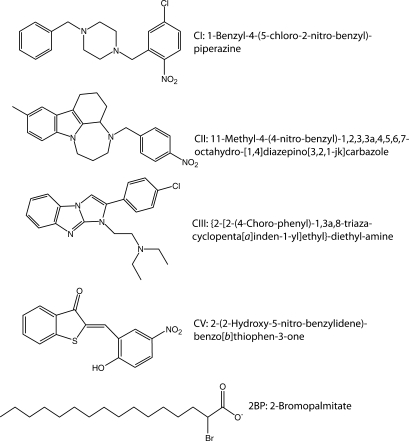
Abstract
Pharmacologic approaches to studying palmitoylation are limited by the lack of specific inhibitors. Recently, screens have revealed five chemical classes of small molecules that inhibit cellular processes associated with palmitoylation (Ducker, C. E., L. K. Griffel, R. A. Smith, S. N. Keller, Y. Zhuang, Z. Xia, J. D. Diller, and C. D. Smith. 2006. Discovery and characterization of inhibitors of human palmitoyl acyltransferases. Mol. Cancer Ther. 5: 1647–1659). Compounds that selectively inhibited palmitoylation of N-myristoylated vs. farnesylated peptides were identified in assays of palmitoyltransferase activity using cell membranes. Palmitoylation is catalyzed by a family of enzymes that share a conserved DHHC (Asp-His-His-Cys) cysteine-rich domain. In this study, we evaluated the ability of these inhibitors to reduce DHHC-mediated palmitoylation using purified enzymes and protein substrates. Human DHHC2 and yeast Pfa3 were assayed with their respective N-myristoylated substrates, Lck and Vac8. Human DHHC9/GCP16 and yeast Erf2/Erf4 were tested using farnesylated Ras proteins. Surprisingly, all four enzymes showed a similar profile of inhibition. Only one of the novel compounds, 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one [Compound V (CV)], and 2-bromopalmitate (2BP) inhibited the palmitoyltransferase activity of all DHHC proteins tested. Hence, the reported potency and selectivity of these compounds were not recapitulated with purified enzymes and their cognate lipidated substrates. Further characterization revealed both compounds blocked DHHC enzyme autoacylation and displayed slow, time-dependent inhibition but differed with respect to reversibility. Inhibition of palmitoyltransferase activity by CV was reversible, whereas 2BP inhibition was irreversible.
Keywords: DHHC, Asp-His-His-Cys; lipidation; fatty acylation; S-acylation; enzyme; inhibitor
Protein palmitoylation is the posttranslational attachment of palmitate or other long-chain fatty acids to cysteine residues in proteins via a thioester linkage (as reviewed in Refs. 1, 2). The functional consequences of protein palmitoylation are diverse and include effects on protein localization, trafficking, and stability. In contrast to other lipid modifications, palmitoylation is reversible. Consequently several cellular processes use palmitoylation-depalmitoylation cycles to affect the localization and function of key proteins.
Protein acyltransferases (PATs) catalyze the addition of palmitate (3). Genetic and biochemical studies in yeast uncovered a family of integral membrane enzymes that mediate palmitate addition to proteins that are modified on the cytoplasmic face of cell membranes. The hallmark of this family of proteins is the DHHC (Asp-His-His-Cys) cysteine-rich domain. A family of seven DHHC proteins is present in S. cerevisiae, and homology searches have identified at least 23 genes that encode DHHC proteins in mammals. PAT activity is dependent upon the DHHC domain and mutation of the cysteine of the DHHC motif abolishes catalytic activity of the enzyme.
Palmitoylated proteins are prominent in cell-signaling pathways and are particularly abundant in the nervous system (4). Signal transducers, receptors, ion channels, and scaffolds are among the targets of palmitoyltransferases. Finding the enzymes responsible for palmitoylation has accelerated our understanding of the role of palmitoylation in native and disease states. An example is HIP14 (DHHC17), a DHHC protein initially identified as huntingtin (htt)-interacting protein 14 (5). HIP14 has been linked to palmitoylation of htt, SNAP-25, cysteine string protein, and other neuronal substrates (6). Palmitoylation of htt by HIP14 influences htt localization and protects it from aggregation (7). In flies, HIP14 is essential for presynaptic function and plays a role in neurotransmitter release, likely through palmitoylation of SNAP-25 and cysteine string protein (8, 9).
Oncogenic Ras proteins are associated with numerous human tumors. Palmitoylation of H- and N-Ras plays a key role in trafficking between the plasma membrane and Golgi apparatus (3), as well as facilitating the oncogenic potential of activating Ras mutations (10). Several DHHC proteins have been linked to cancer. Human HIP14 is oncogenic, promoting colony formation in soft agar and tumor formation in nude mice (11). HIP14 mRNA is upregulated in numerous tumors (12). DHHC9, a PAT for H- and N-Ras in vitro, is upregulated in microsatellite stable tumors (13), whereas DHHC2 is a putative tumor suppressor (14).
The importance of palmitoylation in physiology and pathophysiology suggest that palmitoylation inhibitors could be beneficial for the treatment of diseases, as well as tools for probing the role of palmitoylation in cellular processes. Inhibitors of palmitoylation have been limited to 2-bromopalmitate (2BP), cerulenin, and tunicamycin. The most commonly used, 2BP, inhibits palmitoylation in cells (15) and PAT activity of DHHC proteins in vitro (16). However, 2BP also inhibits fatty acid CoA ligase (17) and other enzymes involved in lipid metabolism (18). Similarly, cerulenin and tunicamycin inhibit palmitoylation within cells but also inhibit other cellular process including fatty acid synthesis (19) and N-glycosylation (20, 21), respectively. Consequently, there is a need to identify specific inhibitors of palmitoylation.
Ducker et al. (12) have developed high throughput screens for palmitoylation inhibitors. The screens yielded compounds that fell into five chemical classes, and a representative compound from each class was further characterized. Cell membranes from MCF-7 cells were used as a source of PAT activity to evaluate the representative compounds' ability to inhibit palmitoylation in vitro. Fluorescently labeled peptides mimicking either N-myristoylated, palmitoylated proteins, such as Gα subunits and Src-related tyrosine kinases, or mimicking C-terminally farnesylated, palmitoylated proteins, like N- and H-Ras, were used as substrates. Four of the representative compounds showed a preference for inhibiting palmitoylation of the Ras-like peptide (average of 76% inhibition) but not the myristoylated peptide (15% inhibited). Conversely, the fifth compound displayed the reverse, inhibiting palmitoylation of the myristoylated peptide (74%) but not the Ras-like peptide (17%). Thus, it appears these compounds not only inhibit palmitoylation but also display substrate specificity. Additionally, IC50 values were reported in low- to submicromolar range (11.8–0.5 μM) (12).
