Abstract
BACKGROUND
Rheumatoid arthritis has a complex mode of inheritance. Although HLA-DRB1 and PTPN22 are well-established susceptibility loci, other genes that confer a modest level of risk have been identified recently. We carried out a genomewide association analysis to identify additional genetic loci associated with an increased risk of rheumatoid arthritis.
METHODS
We genotyped 317,503 single-nucleotide polymorphisms (SNPs) in a combined case-control study of 1522 case subjects with rheumatoid arthritis and 1850 matched control subjects. The patients were seropositive for autoantibodies against cyclic citrullinated peptide (CCP). We obtained samples from two data sets, the North American Rheumatoid Arthritis Consortium (NARAC) and the Swedish Epidemiological Investigation of Rheumatoid Arthritis (EIRA). Results from NARAC and EIRA for 297,086 SNPs that passed quality-control filters were combined with the use of Cochran-Mantel-Haenszel stratified analysis. SNPs showing a significant association with disease (P<1×10-8) were genotyped in an independent set of case subjects with anti-CCP-positive rheumatoid arthritis (485 from NARAC and 512 from EIRA) and in control subjects (1282 from NARAC and 495 from EIRA).
RESULTS
We observed associations between disease and variants in the major-histocompatibility-complex locus, in PTPN22, and in a SNP (rs3761847) on chromosome 9 for all samples tested, the latter with an odds ratio of 1.32 (95% confidence interval, 1.23 to 1.42; P = 4×10-14). The SNP is in linkage disequilibrium with two genes relevant to chronic inflammation: TRAF1 (encoding tumor necrosis factor receptor-associated factor 1) and C5 (encoding complement component 5).
CONCLUSIONS
A common genetic variant at the TRAF1-C5 locus on chromosome 9 is associated with an increased risk of anti-CCP-positive rheumatoid arthritis.
Rheumatoid arthritis is a common inflammatory arthritis of unknown cause, in which both genetic and environmental risk factors have been implicated.1-3 The genetic contribution to a susceptibility to rheumatoid arthritis has been shown in studies of twins4 and families5 and in genomewide linkage scans.6-11
Two genes have shown a strong association with susceptibility: PTPN2212,13 and HLA-DRB1.14 Variants of each gene elevate the risk primarily for a subgroup of severe rheumatoid arthritis characterized by the presence of autoantibodies against cyclic citrullinated peptide (anti-CCP-positive).12,15,16 We have recently reported a significant association at STAT4 on chromosome 2q.17 Several other promising candidate genes have been reported in the literature (e.g., CTLA4 and PADI4), but these genes have had more modest statistical evidence of association.18,19 All of the alleles associated with rheumatoid arthritis are common in healthy persons of European ancestry (allele frequency, >5%). Therefore, it seems likely that additional common genetic variants with a modest effect size (e.g., odds ratio, <1.5 per copy) remain to be discovered.20
Statistical tests based on allele frequencies in case-control studies (association analyses) have more power to identify common alleles that confer a modest risk than do tests based on chromosomal segregation in families (linkage analyses).21 Until recently, genetic association studies were limited to small regions of the genome containing biologic candidate genes or those identified through family-based linkage studies. Recent developments in understanding patterns of human genetic variation,22 together with cost-effective genotyping techniques and statistical methodology,23,24 have made it possible to test, in an unbiased manner, common variants across the entire genome for the risk of disease. Current genotyping platforms are estimated to represent more than two thirds of known common genetic variation throughout the genome, encompassing more than 20,000 human genes.25 By comparison, fewer than 100 candidate genes have been tested for an association with the risk of rheumatoid arthritis.18
Our genomewide association study involved two groups of case subjects with anti-CCP-positive rheumatoid arthritis: one group who had been treated at rheumatology clinics across North America, the North American Rheumatoid Arthritis Consortium (NARAC), and another group from a Swedish population-based study, the Epidemiological Investigation of Rheumatoid Arthritis (EIRA). In the NARAC study, case subjects were matched with control subjects according to self-reported ethnic background; in the EIRA study, case subjects were matched with control subjects according to age, sex, and geographic location.
METHODS
SUBJECTS
We refer to stage 1 as the initial genotyping of samples in the genomewide scan of single-nucleotide polymorphisms (SNPs) (called NARAC-1 and EIRA-1) and stage 2 as the replication genotyping (NARAC-2 and EIRA-2). Patients were drawn from rheumatology clinics across North America (NARAC) and Sweden (EIRA). All patients were anti-CCP-positive and met the criteria for rheumatoid arthritis adopted by the American College of Rheumatology in 198726 (Table 1).
Table 1.
Sources of Subjects for the Original Genomewide Association Scan and the Replication Scan.*
| Study and Collection | Description | Genomewide Association Scan |
Replication Scan |
|---|---|---|---|
| no. of samples | |||
| NARAC | |||
| Case subjects | |||
| NARAC family collection | Erosive disease, familial clustering | 464 | 0 |
| National Data Bank for Rheumatic Diseases |
Sporadic cases with long-standing disease | 168 | 147 |
| National Inception Cohort of Rheumatoid Arthritis Patients |
New-onset cases (<6 mo) | 162 | 157 |
| Study of New Onset Rheumatoid Arthritis |
New-onset cases (<12 mo) | 114 | 181 |
| Control subjects | |||
| New York Cancer Project | Population-based cohort from New York, matched with case subjects according to self-reported ethnic background |
1260 | 1282 |
| EIRA | |||
| Case subjects | New-onset cases (<2yr) from population-based survey | 676 | 568 |
| Control subjects | Population-based samples matched with case subjects according to age, sex, and geographic location |
673 | 516 |
Samples that were genotyped as part of the genomewide association study are categorized as stage 1, including the combined data sets from the North American Rheumatoid Arthritis Consortium (NARAC) and the Swedish Epidemiological Investigation of Rheumatoid Arthritis (EIRA); the replication samples from both data sets are categorized as stage 2. Samples from the NARAC-1 case subjects were genotyped with the Illumina HumanHap550 array; samples from the NARAC-1 control subjects were genotyped with the HumanHap550 array or the HumanHap300+240 arrays. Samples from the EIRA-1 case and control subjects were genotyped with the Illumina HumanHap300 array. Samples from NARAC-2 and EIRA-2 were genotyped with the Sequenom iPLEX platform.
