful to Denis Lee and Paul Lambert for expert advice on the organotypic raft system, as well as to Maureen Hoatlin for the FANCA and to James Lessard for the monoclonal actin antibody. We thank Christopher Baum for the retroviral SF91 expression vector, and Neeraj Singh for generation of the SF91-18E6/E7 retroviral construct. We thank the flow cytometry core facility and Dan Marmer for assistance with flow sorting. Vector supernatant was provided by the CCHMC Viral Vector Core and Translational Trials Development and Support Lab. This research was supported by Public Health Service grant CA102357, a grant from the Translational Research Initiative at CCHMC, and a grant from the Fanconi Anemia Research Fund to S. I. W. S.D. and N.S. are supported by NIH/NCI R01 CA112598. D. A. W was supported by Public Health Service grant HL081499. K. A. W.-B. was supported by HL079193.
References cited
- Alter BP. Radiosensitivity in Fanconi's anemia patients. Radiother Oncol. 2002;62:345–347. doi: 10.1016/s0167-8140(01)00474-1. [DOI] [PubMed] [Google Scholar]
- Alter BP. Cancer in Fanconi anemia, 1927–2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43:147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Bremer M, Schindler D, Gross M, Dork T, Morlot S, Karstens JH. Fanconi's anemia and clinical radiosensitivity report on two adult patients with locally advanced solid tumors treated by radiotherapy. Strahlenther Onkol. 2003;179:748–753. doi: 10.1007/s00066-003-1099-8. [DOI] [PubMed] [Google Scholar]
- Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, et al. A Rapid Method for Retrovirus-Mediated Identification of Complementation Groups in Fanconi Anemia Patients. Mol Ther. 2005;12:976–984. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley v, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing S, Munger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int J Cancer. 2004;109:157–162. doi: 10.1002/ijc.11691. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Guardiola P, Socie G, Li X, Ribaud P, Devergie A, Esperou H, et al. Acute graft-versus-host disease in patients with Fanconi anemia or acquired aplastic anemia undergoing bone marrow transplantation from HLA-identical sibling donors: risk factors and influence on outcome. Blood. 2004;103:73–77. doi: 10.1182/blood-2003-06-2146. [DOI] [PubMed] [Google Scholar]
- Hanenberg H, Batish SD, Pollok KE, Vieten L, Verlander PC, Leurs C, et al. Phenotypic correction of primary Fanconi anemia T cells with retroviral vectors as a diagnostic tool. Exp Hematol. 2002;30:410–420. doi: 10.1016/s0301-472x(02)00782-8. [DOI] [PubMed] [Google Scholar]
- Hoskins EE, Gunawardena RW, Habash KB, Wise-Draper TM, Jansen M, Knudsen ES, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008 doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr., Gorgoulis VG, Zacharatos P, Petty TJ, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003a;129:106–112. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003b;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- Kutler DI, Wreesmann VB, Goberdhan A, Ben-Porat L, Satagopan J, Ngai I, et al. Human papillomavirus DNA and p53 polymorphisms in squamous cell carcinomas from Fanconi anemia patients. J Natl Cancer Inst. 2003c;95:1718–1721. doi: 10.1093/jnci/djg091. [DOI] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcou Y, D'Andrea A, Jeggo PA, Plowman PN. Normal cellular radiosensitivity in an adult Fanconi anaemia patient with marked clinical radiosensitivity. Radiother Oncol. 2001;60:75–79. doi: 10.1016/s0167-8140(01)00370-x. [DOI] [PubMed] [Google Scholar]
- Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25:5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003a;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003b;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- Nakahara T, Peh WL, Doorbar J, Lee D, Lambert PF. Human papillomavirus type 16 E1circumflexE4 contributes to multiple facets of the papillomavirus life cycle. J Virol. 2005;79:13150–13165. doi: 10.1128/JVI.79.20.13150-13165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Pagano G, Degan P, d'Ischia M, Kelly FJ, Nobili B, Pallardo FV, et al. Oxidative stress as a multiple effector in Fanconi anaemia clinical phenotype. Eur J Haematol. 2005;75:93–100. doi: 10.1111/j.1600-0609.2005.00507.x. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2006 doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Schambach A, Wodrich H, Hildinger M, Bohne J, Krausslich HG, Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, et al. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, et al. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi Anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol. 2007 doi: 10.1128/JVI.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamary H, Alter BP. Current diagnosis of inherited bone marrow failure syndromes. Pediatr Hematol Oncol. 2007;24:87–99. doi: 10.1080/08880010601123240. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- van Zeeburg HJ, Snijders PJ, Joenje H, Brakenhoff RH. Re: Human papillomavirus DNA and p53 polymorphisms in squamous cell carcinomas from Fanconi anemia patients. J Natl Cancer Inst. 2004;96:968. doi: 10.1093/jnci/djh178. author reply 968-9. [DOI] [PubMed] [Google Scholar]
- van Zeeburg HJ, Snijders PJ, Pals G, Hermsen MA, Rooimans MA, Bagby G, et al. Generation and molecular characterization of head and neck squamous cell lines of fanconi anemia patients. Cancer Res. 2005;65:1271–1276. doi: 10.1158/0008-5472.CAN-04-3665. [DOI] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]