We sought to further investigate the properties of these compounds. Because cell membranes were used as a source of PAT activity, it is unclear whether the inhibitors reduced palmitoylation by directly blocking DHHC proteins. Here we tested if these compounds inhibit DHHC-mediated palmitoylation of protein substrates. Additionally, by using DHHC proteins that display different substrate preferences, the ability of these inhibitor compounds to specifically inhibit palmitoylation of farnesylated substrates vs. N-myristoylated substrates was examined. Compounds that inhibited DHHC-mediated palmitoylation were further characterized.
MATERIALS AND METHODS
Inhibitor compounds and reagents
Inhibitor compounds (Fig. 1) were purchased from ChemBridge Corporation (San Diego, CA). [3H]palmitoyl-CoA ([3H]palmCoA) was synthesized using [3H]palmitate (45 Ci/mmol, PerkinElmer Life Sciences), CoA (Sigma), and acyl CoA synthase (Sigma) as described (22) with the following modification. Following synthesis, [3H]palmCoA was separated from palmitate by chloroform/methanol extraction (23) and subsequently purified on a C8 reversed phase cartridge (22).
Fig. 1.
Structure and names of inhibitor compounds used in this study.
Plasmids
Standard molecular biology techniques were used to manipulate DNA. All plasmid constructs were verified by DNA sequence analysis. DHHC2 was expressed using recombinant baculovirus. DHHC2 was prone to proteolysis and several constructs were made with affinity tags at both ends to generate a virus that would yield intact DHHC2 protein. Oligonucleotide sequences used to generate plasmids are available upon request. Human DHHC2 was amplified from Image Clone 4398300 (GenBank ID BF938682) and subcloned as a KpnI fragment into pBlueBacHis2B (Invitrogen; Carlsbad, CA). This yielded pML850, encoding His6Express-DHHC2. pML850 was digested with HindIII and SalI to permit ligation with a double-stranded oligonucleotide linker that encodes a HindIII site, the FLAG epitope, an EcoRI site, a His6 epitope, a stop codon, and a SalI site. This yielded pML892, encoding His6Express-DHHC2-FLAG-His6. pML892 was cut with BglII and AgeI, purified, and ligated with double-stranded oligonucleotides encoding the calmodulin binding peptide (CBP) followed by an XbaI site to generate His6Express-CBP-DHHC2-FLAG-His6 (pML943). Catalytically inactive DHHS2 (pML1023) was generated by site-directed mutagenesis (Stratagene) of pML943.
Plasmids expressing the N-terminal 226 amino acids of Lck (LckNT) and LckNT(C3,5S) were constructed as follows. The murine Lck cDNA encoding residues 1-226 was amplified as a NdeI-XhoI fragment and subcloned into the bacterial expression vector pET23a(+) yielding pML1008. This construct was used as template to generate the C3,5S mutant (pML1175) using a mutagenic primer to amplify the 5′ end of the coding sequence.
Yeast N-myristoyltransferase was expressed using pBB131 (24). The plasmid pML1067 was constructed to express human NMT1 by excising the yeast NMT1 gene from pBB131 with BglII and EcoR1 and replacing it with human NMT1 flanked by pBB131 vector sequences. Human NMT1 was amplified from pBB218 and overlap extension PCR was used to generate flanking pBB131 vector sequences (25).
Recombinant baculovirus and insect cell culture
Sf9 insect cells were purchased from ATCC and grown in suspension culture medium [IPL-41 (Gibco) supplemented with 10% heat inactivated bovine growth serum, yeastolate, Pluronic F68, 50 μg/ml gentamycin, and 250 ng/ml fungizone] at 27°C with rotation at 110 rpm. Recombinant baculoviruses were generated using Invitrogen's Bac-n-Blue™ transfection kit with pML943 and pML1023 and plaque purified.
Purification of DHHC proteins
For DHHC2, Sf9 insect cells were inoculated with baculovirus expressing human DHHC2 N-terminally tagged with His6-Express-CBP and C-terminally tagged with FLAG-His6. Infected cells were collected by centrifugation and washed with cold PBS 61 h post infection. Cell pellets were stored at −80°C until purification. All purification steps were performed at 4°C, and all buffers contained the protease inhibitors 1 mM PMSF, 1–5 μg/ml pepstatin A, 1.4 μg/ml aprotinin, 1.6 μg/ml leupeptin, and 1.6 μg/ml lima bean trypsin inhibitor. A cell pellet of ∼5 ml (from 335 ml Sf9 culture) was quickly thawed at 37°C and suspended in 35 ml cavitation buffer (50 mM Tris pH 7.4, 150 mM NaCl, 10 mM β-ME, 1 mM EDTA). Cells were lysed by nitrogen cavitation (30 min at 700 psi). The lysate was centrifuged at 700 g for 10 min to remove nuclei and unbroken cells. The postnuclear supernatant was centrifuged at 100,000 g for 30 min to generate P100 and S100 fractions. P100 membranes were suspended in 9 ml extract buffer (50 mM Tris pH 7.4, 200 mM NaCl, 10 mM β-ME, and 10% glycerol) by sequential passage through 14, 18, and 25 gauge needles. Total protein concentration was determined using Bio-Rad's Bradford protein assay (Hercules, CA). Membranes were diluted with extract buffer and 10% n-dodecyl-β-D-maltoside detergent (DDM; Dojindo Laboratories, Japan) to give a final protein concentration of 2 mg/ml in 1% DDM. The extract was again passed through a 25 G needle and incubated 80 min with end-over-end rotation. The extract was cleared at 100,000 g for 30 min, diluted 1:1 with extract buffer (no DDM), and gravity-flowed twice through a column of 3.4 ml Ni2+-nitrilotriacetic acid-agarose resin (Ni-NTA; Qiagen) equilibrated in wash buffer (50 mM Tris pH 7.4, 100 mM NaCl, 3 mM β-ME, 10% glycerol, 0.1% DDM, and 20 mM imidazole). The resin was washed with 60 ml wash buffer and eluted with wash buffer containing 200 mM imidazole (2 × 3 ml) and wash buffer containing 500 mM imidazole (3 × 3 ml). Ni elutions 1–4 were pooled, diluted 1:1 with buffer A (50 mM Tris pH 7.4, 100 mM NaCl, 10% glycerol, 0.1% DDM, 1 mM EDTA, and 0.2 mM β-ME), and passed thrice through a column of 300 μl ANTI-FLAG® M2-agarose affinity gel (Sigma) equilibrated in buffer A. The resin was washed with 14 ml buffer A and eluted with 5 × 250 μl buffer A containing 0.23 mg/ml FLAG peptide (Sigma) with a 10 min incubation for each elution. The concentration of enzyme was determined by extrapolation from a linear curve with known concentrations of BSA using Sypro Ruby protein gel stain (Molecular Probes) and quantitation with a Storm™ 860 (Amersham Biosciences).