The NARAC “family collection” samples were from multiplex families (primarily affected sibling pairs) in which at least one sibling had documented erosions, as seen on radiography of the hand, and at least one sibling (most often the same patient) had an onset of disease between the ages of 18 and 60 years.8 The other collections that make up NARAC-1 included samples from the National Data Bank for Rheumatic Diseases (mean disease duration, 10 years),27 the National Inception Cohort of Rheumatoid Arthritis (with patients enrolled within 6 months after clinical diagnosis),27,28 and the Study of New Onset Rheumatoid Arthritis (with patients enrolled within 12 months after clinical diagnosis).29
An initial set of samples from case subjects of self-reported white ancestry was randomly drawn from all four collections (464 from NARAC, 168 from the National Data Bank for Rheumatic Diseases, 162 from the National Inception Cohort of Rheumatoid Arthritis, and 114 from the Study of New Onset Rheumatoid Arthritis) (see the Methods section in the Supplementary Appendix, available with the full text of this article at www.nejm.org). Control subjects were selected on the basis of similar self-reported ancestry from 20,000 persons who were part of the New York Cancer Project. Replication samples (NARAC-2) were randomly drawn from the same collections (except that no cases were drawn from the NARAC family collection) and included 485 patients with anti-CCP-positive rheumatoid arthritis and 1282 control subjects from the New York Cancer Project.
Data on participation rates are not available for any of the NARAC collections of patients with rheumatoid arthritis, since recruitment of patients was performed by diverse methods, including advertising, direct mail, and physician-based enrollment. Control subjects from the New York Cancer Project were enrolled during a 2-year period by means of general advertising and point-of-service solicitation, as described previously.30 Written informed consent was obtained from all subjects who provided blood samples in accordance with protocols approved by the local institutional review boards.
EIRA is a population-based case-control study comprising residents of south and central Sweden who were between the ages of 18 and 70 years during the period from May 1996 to December 2005.31 Enrollment was limited to patients who had recently received the diagnosis of rheumatoid arthritis (within 1 year after the first onset of symptoms for 85% of patients). For each patient, a control subject was randomly selected from the study base; control subjects were matched for age, sex, and residential area. Most subjects were born in Sweden, and 97% reported having white ancestry.
We randomly selected 676 patients with anti-CCP-positive rheumatoid arthritis and 673 control subjects for genomewide genotyping (EIRA-1). Replication subjects (EIRA-2) were randomly drawn from the same study base and included 568 anti-CCP-positive case subjects and 516 control subjects. The participation rate was 96% for case subjects after recruitment from the population-based early surveillance system for rheumatoid arthritis in Sweden. Written informed consent was obtained from all subjects, and the ethics review board at the Karolinska Institutet approved the study.
GENOTYPING AND QUALITY-CONTROL FILTERING
Genotyping for stage 1 was performed at the Feinstein Institute for Medical Research for the NARAC scan and at the Genome Institute of Singapore for the EIRA scan, both according to the Illumina Infinium 2 assay manual (Illumina), as previously described.32 The NARAC scan included 545,080 SNPs genotyped in samples from 908 case subjects and 1260 control subjects. Samples from all the NARAC case subjects were genotyped by SNP assay with Infinium HumanHap550, version 1.0 (Illumina); 601 of the controls were genotyped on the same platform, 411 on Human-Hap550 (version 3.0) and 248 on Infinium Human-Hap300 and HumanHap240S arrays. The EIRA scan included genotypes of 317,503 SNPs from the HumanHap300 (version 1.0) array. The data sets were filtered individually on the basis of SNP genotype call rates (>95% completeness), minor allele frequency (>0.01), and the Hardy-Weinberg equilibrium (P<1×10-5). We removed subjects whose percentage of missing genotypes was more than 5%, who had non-European ancestry, who had evidence of relatedness, and who had evidence of possible DNA contamination (see the Supplementary Appendix for more details). The 297,086 SNPs that passed filters in both the NARAC and EIRA sample collections were merged into a single file for analysis.
Stage 2 genotyping of nine TRAF1-C5 haplo-type SNPs was performed with the use of Sequenom iPLEX33 at the Broad Institute of Harvard and the Massachusetts Institute of Technology (for the NARAC-2 samples) and at the Genome Institute of Singapore (for the EIRA-2 samples), both according to the manufacturer's specifications (see the Supplementary Appendix for additional details).
STATISTICAL ANALYSIS
We initially analyzed the NARAC-1 and EIRA-1 data separately and then combined the two data sets for joint analysis. Our primary analyses were performed on the combined data set from NARAC and EIRA with the use of structured association within homogeneous clusters derived through identity-by-state similarity, implemented in the PLINK tool set as a Cochran-Mantel-Haenszel stratified analysis,24 a method we refer to here as structured association analysis (see the Methods section in the Supplementary Appendix). A complete listing of results of the combined NARAC-1 and EIRA-1 data can also be found in the Supplementary Appendix. Additional data on NARAC-1 are available in the Database of Genotype and Phenotype (dbGaP) (accession number phs000099.v1.p1).