DHHS2 purification paralleled that of DHHC2 through nickel affinity chromatography. The identity of purified DHHC2 or DHHS2 was confirmed by immunoblots using antibodies at the following dilutions: anti-FLAG 1:3,000 (Stratagene), anti-Express 1:1,500 (Invitrogen), and goat anti-mouse IgG secondary conjugated to HRP at 1:2,000 (MP Biomedicals, OH).
For human DHHC9/GCP16, Sf9 cells were coinfected with baculoviruses expressing DHHC9-myc-His6 and FLAG-GCP16 (26) and cultured for 73 h before harvesting. DHHC9/GCP16 was purified similarly to DHHC2.
The purification of Erf2/Erf4p was performed as previously described (27).
For partially purified yeast Pfa3, YPH499 was transformed with a Pfa3-His6-Flag pESC expression construct. Yeast were cultured and cell membranes prepared as described (28). Pfa3-His6-Flag was extracted from cell membranes in 1% Triton X-100 at 5 mg/ml total protein in extraction buffer (50 mM Tris pH 8.0, 100 mM NaCl, 10% glycerol, 5 mM β-ME, and 0.1 mM PMSF) rotating for 1 h at 4°C. Insoluble material was pelleted at 200,000 g for 20 min. The detergent extract (7.5 mg total protein) was diluted 1:1 in extraction buffer and allowed to bind to 2 ml Ni-NTA resin for 1 h at 4°C. The resin was reconstituted in a column and washed with 20 ml wash buffer [50 mM Tris pH 8.0, 100 mM NaCl, 10% glycerol, 5 mM β-ME, 0.1% Triton X-100, 0.1 mg/ml bovine liver lipids (Avanti Polar Lipids), and 0.1 mM PMSF]. Bound proteins were eluted with elution buffer (50 mM Tris pH 8.0, 100 mM NaCl, 10% glycerol, 200 mM imidazole, 1 mM β-ME, 0.1% Triton X-100, and 0.1 mg/ml bovine liver lipids), and four 2 ml fractions were collected. Fractions one and two were pooled, aliquoted, and stored at −80°C.
Purification of PAT substrates
Mouse N-myristoylated LckNT (myrLckNT) was purified from ER2566 E.coli (New England Biolabs) harboring pBB131 (24) and pML1008. Cells were grown in Luria-Bertani broth containing 50 μg/ml kanamycin and 50 μg/ml ampicillin at 37°C with shaking at 250 rpm for 4 h. Isopropyl β-D-1-thiogalactopyranoside was added to 0.6 mM final concentration, and the temperature was reduced to 30°C for 5 h. Cells were harvested by centrifugation at 3,080 g for 20 min at 4°C. Cell pellets were washed with cold PBS, collected by centrifugation at 3,800 g for 17 min at 4°C, flash frozen in liquid N2, and stored at −80°C until needed. Purification steps were performed on ice or in a 4°C cold room. Cell pellets representing 400 ml culture were quickly thawed at 30°C, suspended in 11 ml lysis buffer (50 mM Tris pH 7.4, 8 mM β-ME, 150 mM NaCl, 1 μg/ml pepstatin A, 1.6 μg/ml leupeptin, 1.6 μg/ml lima bean trypsin inhibitor, and 1.4 μg/ml aprotinin), and passed thrice through a French press cell. The soluble fraction was collected by centrifugation at 76,500 g for 30 min. To enrich for myristoylated-LckNT, this soluble fraction was extracted twice with 1.3 ml 10% Triton X-114 detergent (Sigma) essentially as described (29) except incubations were for 5 min at 37°C. The detergent phases were pooled, diluted to 0.2% Triton X-114 (135 ml total), and gravity loaded onto a 3 ml Ni-NTA column equilibrated in wash buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, and 8 mM β-ME). The column was washed with 150 ml wash buffer and eluted 5 × 3 ml with elution buffer (50 mM Tris pH 7.4, 100 mM NaCl, 300 mM imidazole, 8 mM β-ME, and 10% glycerol). Elution 2 was used for subsequent PAT assays. The concentration of myrLckNT was determined by extrapolation from a linear curve with known concentrations of BSA using Coomassie Blue gel stain and quantitation with ImageJ software (NIH). C3,5S myrLckNT was expressed in BL21(DE3) E.coli transformed with pML1067 and pML1175 and purified as described for wild-type. The identity of purified protein was confirmed by immunoblotting with mouse monoclonal anti-Lck (3A5, sc-433) purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Yeast N-myristoylated Vac8 was purified by ion-exchange and hydroxylapatite chromatography as described (27). Prenylated Ras proteins were expressed in yeast from the galactose-inducible vector pEG(KG) and purified as described (27, 30). The glutathione S-transferase (GST)-Ras2(HV) construct consists of the C-terminal 35 amino acid residues of yeast Ras2 fused to GST. Full-length mammalian H-Ras was fused to GST.
In vitro PAT assays
Inhibitors were dissolved and serially diluted to 100× stocks in 100% DMSO. Stocks were diluted to 5× with buffer containing 20 mM Tris pH 7.4, 1 mM EDTA, and 1 mM 1,4- dithiothreitol (DTT). Equal volumes (5 μl) of DHHC enzyme and 5× inhibitor solutions were preincubated for various times and temperatures as indicated in figure legends. Protein substrates diluted in buffer containing 20 mM Tris pH 7.4, 1 mM EDTA, and 1 mM DTT (5 μl) were premixed with reaction hot mix [RHM; 10 μl; 250 mM MES pH 6.4, 1 mM DTT, 1.2-3.4 μM [3H]palmCoA and incubated at the same temperature as enzyme/inhibitor mix. This substrate mixture (15 μl) was added to the enzyme/inhibitor mix to start the reaction and incubated as indicted for each figure. The reactions (25 μl) were stopped by addition of 5× gel loading buffer with final 2 mM DTT (6.25 μl). Except for assays of reversibility, all inhibitor concentrations shown are for the final 25 μl reaction. Stopped reactions were heated for 60 s at 95°C before resolution by SDS-PAGE. Gels were stained with Coomassie Blue, destained, and scanned. Protein substrate bands were excised from the gel, placed into scintillation vials, solubilized in 500 μl Soluene 350 (PerkinElmer) at 50°C for at least 3 h, and quantitated by liquid scintillation spectroscopy.
Enzyme autoacylation assays without inhibitors (Fig. 2, right panel) were performed similar to inhibitor profile assays described above. Briefly, 500 fmol of enzyme was incubated in 0.82 μM [3H]palmCoA for 10 min at 25°C in RHM. Reactions were stopped with gel loading buffer and resolved by SDS-PAGE. Gels were stained, destained, soaked in 1 M sodium salicylate/15% methanol for 20 min, dried onto filter paper, and exposed to film at −80°C for 8 days. Enzyme autoacylation assays with inhibitors (Fig. 5) were performed similar to inhibitor profile assay except without protein substrate. Briefly, 250 fmol of DHHC2 was preincubated with inhibitor for 8 min at 25°C. Prewarmed RHM was added to give a final concentration of 1.1 μM [3H]palmCoA in 25 μl and the reaction proceeded for 2 min before stopping with 5× gel loading buffer. Reactions were processed similar to those in Fig. 2.