After we identified the TRAF1-C5 region through the genomewide scans of subjects from NARAC-1 and EIRA-1, we selected nine SNPs that lie in a 100-kb block of linkage disequilibrium to test for association with disease in NARAC-2 and EIRA-2. We selected these SNPs on the basis of linkage disequilibrium patterns within European samples from the International HapMap Project (the Centre d'Etude du Polymorphisme Humain from Utah [CEU] HapMap34) with the use of the software program Tagger.35 We performed association analysis with the use of 2-by-2 contingency tables of allele frequencies and Fisher's exact test. For the NARAC-2 replication samples, we performed a secondary analysis, correcting for population stratification by applying the software program EIGENSTRAT23 to a set of 704 SNPs informative about European ancestry36 and corrected along the first principal component.
Results were combined across all samples (NARAC-1, NARAC-2, EIRA-1, and EIRA-2) in three ways (see the Methods section of the Supplementary Appendix). We also carried out association analysis conditional on each SNP and haplotype with the use of combined genotype data from all four sample collections. These analyses were performed with the software program WHAP,37 which also provided an omnibus test for haplotype association.
To estimate power in the combined NARAC-1 and EIRA-1 scan, we considered a variety of effect sizes (as estimated by odds ratio) and allele frequencies with the use of an online genetics power calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/). The study had a power of about 90% to detect a disease-associated allele with a population frequency of 0.20 and an odds ratio of 1.5 (at P = 5×10-8 under a multiplicative genetic model) but a power of only 13% to detect the same allele with an odds ratio of 1.3.
RESULTS
GENOMEWIDE ASSOCIATION ANALYSIS
We identified a set of 297,086 polymorphic SNPs genotyped in samples from 1522 case subjects with anti-CCP-positive rheumatoid arthritis and from 1850 control subjects in the combined NARAC-1 and EIRA-1 analysis that passed our quality-control filters (see the Methods section of the Supplementary Appendix). The average call rate for these SNPs was 99.71%.
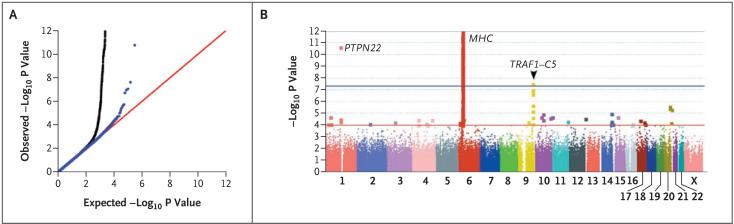
To combine results between NARAC-1 and EIRA-1 while minimizing bias caused by population stratification, we conducted a structured analysis within homogeneous clusters defined with the use of genomewide SNP data. Advantages of this approach include the ability to match case-control clusters within each collection (i.e., NARAC case and control subjects are clustered together, as are EIRA case and control subjects) and the ability to calculate odds ratios that account for population stratification. To determine whether we observed more significant results than expected by chance alone, we calculated the genomic control inflation measure,38 which is based on the median chi-square distribution (in which 1.0 signifies no inflation), and plotted the observed distribution, as compared with the expected distribution, of P values. After correcting for residual inflation by genomic control (1.14) and after removing SNPs from the extended major-histocompatibility-complex (MHC) region, we observed a slight excess number of SNPs in the tail of the statistical distribution (Fig. 1A). We obtained a similar result after correcting for population stratification with a principal components method (EIGENSTRAT23; genomic control, 1.08) (see Fig. 1 of the Supplementary Appendix). The inflation in the tail of the distribution could represent true positive associations (e.g., PTPN22) or could reflect the effect of an unknown source of bias in our study.
Figure 1. Results of the Genomewide Association Study.
Panels A and B show results for 297,086 polymorphic SNPs genotyped in samples from 1522 case subjects with rheumatoid arthritis who were seropositive for autoantibodies against cyclic citrullinated peptide (anti-CCP-positive) and 1850 control subjects from the combined data sets from the North American Rheumatoid Arthritis Consortium (NARAC) and the Swedish Epidemiological Investigation of Rheumatoid Arthritis (EIRA). Panel A shows a comparison of distributions of observed versus expected P values generated by Cochran-Mantel-Haenszel stratified analysis and corrected for residual inflation by genomic control. Black data points represent the inclusion of SNPs from the major-histocompatibility-complex (MHC) locus; blue data points represent the exclusion of MHC. The most significant non-MHC SNPs are at PTPN22 and the TRAF1-C5 locus. Panel B shows SNPs plotted according to chromosomal location, with the -log10 P values corrected with the use of genomic control. The blue horizontal line indicates SNPs that are significant at a genomewide level (P = 5×10-8). SNPs at PTPN22 and within the MHC locus, where the HLA-DRB1 gene resides, reach genomewide significance. Multiple SNPs across the TRAF1-C5 locus on chromosome 9 (e.g., rs3761847; P = 2×10-8 have a significant association.
A graphical summary of the results of our genomewide association scan is shown in Figure 1B. We clearly identified SNPs in linkage disequilibrium with known susceptibility variants at HLA-DRB1 (P<1×10-100) and PTPN22 (P = 2×10-11). Common variants within these two regions contributed the strongest statistical signal of risk for anti-CCP-positive rheumatoid arthritis in our study of patients of European ancestry.
In the combined stage 1 analysis (data not shown), a single SNP within a region on chromosome 9 in the TRAF1-C5 locus reached genomewide significance (defined here as P<5×10-8), and several neighboring SNPs also showed a highly significant association with a diagnosis of rheumatoid arthritis. The strongest association was seen for SNP rs3761847 (P = 2.8×10-8). The minor G allele frequency was higher in case subjects (0.49) than in control subjects (0.41), providing an allelic odds ratio of 1.36 (95% confidence interval [CI], 1.23 to 1.50). There were several other regions with intermediate levels of significance (between P>5×10-8 and P<1×10-4) that contained candidate genes of known biologic relevance to rheumatoid arthritis, including CD40 (P = 3×10-6), bradykinin receptor 1 (BDKR1) (P = 1×10-5), and the 17q chemokine gene cluster containing CCL1, CCL8, and CCL13 (P = 4×10-5). These SNPs require further study to determine which, if any, may represent true associations with anti-CCP-positive rheumatoid arthritis. A complete list of all SNPs with P<1×10-4 can be found in Table 1 of the Supplementary Appendix.
REPLICATION OF TRAF1-C5 HAPLOTYPE TAG SNPs
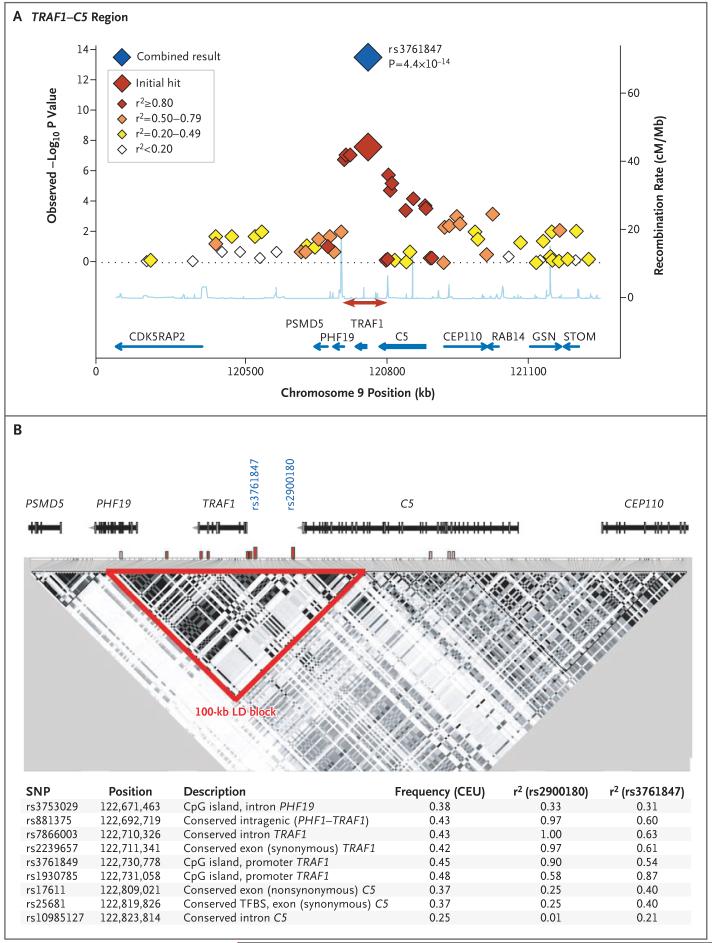
Replication of a specific genetic model in additional samples is critical to differentiate true positive associations from statistical fluctuations. Although the combined NARAC-1 and EIRA-1 result at TRAF1-C5 was highly significant (P = 2.8×10-8), we genotyped the most strongly associated SNPs, in addition to tag SNPs across the region, in an independent set of samples from 485 anti-CCP-positive case subjects and 1282 control subjects in NARAC-2 and from 568 anti-CCP-positive case subjects and 516 control subjects in EIRA-2 to obtain additional support for our finding, as well as to perform fine mapping of the causal allele. A summary of the replication results for nine SNPs that capture haplotypes at TRAF1-C5 (haplotype tag SNPs) is presented in Table 2. These nine tag SNPs capture 74 of 76 (97%) of the common HapMap CEU SNPs across a 100-kb block of linkage disequilibrium that includes the TRAF1 and C5 genes, as well as another gene, PHF19 (Fig. 2).
Table 2.
Results of Case-Control Association Analysis for Nine TRAF1-C5 Tag SNPs in all NARAC and EIRA Samples.*
| SNP | Position | NARAC-1 | NARAC-2 | EIRA-1 | EIRA-2 | Combined Group | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor Allele Frequency |
P Value |
Odds Ratio |
Minor Allele Frequency |
P Value |
Odds Ratio |
Minor Allele Frequency |
P Value |
Odds Ratio |
Minor Allele Frequency |
P Value |
Odds Ratio |
P Value † |
Odds Ratio (95% Cl) |
||||||
| Case | Control | Case | Control | Case | Control | Case | Control | ||||||||||||
| rs3761847 | 120769793 | 0.458 | 0.371 | 4×10-8 | 1.43 | 0.463 | 0.381 | 7×10-6 | 1.40 | 0.503 | 0.443 | 0.004 | 1.27 | 0.472 | 0.449 | 0.31 | 1.10 | 4×10-14 | 1.32 (1.23-1.42) |
| rs10985095 | 120778637 | 0.010 | 0.016 | 0.12 | 1.58 | 0.016 | 0.022 | 0.24 | 1.40 | 0.008 | 0.011 | 0.47 | 1.37 | 0.019 | 0.027 | 0.28 | 1.41 | 0.01 | 1.45 (1.06-1.98) |
| rs12338903 | 120783240 | 0.077 | 0.059 | 0.03 | 1.31 | 0.072 | 0.068 | 0.66 | 1.07 | 0.083 | 0.092 | 0.48 | 1.11 | 0.096 | 0.084 | 0.35 | 1.17 | 0.01 | 1.17 (1.02-1.35) |
| rs10985097 | 120783448 | 0.010 | 0.013 | 0.42 | 1.28 | 0.016 | 0.019 | 0.54 | 1.19 | 0.008 | 0.011 | 0.47 | 1.37 | 0.019 | 0.027 | 0.28 | 1.41 | 0.06 | 1.30 (0.94-1.78) |
| rs2900180 | 120785936 | 0.396 | 0.304 | 2×10-9 | 1.50 | 0.376 | 0.303 | 2×10-5 | 1.39 | 0.433 | 0.379 | 0.01 | 1.25 | 0.404 | 0.379 | 0.26 | 1.11 | 8×10-14 | 1.34 (1.24-1.45) |
| rs7035682 | 120807548 | 0.069 | 0.076 | 0.40 | 0.90 | 0.101 | 0.095 | 0.61 | 1.06 | 0.082 | 0.076 | 0.58 | 1.09 | 0.084 | 0.085 | 0.94 | 0.99 | 0.49 | 1.00 (0.87-1.15) |
| rs10985112 | 120810962 | 0.062 | 0.065 | 0.65 | 0.94 | 0.091 | 0.080 | 0.26 | 1.16 | 0.073 | 0.065 | 0.46 | 1.13 | 0.063 | 0.055 | 0.46 | 1.16 | 0.16 | 1.08 (0.93-1.25) |
| rs7O26551 | 120812687 | 0.208 | 0.161 | 2×10-4 | 1.36 | 0.214 | 0.171 | 0.003 | 1.32 | 0.255 | 0.190 | 2×10-4 | 1.45 | 0.199 | 0.196 | 0.90 | 1.01 | 2×10-8 | 1.31 (1.19-1.44) |
| rs2269066 | 120816572 | 0.120 | 0.089 | 0.002 | 1.39 | 0.106 | 0.087 | 0.08 | 1.24 | 0.142 | 0.098 | 0.001 | 1.53 | 0.102 | 0.118 | 0.29 | 0.85 | 7×10-5 | 1.27 (1.12-1.43) |