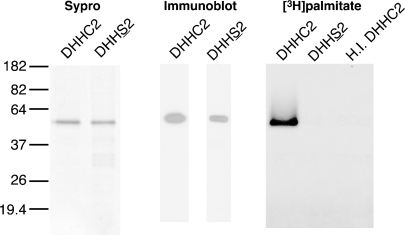
Fig. 2.
Purification of human DHHC2 and catalytically inactive DHHS2. DHHC2 was expressed in Sf9 cells and purified sequentially by nickel chelate and FLAG affinity chromatography. Catalytically inactive DHHS2 was purified by nickel affinity chromatography; the fifth elution is shown. Purified DHHC2 and DHHS2 were detected by Sypro staining (left) and immunoblotting with anti-Express antibody (center). DHHC2, but not DHHS2, autoacylates (right). DHHC2, DHHS2, or heat inactivated DHHC2 (500 fmol each) were incubated in 0.8 μM [3H]palmCoA at pH 6.4 for 10 min. The reactions were stopped with SDS sample buffer and processed for fluorography. The film was exposed for 8 days.
Fig. 5.
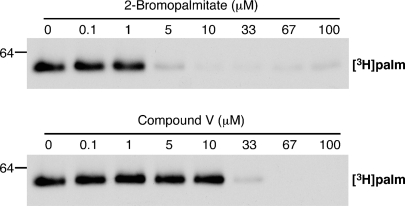
Inhibitor effect on enzyme autoacylation. DHHC2 (250 fmol) was preincubated with inhibitor and assayed as described in Materials and Methods. The film was exposed for 8 days.
The assay for reversibility was similar to those above, except the initial reaction volume was 200 μl and 25 μl aliquots were removed at the indicated times. DHHC2 was preincubated for 30 min at 25°C with inhibitor at the first concentration indicated. Equilibrated substrates in reaction hot mix with either DMSO or inhibitor were used to dilute the enzyme/inhibitor 40-fold to the second concentration indicated and to start the reaction, which was maintained at 25°C. At the indicated times, aliquots (25 μl) were removed from the reaction, stopped with gel loading buffer, and processed as above. In experiments to measure time-dependent inhibition, DHHC2 (100 fmol) was preincubated 30 min at 25°C with inhibitor or a DMSO control. Reactions were started by addition of either both substrates in RHM to enzyme/inhibitor mix (preincubation) or both substrates and inhibitor to enzyme/DMSO mix (no preincubation). Reactions (25 μl, 2% DMSO final) were incubated 7 min at 25°C before stopping and processing as above. Data were graphed and analyzed using Microsoft Excel and GraphPad Prism 5.0.
RESULTS
Purification of active DHHC2
The predicted targets of the palmitoylation inhibitors identified by Ducker et al. (12) are the DHHC proteins (Fig. 1). Furthermore, the pattern of specificity shown by the inhibitors in assays using cell membranes as a source of palmitoyltransferase activity suggests that some of the inhibitors would more potently inhibit DHHC proteins that modify farnesylated substrates, whereas others would be more effective inhibitors of PATs for N-myristoylated substrates. To test these predictions, we purified mammalian and yeast DHHC PATs that display the appropriate substrate specificity. Yeast Erf2/Erf4 is a PAT for yeast Ras (30), but displays little activity toward Vac8, an N-myristoylated protein (27). Conversely, yeast Pfa3 palmitoylates Vac8 (28) but not Ras2 (27). Mammalian DHHC9/GCP16 palmitoylates H-Ras in vitro, but has little or no activity for N-myristoylated Giα1 (26).
We sought to identify a mammalian DHHC protein that has robust activity for an N-myristoylated substrate. Preliminary experiments revealed that the nonreceptor tyrosine kinase Lck, a protein palmitoylated at cysteine residues near the myristoylated N-terminus, was a good substrate for DHHC2 (data not shown). To further characterize this interaction, we expressed human DHHC2 with epitope tags at the N- and C termini to aid in the purification of full-length enzyme. Detergent extracts from membranes were passed sequentially over nickel and FLAG affinity resins to generate purified enzyme (Fig. 2, left). Immunoblotting revealed full-length protein was isolated (Fig. 2, center).
In all DHHC proteins characterized to date, PAT activity has been coupled with incorporation of palmitate into the DHHC protein itself, a process termed enzyme autoacylation (3). Although it is uncertain whether this process represents an acyl-enzyme transfer intermediate, autoacylation serves as a read out for functional enzyme. Purified DHHC2 incorporated [3H]palmitate derived from [3H]palmCoA. This reaction was specific as neither heat-treated DHHC2 nor DHHS2, a mutant predicted by homology to be catalytically inactive (3), showed radiolabeling (Fig. 2, right). Thus, we have purified DHHC2 and shown it to be functionally active as monitored by autoacylation.
DHHC2 palmitoylates myrLckNT at cysteines 3 and 5
To test Lck as a substrate for DHHC2, we used a model substrate consisting of the N-terminal myristoylation, palmitoylation motif (SH4 domain) and the SH3 and SH2 domains (LckNT). This fragment is more easily purified than the full-length protein that includes the kinase domain. To confirm that in vitro palmitoylation of LckNT was occurring at cysteine residues palmitoylated in vivo, we also expressed and purified C3,5S myrLckNT. Lck is palmitoylated in vivo at Cys3 and Cys5, which may differentially regulate Lck localization (31). Lck has two additional cysteines near the N terminus (Cys20 and Cys23) that are required for binding coreceptors CD4 and CD8 (32). Because myristoylation facilitates subsequent palmitoylation both in vivo and in vitro (22, 33), we purified wild-type and mutant LckNT from E. coli coexpressing human N-myristoyltransferase to produce N-myristoylated substrates. Metabolic labeling with [3H]myristic acid indicated that both proteins were myristoylated in this system and that mutation of the palmitoylation sites did not affect myristoylation (see supplementary Fig. I).
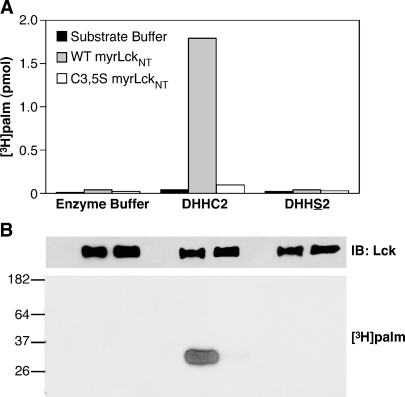
Wild-type and C3,5S myrLckNT were purified and assayed for their ability to be acylated by DHHC2 in an in vitro PAT assay. Incubation of either form of myrLckNT with [3H]palmCoA and enzyme buffer did not result in incorporation of radiolabel in myrLckNT as monitored by scintillation counting or fluorography (Fig. 3, first 3 lanes). When 200 fmol of DHHC2 was included in the reaction, greater than 1,500 fmol of [3H]palmitate was incorporated into wild-type myrLckNT, indicating that transfer is catalytic. Less than 100 fmol of [3H]palmitate was incorporated into C3,5S myrLckNT by DHHC2. When autoacylation-defective DHHS2 was assayed, no radiolabel was incorporated into myrLckNT (Fig. 3, last 3 lanes). The low levels of radiolabel in C3,5S myrLckNT may indicate that DHHC2 weakly palmitoylates myrLckNT at other cysteine residues nonspecifically.
Fig. 3.
DHHC2 palmitoylates myrLckNT at cysteines 3 and 5. Purified DHHC2, DHHS2 (200 fmol), or enzyme buffer was mixed with purified myrLckNT (25 pmol) on ice. Reaction buffer was added to give a final concentration of 0.8 μM [3H]palmCoA in 50 μl and incubated at 25°C for 7 min. The reaction was stopped with SDS sample buffer and divided between two SDS-PAGE gels, which were stained and destained. A: The region containing myrLckNT was excised from one gel, solubilized with Soluene, and the amount of [3H]palmitate present determined by scintillation counting. B: The second gel was processed for fluorography and exposed to film at −80°C for 90 h. MyrLckNT was detected by immunoblotting reactions run without radioactive palmCoA.
Evaluation of potential PAT inhibitors using enzyme-substrate pairs
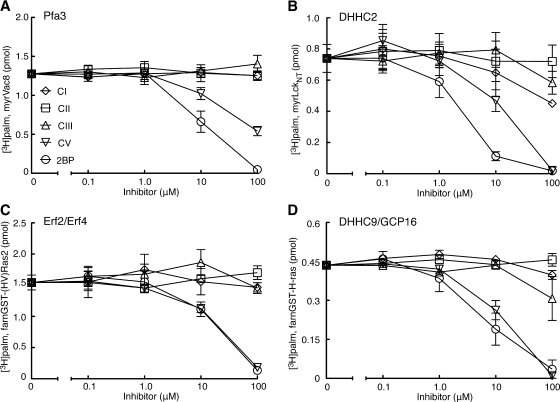
Using the three established enzyme-substrate pairs described above along with DHHC2 and myrLckNT, we sought to determine if the PAT inhibitors identified by Ducker et al. (12) blocked DHHC-mediated palmitoylation and displayed the predicted substrate specificity. We obtained and tested four of the five compounds along with 2BP (Fig. 1). Compound IV (2-(2-fluoro-benzyl)-1-phenyl-1,2,3,4-tetrahydro-pyrrolo[1,2,-a]pyrazine) was not available from commercial sources and was not tested here. For inhibitor PAT assays, the inhibitor was preincubated with DHHC protein, followed by the addition of protein substrate and [3H]palmCoA that had been premixed and equilibrated at the assay temperature. To remain in the linear range for each reaction, concentrations, temperature, and lengths of incubations varied for each pair and are listed in the legend to Fig. 4.
Fig. 4.
Inhibitor profiles for DHHC proteins. Inhibitor profiles for each enzyme-substrate pair were carried out as described in the text. A: Partially purified yeast Pfa3 (5 μl) was preincubated with inhibitor for 8 min at 30°C. MyrVac8 and [3H]palmCoA were mixed, warmed to 30°C, and added to the reaction at final concentrations of 0.5 μM and 0.8 μM, respectively. The reaction proceeded for 10 min at 30°C before stopping with SDS sample buffer. B: Purified human DHHC2 (100 fmol) was preincubated 8 min at 25°C with inhibitors. MyrLckNT and [3H]palmCoA were added to give final concentrations of 0.5 μM and 0.8 μM, respectively, and the reaction proceeded for 7 min at 25°C before stopping. C: Purified yeast Erf2/Erf4 (100 fmol) was preincubated 8 min at 25°C with inhibitors. [3H]palmCoA and the farnesylated hypervariable region of Ras2 fused to glutathione S-transferase (GST) were added to give final concentrations of 0.9 μM and 1.0 μM, respectively, and the reaction proceeded for 10 min at 25°C before stopping. D: Purified human DHHC9/GCP16 (250 fmol) was preincubated 10 min on ice with inhibitors. [3H]palmCoA and farnesylated H-Ras fused to GST were added to give final concentrations of 0.9 μM and 1.0 μM, respectively, and the reaction proceeded for 7 min at 25°C before stopping. Values represent the means ± SEM determined from three independent experiments.
Overall, less potent and specific inhibition was observed than expected (Fig. 4). For all pairs tested, 2BP was the best inhibitor displaying an average IC50 of ∼10 μM. This result confirmed that 2BP inhibits DHHC-mediated palmitoylation in vitro as shown previously for DHHC15 (16). Compound V (CV) was initially described as a selective inhibitor of N-myristoylated, palmitoylated protein but not farnesylated, palmitoylated proteins, with an IC50 of 0.5 μM for an N-myristoylated peptide (12). As shown here, CV inhibited all DHHCs tested but was less potent than 2BP, displaying a higher IC50 for three of the four enzyme-substrate pair. The remaining compounds (CI, CII, and CIII) were previously demonstrated to inhibit PAT activity toward a farnesylated peptide mimicking Ras, with IC50 values of 4 to 12 μM. When tested against the DHHC Ras PATs, DHHC9/GCP16 and Erf2/Erf4, little or no inhibition was observed for any of the enzymes, even at the highest concentration tested (100 μM, Fig. 4). Thus with the DHHC enzyme-substrate pairs tested, the predicted selectivity of these compounds was not observed.
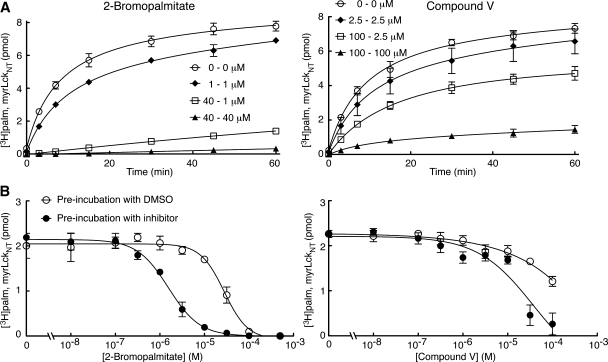
2BP and CV inhibit enzyme autoacylation
2BP and CV reduced DHHC-mediated palmitoylation. Accordingly, we characterized these compounds further. Because 2BP and CV affected all four enzyme-substrates pairs equally, DHHC2 and myrLckNT were chosen as a representative pair for further experiments. To date all DHHC proteins that display PAT activity also autoacylate. Thus, we determined the effect of 2BP and CV on enzyme autoacylation. As seen in Fig. 5, both compounds inhibited enzyme autoacylation, with 2BP being more potent at blocking enzyme radiolabeling than CV.