The minor allele frequency and odds ratio are shown for case subjects with rheumatoid arthritis and for unrelated control subjects. P values are for the comparison in allele frequency between case subjects and control subjects and were calculated by a two-tailed Pearson's chi-square test, except as indicated.
P values and odds ratios for all samples were calculated with a Mantel-Haenszel method of combining allele frequency counts. For rs3761847, we also calculated a combined P value with Fisher's meta-analysis (P = 3.6×10-13). The omnibus haplotype test for the six common haplotypes generated by these nine SNPs was also highly significant (P = 2×10-16). The odds ratio we obtain by this method for rs3761847, 1.32 (95% CI, 1.23 to 1.42), is nearly identical to the odds ratio, 1.36 (95% CI, 1.23 to 1.50), that was obtained from structured association analysis of the genome scan.
Figure 2. (facing page) Case-Control Association Results and Linkage Disequilibrium Structure in the> TRAF1-C5 Locus.
Panel A shows results for SNPs genotyped across 1 Mb as part of the original genomewide association scan in samples from 1522 case subjects with anti-CCP-positive rheumatoid arthritis and 1850 control subjects. Each diamond indicates a genotyped SNP; the color of each diamond is based on the correlation coefficient (r2) with the CEU HapMap with the most significant SNP in our study (rs3761847). The blue diamond indicates the P value for all samples in our study (the original scan plus replication samples), as determined by the Cochran-Mantel-Haenszel method in both NARAC and EIRA samples. The recombination rate (in centimorgans per megabase) with the CEU HapMap is shown in light blue along the x axis; the red arrow indicates the block of linkage disequilibrium shown in Panel B. The blue arrows indicate gene location. Panel B shows the linkage-disequilibrium (LD) structure across 200 kb of the TRAF1-C5 locus, based on pairwise r2 with the CEU HapMap. The intron-exon structure of each gene is at the top of the figure. Putative functional SNPs in linkage disequilibrium with either rs3761847 or rs2900180 are indicated by hatched bars, in which red indicates r2>0.80 and pink indicates r2 = 0.20 to 0.80; the specific SNPs, frequency, pairwise r2 with the CEU HapMap, and the putative annotated function are listed at the bottom of the figure. CpG denotes cytidine and guanosine joined by a phosphodiester bond.
The most significant result from the genome scan, at rs3761847, was significant in the NARAC-2 samples under the same genetic model (P = 1×10-5), with an odds ratio of 1.37 (95% CI, 1.18 to 1.58) and showed a nonsignificant trend toward an association in the EIRA-2 replication samples, with an odds ratio of 1.11 (95% CI, 0.93 to 1.32). To provide an additional test against population stratification in the NARAC-2 replication samples, we implemented a principal components method23 using 704 European-derived ancestry informative markers36 and saw continued evidence of an association at rs3761847 (P = 0.003).
Combining the allele counts among all 2575 samples from case subjects with anti-CCP-positive rheumatoid arthritis and 3648 samples from control subjects and calculating the significance with a Mantel-Haenszel statistic, we observe a highly significant result at rs3761847 (P = 4×10-14), with an odds ratio of 1.32 (95% CI, 1.23 to 1.42) per copy of the risk allele (Fig. 2A) or P = 2×10-16 by the omnibus haplotype test. Homozygotes for the susceptibility allele had an odds ratio of 1.87 (95% CI, 1.61 to 2.18), as compared with homozygotes for the protective allele. The attributable risk of disease conferred by the allele was 7%.
FINE MAPPING AT TRAF1-C5
Logistic-regression analyses that were conditional on each of the nine tag SNPs (and haplotypes defined by these SNPs) in all case-control samples showed that the most significant SNP from the genome scan (rs3761847) could explain the majority of the association signal across the locus. A neighboring SNP, rs2900180, in strong linkage disequilibrium with a correlation coefficient (r2) of 0.62 with our most significant SNP, rs3761847, was also highly significant; the two SNPs could not be distinguished statistically in our combined sample collection. We thus considered one of these variants, or an untyped variant in strong linkage disequilibrium with rs3761847 and rs2900180, to be the causal variant.