Evaluation of reversibility and time-dependent inhibition
We next wanted to assess whether the inhibition of PAT activity by 2BP and CV was reversible or irreversible. DHHC2 was preincubated with either 2BP or CV at the first predilution concentration indicated in Fig. 6A. This preformed enzyme-inhibitor complex was diluted 40-fold with buffer containing protein and lipid substrates to give the second inhibitor concentration indicated. The lack of recovery of PAT activity following dilution of 2BP compared with the inhibited controls maintained at persistent concentrations of inhibitor indicates that 2BP is an irreversible inhibitor (Fig. 6A, left side; square symbols relative to filled symbols). In contrast, when CV was similarly diluted, approximately 70 percent of PAT activity was recovered compared with the inhibited control maintained at the diluted concentration of CV (Fig. 6A, right side). This indicates that the inhibitory effect of CV is reversible.
Fig. 6.
Reversibility and time-dependent inhibition. Experiments were performed as described in Materials and Methods. A: Reversibility of 2-bromopalmitate (2BP) and Compound V (CV) was tested by incubating DHHC2 (100 fmol) with the first concentration indicated for 30 min at 25°C. This mix was diluted 40-fold with both substrates to the postdilution concentration indicated, incubated at 25°C, and aliquots removed at the indicated time points. B: Time-dependent inhibition was evaluated by preincubating enzyme with inhibitors at the indicated concentrations or dimethylsulfoxide (DMSO) for 30 min at 25°C. Both substrates without or with inhibitor were added to start the reaction, which was incubated at 25°C for 7 min before stopping. Values represent the means ± SEM determined from three independent experiments.
Finally, because inhibitor profiles for each enzyme-substrate pairs were done following preincubation with inhibitor, we wanted to assess whether 2BP and CV displayed time-dependent inhibition (also called slow-binding inhibition; Fig. 6B). During one set of conditions (without inhibitor preincubation) substrates and 2BP or CV were premixed and the assay started by addition of DHHC2 preincubated with a DMSO control. Alternatively, under a second set of conditions (with inhibitor preincubation), DHHC2 and 2BP or CV were preincubated 30 min before addition of both substrates to initiate the reaction. For both compounds, an increase in inhibitor potency following preincubation was observed with the curves shifting to the left (Fig. 6B). This suggests that CV and 2BP are time-dependent inhibitors of DHHC2 PAT activity. Higher concentrations of CV were not tested because of limitations in the solubility of the compound.
DISCUSSION
As advances are made in the palmitoylation field, it is becoming increasingly clear that DHHC proteins play an important role in native and disease states. DHHC proteins have been identified that are responsible for palmitoylating proteins involved in cell growth (11, 26), maintaining vascular homeostasis (34), neurotransmitter receptor trafficking (16), and cell-cell adhesion (35). DHHC proteins have also been linked to several diseases including cancer (11–13), Huntington's disease (7), and mental retardation (36). Identification of potent and specific palmitoylation inhibitors that target DHHC proteins could lead to novel therapies, as well as provide tools to investigate palmitoylation's roles in cellular processes.
With that in mind, we sought to determine whether DHHC proteins were targets of the palmitoylation inhibitor compounds described by Ducker et al. (12). We found that one of the four compounds, CV, behaved similarly to 2BP, in that it inhibited all four of the DHHC proteins tested. 2BP and CV inhibited autoacylation of the enzyme, which is tightly correlated with the ability to transfer palmitate to substrate. It is unknown whether autoacylation represents an acyltransfer intermediate. However, the loss of autoacylation with inhibitors that also block transfer to substrate is consistent with such a mechanism. Furthermore, the ability of CV to inhibit autoacylation, a property common to all DHHC proteins, suggests that it will not be selective for different DHHC proteins.
Inhibition of DHHC2 PAT activity by 2BP was irreversible. This was not unexpected as 2BP has been shown to irreversibly inhibit fatty acid metabolism (37, 38), as have other halogenated fatty acid analogs (39). Additionally, others have previously hypothesized that 2BP forms an irreversible bond with PATs (40). In contrast, the inhibition by CV was mostly reversible. This is not unexpected in that CV does not contain any highly reactive centers (Fig. 1). 2BP and CV displayed time-dependent inhibition, albeit to different degrees. Time-dependent inhibition is considered an advantage in a pharmacological setting because the enzyme is inhibited for longer periods of time (41). This, plus the fact that 2BP inhibits other enzyme families (17, 18), suggest that CV may be a good candidate for future structure-activity relationship studies with substituent side-chain replacement focused on enhancing potency and specificity.
We were unable to detect significant inhibition of DHHC proteins by three of the compounds tested, nor did CV display selective inhibition of DHHC PAT activity for N-myristoylated substrates. Several possible explanations could account for the differences between the original report describing these compounds and those documented here. Ducker et al. (12) used MCF-7 cell membranes as a source of PAT activity. DHHC proteins have not been studied in these cells, but given the large family size, it is likely that multiple DHHC proteins are expressed. Reverse transcriptase-PCR has detected mRNA for DHHC2 and DHHC9 in MCF-7 cells (unpublished observations). However, it is possible that DHHC proteins other than DHHC2 and DHHC9/GCP16 are possibly more sensitive to these compounds. Phylogenetic analysis of DHHC sequences from humans and yeast place DHHC9 and Erf2 within a larger subfamily (DHHC5, 8, 9, 14, 18, 19, and Erf2). Similarly, DHHC2 and Pfa3 cluster with a different group of DHHC proteins (DHHC2, 3, 7, 15, 20, and Pfa3) (16). DHHC proteins less closely related to these two groups were not tested.
An important difference between the two studies is the substrates that were tested. Ducker et al. (12) assayed short fluorescent, lipidated peptides (myristoyl-Gly-Cys and Cys-Leu-Cys-farnesyl,O-methyl). We assayed full-length (myristoyl-Vac8 and farnesyl-H-Ras) or larger protein fragments (HVRas2, the farnesylated hypervariable domain of Ras, and myristoyl-LckNT). It remains to be determined whether Lck and H-Ras are in vivo substrates, as well as in vitro substrates of DHHC2 and DHHC9/GCP16. However, the yeast enzymes and their substrates have been validated genetically and biochemically (28, 30, 42), and display strong preferences for their respective substrates relative to the other lipidated substrates (27). Accordingly, these yeast enzymes and their substrates represented an excellent system to test the inhibitors for substrate specificity.