To determine whether any putative functional variant from the public database could explain the association signal, we identified and genotyped such SNPs in the CEU HapMap and determined the extent of linkage disequilibrium between each SNP and both rs3761847 and rs2900180. (We defined putative functional motifs as those within coding exons, transcription-factor-binding sites, highly conserved regions, CpG islands, 5′ and 3′ untranslated regions, intron-exon boundaries, and microRNA binding sites.) Although no missense SNP from the database was in strong linkage disequilibrium with rs3761847, we identified a synonymous SNP in TRAF1 (rs2239657, r2 = 0.97 with rs2900180) and several SNPs within highly conserved motifs for both TRAF1 and C5 (Fig. 2B).
DISCUSSION
This comprehensive genetic analysis of rheumatoid arthritis has led to the identification of a novel association with a 100-kb region on chromosome 9 that contains the TRAF1 and C5 genes. Our study adds to the small but growing list of validated susceptibility genes for rheumatoid arthritis that includes HLA-DRB1, PTPN22, and STAT4.
Since the most highly associated SNPs (rs376147 and rs2900180) are in linkage disequilibrium with both genes, it is not clear whether the causal alleles or group of alleles influences TRAF1 or C5 (or both) to increase susceptibility for rheumatoid arthritis. There is no known or obvious functional allele that explains these associations. In theory, the causal allele could exert its effect through a neighboring gene (e.g., PHF19), although the weight of the biologic evidence supports a role for either TRAF1 or C5. Identification of the causal allele will ultimately require the resequencing and genotyping of samples from a large number of patients with rheumatoid arthritis, together with functional studies stratified according to genotype.
The TRAF1 gene encodes an intracellular protein that mediates signal transduction through tumor necrosis factor (TNF) receptors 1 and 2 and through CD40. TNF is a critical cytokine in the pathogenesis of rheumatoid arthritis,1 and TNF antagonists are an effective treatment for rheumatoid arthritis.39,40 TRAF1 knockout mice have exaggerated T-cell proliferation and activation in response to TNF or when stimulated through the T-cell-receptor complex, suggesting that TRAF1 acts as a negative regulator of these signaling pathways.41 TRAF1 binds several intra-cellular proteins, including the nuclear factor-κB inhibitory protein TNFAIP3.42
The clinical and biologic data for C5 are equally compelling. The complement pathway has been implicated in the pathogenesis of rheumatoid arthritis for more than 30 years.43,44 Complement activation leading to significant depletion of complement components has been shown in synovial fluid of patients with rheumatoid arthritis. C5 cleavage generates the proinflammatory anaphylatoxin C5a, as well as C5b, which initiates the generation of the membrane-attack complex. C5-deficient mice are resistant to inflammatory arthritis in models with a dominant humoral component.45-47 If the causal allele acts through C5, it may do so by amplifying complement activation in joints of patients with rheumatoid arthritis.
Our initial genomewide scan (stage 1) was powered to detect moderate genetic effect sizes (odds ratio, >1.50), and only the MHC locus, PTPN22, and TRAF1-C5 achieved a P value of less than 5×10-8. Integration of data from NARAC-1 and EIRA-1 was critical in choosing SNPs for replication at TRAF1-C5, since neither study achieved a P value of less than 5×10-8. The recently published study by the Wellcome Trust Case Control Consortium (WTCCC)48 involving approximately 2000 case subjects with rheumatoid arthritis and approximately 3000 control subjects did not highlight the TRAF1-C5 region, and a SNP in near complete linkage disequilibrium with rs3761847 (WTCCC SNP rs10118357; r2 = 0.97 in the CEU HapMap) was not significant at a P value of less than 0.05 in the WTCCC study. In a similar manner, our replication samples (stage 2) had limited statistical power, as made evident by a nonsignificant trend toward an association for our most significant SNP, rs3761847, in EIRA-2 (which had a power of approximately 70% at P = 0.05). Together, these findings emphasize that even rather large-scale genomewide association studies together with independent replication may have limited power to detect common risk variants of modest effect. However, we note that a recent candidate-gene study of rheumatoid arthritis supports our findings.49
There are undoubtedly additional risk variants with modest effect sizes that have yet to be discovered. For example, we integrated our data with those from the WTCCC study. Among 11 SNPs that were not within the MHC locus with moderate evidence of association (P<1×10-5) reported in the WTCCC study, we found evidence that at least 1 SNP is significant in our study (WTCCC SNP rs6920220, P = 13×10-5 in the combined NARAC-1 and EIRA-1 analysis) (Table 3 of the Supplementary Appendix). This SNP, which is located on chromosome 6q23 near the gene TNFAIP3, has been identified in a completely independent genomewide association analysis (unpublished data). Beyond the simple identification of causative alleles, it is important to recognize that genes may act at multiple different stages of disease, from early breakage of immune tolerance to the regulation of tissue destruction and the response to therapy. Therefore, continuing international collaborative studies of large cohorts of patients will be essential to understand fully the clinical significance of the wealth of genetic information that is now emerging on rheumatoid arthritis and related autoimmune disorders.
Supplementary Material
Acknowledgments
The NARAC study was supported by grants (RO1-AR44422 and NO1-AR22263, to Dr. Gregersen; R01-AR050267, to Dr. Seldin; K24-AR02175 and R01-AI065841, to Dr. Criswell; and K08-AI55314-3, to Dr. Plenge) from the National Institutes of Health; a grant from Biogen Idec; a grant from the National Arthritis Foundation; grants from the Boas Family and the Eileen Ludwig Greenland Center for Rheumatoid Arthritis (to Dr. Gregersen); a grant (5-M01-RR-00079) to the General Clinical Research Center, Moffitt Hospital, University of California at San Francisco, and a grant (M01-RR018535) to the General Clinical Research Center, Feinstein Institute for Medical Research, from the National Center for Research Resources; a grant from the Rosalind Russell Medical Research Center for Arthritis and the Kirkland Scholar Award (to Dr. Criswell); and a grant from the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. The EIRA study was supported by grants from the Swedish Medical Research Council, the Swedish Council for Working Life and Social Research, King Gustaf V's 80-Year Foundation, the Swedish Rheumatism Foundation, the Stockholm County Council, the AFA insurance company, and the Agency for Science Technology and Research, Singapore.
Dr. Plenge reports receiving consulting fees from Biogen Idec and lecture fees from Genentech; Drs. Liu, Carulli, and Beckman, being employees of Biogen Idec; Dr. Altshuler, receiving consulting fees from Rosetta Inpharmatics (a subsidiary of Merck) and serving on the advisory board of Medical Portfolio Management; Dr. Criswell, receiving consulting fees from Celera Diagnostics; Dr. Klareskog, receiving research grants and serving on advisory boards for Roche Pharmaceuticals, Bristol-Myers Squibb, Schering-Plough, Abbott, and Wyeth; and Dr. Gregersen, serving on the Abbott Scholar Award Advisory Committee and receiving honoraria from Biogen Idec, Genentech, and Roche Pharmaceuticals. No other potential conflict of interest relevant to this article was reported.
We thank the large number of investigators, practicing physicians, and research nurses who identified and enrolled subjects and played a critical role in assembling the various samples we used in our studies, including Drs. Elena Massarotti, Claire Bombardier, and Michael Weisman for the Study of New Onset Rheumatoid Arthritis; Marlena Kern, R.N., for NARAC; and Dr. Frederick Wolfe for the National Data Bank for Rheumatic Diseases; Kian Mun Chan, Boon Yeong Goh, Wee Yang Meah, Jameelah B.S. Mohamed, Jason Ong, Eileen Png, and Sigeeta Rajaram for their invaluable laboratory assistance; Ingeli Andréasson, Landvetter, for assistance in the recruitment of patients; Eva Baecklund, Akademiska Hospital; Ann Bengtsson and Thomas Skogh, Linköping Hospital; Birgitta Nordmark, Johan Bratt, and Ingiäld Hafström, Karolinska University Hospital; Kjell Huddénius, Rheumatology Clinic in Stockholm City; Shirani Jayawardene, Bollnäs Hospital; Ann Knight, Hudiksvall Hospital and Uppsala University Hospital; Ido Leden, Kristianstad Hospital; Göran Lindahl, Danderyd Hospital; Bengt Lindell, Kalmar Hospital; Christin Lindström and Gun Sandahl, Sophiahemmet; Björn Löfström, Katrineholm Hospital; Ingmar Petersson, Spenshult Hospital; Christoffer Schaufelberger, Sahlgrenska University Hospital; Patrik Stolt, Västerås Hospital; Berit Sverdrup, Eskilstuna Hospital; Olle Svernell, Västervik Hospital; Tomas Weitoft, Gävle Hospital; and Marie-Louise Serra, Camilla Bengtsson, Eva Jemseby, and Lena Nise, who made invaluable contributions to the collection of data and maintenance of the database; and Ralph Nappi of the Feinstein Institute for his long-standing support.
APPENDIX
The following is a list of the authors' affiliations: the Broad Institute of Harvard and the Massachusetts Institute of Technology — both in Cambridge, MA (R.M.P., L.R.L.D., D.A.); Brigham and Women's Hospital, Boston (R.M.P.); the Genome Institute of Singapore, Singapore (M.S., A.K.S.T., C.B., R.T.H.O., A.T., S.P.); Harvard School of Public Health, Boston (M.S.); the Karolinska Institutet, Stockholm (L.P., B.D., S.P., L.A., L.K.); the Feinstein Institute for Medical Research, North Shore-Long Island Jewish Health System, Manhasset, NY (A.T.L., A.L., H.K., A.C., W.L., P.K.G.); the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, MD (E.F.R., D.L.K.); Biogen Idec, Cambridge, MA (C.L., J.P.C., E.M.B.); the Rowe Program, University of California Davis, Davis (C.T., M.F.S.); Massachusetts General Hospital, Boston (D.A.); the University of Texas M.D. Anderson Cancer Center, Houston (W.V.C., C.I.A.); and the University of California San Francisco, San Francisco (L.A.C.).