Inhibition of PAT activity in MCF-7 membranes by the compounds may be dependent on a membrane environment. DHHC proteins are integral membrane proteins and the surrounding lipid environment can affect rate and specificity of such enzymes (43). The hydrophobic compounds tested here may act indirectly, disrupting the membrane surrounding DHHC proteins rather than directly interacting with them. Alternatively, DHHC proteins within a lipid bilayer may adapt a conformation that makes them more susceptible to these compounds. Because the DHHC proteins tested here were purified and assayed in detergent micelles, effects of the compounds on DHHC proteins that require a lipid membrane would have been missed. Initial attempts at reconstituting DHHC proteins into liposome were unsuccessful and more work is needed in this area.
Finally, it is possible that proteins other than DHHC proteins are targets of these compounds. A study of the palmitoylproteome in yeast suggests that DHHC PATs account for most cellular palmitoylation events (44). In mammals, secreted proteins such as the morphogens Hedgehog and Wnt, as well as the peptide hormone ghrelin, are posttranslationally modified with fatty acids through the action of membrane-bound O-acyltransferases (MBOAT) (1, 45). Modification of these secreted proteins occurs in the lumen of the secretory pathway. To date only one MBOAT protein has been characterized biochemically (46). It is unknown if these activities could account for palmitoylation of the peptides in the MCF-7 cell membranes.
The finding that CV is an inhibitor of DHHC PATs and the antitumor activity reported for all of the compounds makes them interesting reagents for further study. Future investigations into DHHC enzymology and mechanism will likely aid the search for specific and potent PAT inhibitors.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Charles Smith for providing the compounds studied here and Wendy Greentree for technical support.
Abbreviations
2BP, 2-bromopalmitate
β-ME, β-mercaptoethanol
CBP, calmodulin binding peptide
CV, Compound V, 2-(2-Hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one
DDM, n-dodecyl-β-D-maltoside detergent
DHHC, Asp-His-His-Cys
DMSO, dimethylsulfoxide
DTT, 1,4-dithiothreitol
GST, glutathione S-transferase
htt, huntingtin
MBOAT, membrane-bound O-acyltransferase
myrLckNT, N-myristoylated lymphocyte specific kinase, N-terminal residues 1-226
[3H]palmCoA, [3H]9,10-palmitoyl coenzyme A
PAT, protein acyltransferase
PMSF, phenylmethylsulphonyl fluoride
Published, JLR Papers in Press, September 30, 2008.
Footnotes
B.C.J. and M.J.N. were supported by American Heart Association pre-doctoral fellowships (Midwest Affiliate). Work in the authors' laboratories is supported by NIH grants GM51466 and NS057112 (M.E.L.), CA050211 and GM073977 (R.J.D.), and GM48099 (M.L.H).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of a figure.
References
- 1.Resh M. D. 2006. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE. 2006 re14. [DOI] [PubMed] [Google Scholar]
- 2.Linder M. E., and R. J. Deschenes. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8 74–84. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell D. A., A. Vasudevan, M. E. Linder, and R. J. Deschenes. 2006. Protein pamitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47 1118–1127. [DOI] [PubMed] [Google Scholar]
- 4.el-Husseini Ael D., and D. S. Bredt. 2002. Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3 791–802. [DOI] [PubMed] [Google Scholar]
- 5.Singaraja R. R., S. Hadano, M. Metzler, S. Givan, C. L. Wellington, S. Warby, A. Yanai, C. A. Gutekunst, B. R. Leavitt, H. Yi, et al. 2002. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 11 2815–2828. [DOI] [PubMed] [Google Scholar]
- 6.Huang K., A. Yanai, R. Kang, P. Arstikaitis, R. R. Singaraja, M. Metzler, A. Mullard, B. Haigh, C. Gauthier-Campbell, C. A. Gutekunst, et al. 2004. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 44 977–986. [DOI] [PubMed] [Google Scholar]
- 7.Yanai A., K. Huang, R. Kang, R. R. Singaraja, P. Arstikaitis, L. Gan, P. C. Orban, A. Mullard, C. M. Cowan, L. A. Raymond, et al. 2006. Palmitoylation of huntingtin by HIP14is essential for its trafficking and function. Nat. Neurosci. 9 824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stowers R. S., and E. Y. Isacoff. 2007. Drosophila huntingtin-interacting protein 14 is a presynaptic protein required for photoreceptor synaptic transmission and expression of the palmitoylated proteins synaptosome-associated protein 25 and cysteine string protein. J. Neurosci. 27 12874–12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohyama T., P. Verstreken, C. V. Ly, T. Rosenmund, A. Rajan, A. C. Tien, C. Haueter, K. L. Schulze, and H. J. Bellen. 2007. Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles. J. Cell Biol. 179 1481–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willumsen B. M., A. D. Cox, P. A. Solski, C. J. Der, and J. E. Buss. 1996. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene. 13 1901–1909. [PubMed] [Google Scholar]
- 11.Ducker C. E., E. M. Stettler, K. J. French, J. J. Upson, and C. D. Smith. 2004. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 23 9230–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducker C. E., L. K. Griffel, R. A. Smith, S. N. Keller, Y. Zhuang, Z. Xia, J. D. Diller, and C. D. Smith. 2006. Discovery and characterization of inhibitors of human palmitoyl acyltransferases. Mol. Cancer Ther. 5 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansilla F., K. Birkenkamp-Demtroder, M. Kruhoffer, F. B. Sorensen, C. L. Andersen, P. Laiho, L. A. Aaltonen, H. W. Verspaget, and T. F. Orntoft. 2007. Differential expression of DHHC9 in microsatellite stable and instable human colorectal cancer subgroups. Br. J. Cancer. 96 1896–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oyama T., Y. Miyoshi, K. Koyama, H. Nakagawa, T. Yamori, T. Ito, H. Matsuda, H. Arakawa, and Y. Nakamura. 2000. Isolation of a novel gene on 8p21.3–22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes Chromosomes Cancer. 29 9–15. [DOI] [PubMed] [Google Scholar]
- 15.Webb Y., L. Hermida-Matsumoto, and M. D. Resh. 2000. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 275 261–270. [DOI] [PubMed] [Google Scholar]
- 16.Fukata M., Y. Fukata, H. Adesnik, R. A. Nicoll, and D. S. Bredt. 2004. Identification of PSD-95 palmitoylating enzymes. Neuron. 44 987–996. [DOI] [PubMed] [Google Scholar]