REFERENCES
- 1.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–61. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.Seldin MF, Amos CI, Ward R, Gregersen PK. The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum. 1999;42:1071–9. doi: 10.1002/1529-0131(199906)42:6<1071::AID-ANR1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Klareskog L, Stolt P, Lundberg K, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLADR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 4.MacGregor AJ, Snieder H, Rigby AS, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–7. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.Bali D, Gourley S, Kostyu DD, et al. Genetic analysis of multiplex rheumatoid arthritis families. Genes Immun. 1999;1:28–36. doi: 10.1038/sj.gene.6363635. [DOI] [PubMed] [Google Scholar]
- 6.Cornélis F, Fauré S, Martinez M, et al. New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci U S A. 1998;95:10746–50. doi: 10.1073/pnas.95.18.10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiozawa S, Hayashi S, Tsukamoto Y, et al. Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol. 1998;10:1891–5. doi: 10.1093/intimm/10.12.1891. [DOI] [PubMed] [Google Scholar]
- 8.Jawaheer D, Seldin MF, Amos CI, et al. Screening the genome for rheumatoid arthritis susceptibility genes: a replication study and combined analysis of 512 multi-case families. Arthritis Rheum. 2003;48:906–16. doi: 10.1002/art.10989. [DOI] [PubMed] [Google Scholar]
- 9.MacKay K, Eyre S, Myerscough A, et al. Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum. 2002;46:632–9. doi: 10.1002/art.10147. [Erratum, Arthritis Rheum 2002;46:1406.] [DOI] [PubMed] [Google Scholar]
- 10.Amos CI, Chen WV, Lee A, et al. High-density SNP analysis of 642 Caucasian families with rheumatoid arthritis identifies two new linkage regions on 11p12 and 2q33. Genes Immun. 2006;7:277–86. doi: 10.1038/sj.gene.6364295. [DOI] [PubMed] [Google Scholar]
- 11.Etzel CJ, Chen WV, Shepard N, et al. Genome-wide meta-analysis for rheumatoid arthritis. Hum Genet. 2006;119:634–41. doi: 10.1007/s00439-006-0171-8. [DOI] [PubMed] [Google Scholar]
- 12.Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee AT, Li W, Liew A, et al. The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun. 2005;6:129–33. doi: 10.1038/sj.gene.6364159. [DOI] [PubMed] [Google Scholar]
- 14.Stastny P, Fink CW. HLA-Dw4 in adult and juvenile rheumatoid arthritis. Transplant Proc. 1977;9:1863–6. [PubMed] [Google Scholar]
- 15.Irigoyen P, Lee AT, Wener MH, et al. Regulation of anti-cyclic citrullinated peptide antibodies in rheumatoid arthritis: contrasting effects of HLA-DR3 and the shared epitope alleles. Arthritis Rheum. 2005;52:3813–8. doi: 10.1002/art.21419. [DOI] [PubMed] [Google Scholar]
- 16.Huizinga TW, Amos CI, van der Helm-van Mil AH, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–8. doi: 10.1002/art.21385. [DOI] [PubMed] [Google Scholar]
- 17.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plenge RM, Padyukov L, Remmers EF, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–60. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 20.Plenge R, Rioux JD. Identifying susceptibility genes for immunological disorders: patterns, power, and proof. Immunol Rev. 2006;210:40–51. doi: 10.1111/j.0105-2896.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- 21.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 22.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pe'er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–7. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 26.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe F, Michaud K, Gefeller O, Choi HK. Predicting mortality in patients with rheumatoid arthritis. Arthritis Rheum. 2003;48:1530–42. doi: 10.1002/art.11024. [DOI] [PubMed] [Google Scholar]
- 28.Fries JF, Wolfe F, Apple R, et al. HLADRB1 genotype associations in 793 white patients from a rheumatoid arthritis inception cohort: frequency, severity, and treatment bias. Arthritis Rheum. 2002;46:2320–9. doi: 10.1002/art.10485. [DOI] [PubMed] [Google Scholar]
- 29.Weisman M, Bombardier C, Massarotti E, et al. Analysis at one year of an inception cohort of early rheumatoid arthritis: the SONORA study. Arthritis Rheum. 2003;48:5119. abstract. [Google Scholar]
- 30.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–10. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 2004;50:3085–92. doi: 10.1002/art.20553. [DOI] [PubMed] [Google Scholar]
- 32.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragoussis J, Elvidge GP, Kaur K, Colella S. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics Genet research. PLoS. 2006;2(7):e100. doi: 10.1371/journal.pgen.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 36.Seldin MF, Shigeta R, Villoslada P, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2(9):e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–6. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 38.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 39.Elliott MJ, Maini RN, Feldmann M, et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor alpha (cA2) versus placebo in rheumatoid arthritis. Lancet. 1994;344:1105–10. doi: 10.1016/s0140-6736(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 40.Weinblatt ME, Kremer JM, Bankhurst AD, et al. A trial of etanercept, a recombinant tumor necrosis factor receptor:Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N Engl J Med. 1999;340:253–9. doi: 10.1056/NEJM199901283400401. [DOI] [PubMed] [Google Scholar]
- 41.Tsitsikov EN, Laouini D, Dunn IF, et al. TRAF1 is a negative regulator of TNF signaling: enhanced TNF signaling in TRAF1-deficient mice. Immunity. 2001;15:647–57. doi: 10.1016/s1074-7613(01)00207-2. [DOI] [PubMed] [Google Scholar]
- 42.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–91. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 43.Cooke TD, Hurd ER, Jasin HE, Bienen-stock J, Ziff M. Identification of immunoglobulins and complement in rheumatoid articular collagenous tissues. Arthritis Rheum. 1975;18:541–51. doi: 10.1002/art.1780180603. [DOI] [PubMed] [Google Scholar]
- 44.Zvaifler NJ. The immunopathology of joint inflammation in rheumatoid arthritis. Adv Immunol. 1973;16:265–336. doi: 10.1016/s0065-2776(08)60299-0. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Kristan J, Hao L, Lenkoski CS, Shen Y, Matis LA. A role for complement in antibody-mediated inflammation: C5-deficient DBA/1 mice are resistant to collagen-induced arthritis. J Immunol. 2000;164:4340–7. doi: 10.4049/jimmunol.164.8.4340. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Rollins SA, Madri JA, Matis LA. Anti-C5 monoclonal antibody therapy prevents collagen-induced arthritis and ameliorates established disease. Proc Natl Acad Sci U S A. 1995;92:8955–9. doi: 10.1073/pnas.92.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji H, Ohmura K, Mahmood U, et al. Arthritis critically dependent on innate immune system players. Immunity. 2002;16:157–68. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 48.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurreeman F, Padyukov L, Marques R, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. doi: 10.1371/journal.pmed.0040278. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.