- 17.Chase J. F., and P. K. Tubbs. 1972. Specific inhibition of mitochondrial fatty acid oxidation by 2-bromopalmitate and its coenzyme A and carnitine esters. Biochem. J. 129 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman R. A., P. Rao, R. J. Fogelsong, and E. S. Bardes. 1992. 2-Bromopalmitoyl-CoA and 2-bromopalmitate: promiscuous inhibitors of membrane-bound enzymes. Biochim. Biophys. Acta. 1125 203–209. [DOI] [PubMed] [Google Scholar]
- 19.Omura S. 1976. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol. Rev. 40 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeJesus G., and O. A. Bizzozero. 2002. Effect of 2-fluoropalmitate, cerulenin and tunicamycin on the palmitoylation and intracellular translocation of myelin proteolipid protein. Neurochem. Res. 27 1669–1675. [DOI] [PubMed] [Google Scholar]
- 21.Patterson S. I., and J. H. Skene. 1995. Inhibition of dynamic protein palmitoylation in intact cells with tunicamycin. Methods Enzymol. 250 284–300. [DOI] [PubMed] [Google Scholar]
- 22.Dunphy J. T., W. K. Greentree, C. L. Manahan, and M. E. Linder. 1996. G-protein palmitoyltransferase activity is enriched in plasma membranes. J. Biol. Chem. 271 7154–7159. [DOI] [PubMed] [Google Scholar]
- 23.Duncan J. A., and A. G. Gilman. 1996. Autoacylation of G protein alpha subunits. J. Biol. Chem. 271 23594–23600. [DOI] [PubMed] [Google Scholar]
- 24.Duronio R. J., E. Jackson-Machelski, R. O. Heuckeroth, P. O. Olins, C. S. Devine, W. Yonemoto, L. W. Slice, S. S. Taylor, and J. I. Gordon. 1990. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria. Proc. Natl. Acad. Sci. USA. 87 1506–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duronio R. J., S. I. Reed, and J. I. Gordon. 1992. Mutations of human myristoyl-CoA:protein N-myristoyltransferase cause temperature-sensitive myristic acid auxotrophy in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 89 4129–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarthout J. T., S. Lobo, L. Farh, M. R. Croke, W. K. Greentree, R. J. Deschenes, and M. E. Linder. 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 280 31141–31148. [DOI] [PubMed] [Google Scholar]
- 27.Budde C., M. J. Schoenfish, M. E. Linder, and R. J. Deschenes. 2006. Purification and characterization of recombinant protein acyltransferases. Methods. 40 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smotrys J. E., M. J. Schoenfish, M. A. Stutz, and M. E. Linder. 2005. The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J. Cell Biol. 170 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osanai K., K. Takahashi, K. Nakamura, M. Takahashi, M. Ishigaki, T. Sakuma, H. Toga, T. Suzuki, and D. R. Voelker. 2005. Expression and characterization of Rab38, a new member of the Rab small G protein family. Biol. Chem. 386 143–153. [DOI] [PubMed] [Google Scholar]
- 30.Lobo S., W. K. Greentree, M. E. Linder, and R. J. Deschenes. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277 41268–41273. [DOI] [PubMed] [Google Scholar]
- 31.Bijlmakers M. J., M. Isobe-Nakamura, L. J. Ruddock, and M. Marsh. 1997. Intrinsic signals in the unique domain target p56(lck) to the plasma membrane independently of CD4. J. Cell Biol. 137 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner J. M., M. H. Brodsky, B. A. Irving, S. D. Levin, R. M. Perlmutter, and D. R. Littman. 1990. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 60 755–765. [DOI] [PubMed] [Google Scholar]
- 33.Resh M. D. 1994. Myristylation and palmitylation of Src family members: the fats of the matter. Cell. 76 411–413. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Hernando C., M. Fukata, P. N. Bernatchez, Y. Fukata, M. I. Lin, D. S. Bredt, and W. C. Sessa. 2006. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J. Cell Biol. 174 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma C., X. H. Yang, and M. E. Hemler. 2008. DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol. Biol. Cell. 19 3415–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond F. L., P. S. Tarpey, S. Edkins, C. Tofts, S. O'Meara, J. Teague, A. Butler, C. Stevens, S. Barthorpe, G. Buck, et al. 2007. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am. J. Hum. Genet. 80 982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.el-Aleem S. A., and H. Schulz. 1987. Evaluation of inhibitors of fatty acid oxidation in rat myocytes. Biochem. Pharmacol. 36 4307–4312. [DOI] [PubMed] [Google Scholar]
- 38.Fong J. C., S. J. Leu, and S. P. Chai. 1997. Differential inhibition of lipolysis by 2-bromopalmitic acid and 4-bromocrotonic acid in 3T3–L1 adipocytes. Biochim. Biophys. Acta. 1344 65–73. [DOI] [PubMed] [Google Scholar]
- 39.Raaka B. M., and J. M. Lowenstein. 1979. Inhibition of fatty acid oxidation by 2-bromooctanoate. Evidence for the enzymatic formation of 2-bromo-3-ketooctanoyl coenzyme A and the inhibition of 3-ketothiolase. J. Biol. Chem. 254 6755–6762. [PubMed] [Google Scholar]
- 40.Mikic I., S. Planey, J. Zhang, C. Ceballos, T. Seron, B. von Massenbach, R. Watson, S. Callaway, P. M. McDonough, J. H. Price, et al. 2006. A live cell, image-based approach to understanding the enzymology and pharmacology of 2-bromopalmitate and palmitoylation. Methods Enzymol. 414 150–187. [DOI] [PubMed] [Google Scholar]
- 41.Baron R. A., Y. K. Peterson, J. C. Otto, J. Rudolph, and P. J. Casey. 2007. Time-dependent inhibition of isoprenylcysteine carboxyl methyltransferase by indole-based small molecules. Biochemistry. 46 554–560. [DOI] [PubMed] [Google Scholar]
- 42.Hou H., K. Subramanian, T. J. LaGrassa, D. Markgraf, L. E. Dietrich, J. Urban, N. Decker, and C. Ungermann. 2005. The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc. Natl. Acad. Sci. USA. 102 17366–17371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson J. L., H. Frase, S. Michaelis, and C. A. Hrycyna. 2005. Purification, functional reconstitution, and characterization of the Saccharomyces cerevisiae isoprenylcysteine carboxylmethyltransferase Ste14p. J. Biol. Chem. 280 7336–7345. [DOI] [PubMed] [Google Scholar]
- 44.Roth A. F., J. Wan, A. O. Bailey, B. Sun, J. A. Kuchar, W. N. Green, B. S. Phinney, J. R. Yates 3rd, and N. G. Davis. 2006. Global analysis of protein palmitoylation in yeast. Cell. 125 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., M. S. Brown, G. Liang, N. V. Grishin, and J. L. Goldstein. 2008. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 132 387–396. [DOI] [PubMed] [Google Scholar]
- 46.Buglino J. A., and M. D. Resh. 2008. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of sonic hedgehog. J. Biol. Chem. 283 22076–22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.