Abstract
Immortalization of human bronchial epithelial (hBE) cells often entails loss of differentiation. Bmi-1 is a protooncogene that maintains stem cells, and its expression creates cell lines that recapitulate normal cell structure and function. We introduced Bmi-1 and the catalytic subunit of telomerase (hTERT) into three non-cystic fibrosis (CF) and three ΔF508 homozygous CF primary bronchial cell preparations. This treatment extended cell life span, although not as profoundly as viral oncogenes, and at passages 14 and 15, the new cell lines had a diploid karyotype. Ussing chamber analysis revealed variable transepithelial resistances, ranging from 200 to 1,200 Ω·cm2. In the non-CF cell lines, short-circuit currents were stimulated by forskolin and inhibited by CFTR(inh)-172 at levels mostly comparable to early passage primary cells. CF cell lines exhibited no forskolin-stimulated current and minimal CFTR(inh)-172 response. Amiloride-inhibitable and UTP-stimulated currents were present, but at lower and higher amplitudes than in primary cells, respectively. The cells exhibited a pseudostratified morphology, with prominent apical membrane polarization, few apoptotic bodies, numerous mucous secretory cells, and occasional ciliated cells. CF and non-CF cell lines produced similar levels of IL-8 at baseline and equally increased IL-8 secretion in response to IL-1β, TNF-α, and the Toll-like receptor 2 agonist Pam3Cys. Although they have lower growth potential and more fastidious growth requirements than viral oncogene transformed cells, Bmi-1/hTERT airway epithelial cell lines will be useful for several avenues of investigation and will help fill gaps currently hindering CF research and therapeutic development.
Keywords: Bmi-1, cystic fibrosis transmembrane conductance regulator, inflammation, karyotype, Ussing chamber
the airway epithelium is a crucial environmental interface that plays a key role in several respiratory diseases. In vitro models of this system are vital for research and development. Epithelial cells have been cultured from animal (18) and human (21) airways, either as outgrowths from explanted tissue fragments or as protease-dissociated cell suspensions (reviewed in Ref. 14). Primary human bronchial epithelial (hBE) cells on plastic in serum-free, supplemented medium can be grown for 10–15 passages, or even up to 30 passages in the presence of feeder cells (21). Under these conditions, the original cells revert to a poorly differentiated phenotype. However, when early passage hBE cells are grown in heterotopic tracheal grafts in vivo (37), in vitro as three-dimensional spheroids (19), within collagen gels (4), or, most commonly, on porous supports at an air-liquid interface (ALI) (41), the cells recapitulate many features of the native epithelium. The achievement of characteristic respiratory epithelial structure and function in ALI cultures underlies their utility for basic and applied studies of airway disease pathogenesis and treatment.
Practical obstacles inhibit use of ALI hBE cell cultures. Specimens from some diseases, for example, cystic fibrosis (CF), are frequently contaminated with difficult-to-eradicate antibiotic-resistant organisms (31). Only low-passage hBE cells will reliably grow and differentiate at an ALI, and the resulting epithelium can vary dramatically as a function of individual donor and culture stage. ALI cells are difficult to transfect with plasmids or infect with viral vectors, and commercial sources of the cells may be prohibitively expensive. These factors have driven the creation of hBE cell lines. Airway epithelial cell lines originate from human lung cancers (5, 12) and can be produced by chemical or physical mutagenesis of normal airway cells (36) or created by introduction of viral oncogenes, with (23, 43) or without (13, 25) cointroduction of human telomerase reverse transcriptase (hTERT). Cell lines found useful for CF research have been comprehensively reviewed (15).
The viral oncogenes most commonly used for immortalization are E6 and E7 from human papilloma virus (HPVE6/E7) and the early region from simian virus 40 (SV40ER). The viral gene products inhibit the pRb and p53 tumor suppressor pathways, effectors of growth arrest in normal, mortal cells as they age or become damaged (see Refs. 28, 39), and suppression of pRb and p53 increases cell replicative life span. HPVE6/E7 or SV40ER growth-enhanced human epithelial cells commonly undergo a crisis period, likely due to the inevitable shortening of chromosome ends in the absence of telomerase, and a subset of clones emerges from crisis when genomic instability activates hTERT. Cells surviving crisis often display myriad oligoclonal chromosomal abnormalities. The direct actions of the oncogenes and their indirect effect to create aneuploid cells influence cell behavior, especially the ability of the resulting cells to differentiate normally.
In many cell types, replicative senescence is triggered by chromosome shortening that can be prevented by hTERT expression. However, hTERT alone (23) does not immortalize hBE cells, which has been attributed to suboptimal culture conditions by some authors (29). In epidermal keratinocytes, mammary epithelium (20, 32), and hBE cells, hTERT-independent senescence entails induction of the cyclin-dependent kinase inhibitor p16Ink4a. The logical assumption is that cointroduction of hTERT with viral oncogenes would minimize crisis, decreasing genomic instability, and result in cells with fewer chromosomal abnormalities. Zabner et al. (43) created a series of hBE cell lines using both HPVE6/E7 and hTERT. These cells still had chromosomal abnormalities at passage 25, but at passage 18, five of six cell lines were able to develop transepithelial electrical resistance (Rt) and potential difference (PD) when assessed in Ussing chambers. Although viral oncogenes in combination with hTERT potently immortalize cells, the resulting lines often have limited differentiation capacity and other abnormalities, irrespective of whether they have undergone crisis. Thus viral oncogene-independent approaches for cell immortalization have been sought, including overexpression of the cyclin-dependent kinase 4, which competes for p16Ink4a (30), silencing of p16Ink4a mRNA, or expression of Bmi-1 (16).
The Bmi-1 locus in mice was identified as the B-cell Moloney murine leukemia retrovirus-specific integration site 1. When transgenic mice expressing the c-Myc oncogene driven by the immunoglobulin heavy chain enhancer were infected with this virus, leukemia formation was dramatically increased (17, 38), implicating Bmi-1 as a potent cooperating oncogene. Amplification and overexpression of Bmi-1 is associated with human mantle cell lymphoma (2). Bmi-1 is a highly conserved polycomb group protein with domains typical of transcriptional regulators, including a zinc finger motif. Genetic deletion of Bmi-1 in mice results in posterior transformation of the axial skeleton, and conversely, overexpression results in anterior transformation (1). A multiprotein complex that includes Bmi-1 regulates Hoxa9 and Meis1, genes that control homeobox and Hox gene expression (44). Bmi-1-deficient mice become growth restricted, with hematologic and neurological defects, which is consistent with Bmi-1 regulation of hematopoietic (22) and neural (20, 26) stem cells. Very recently, Bmi-1 was identified as an intestinal stem cell marker (35). Bmi-1 is thought to maintain stem cells by repressing the tumor suppressors p19ARF (p14ARF in humans) and p16Ink4a. A high percentage of human non-small cell lung cancers exhibit enhanced staining for Bmi-1, which inversely correlates with levels of p14ARF and p16Ink4a (6, 7, 40). Bmi-1 overexpression preserves stemness of hematopoietic progenitor cells (33), results in proliferation of insulin-producing cells from human pancreatic islets (34), and immortalizes human mammary epithelial cells (10). Significantly, SV40ER-immortalized human muscle satellite cells had multiple chromosomal aberrations and were phenotypically abnormal, whereas Bmi-1 and hTERT immortalization of the same cells yielded diploid, growth factor-dependent cells, variably maintaining their ability to differentiate into myotubes (9).
Our hypothesis was that Bmi-1 and hTERT expression would enhance the growth potential of hBE cells while preserving differentiation capacity. We infected three non-CF and three CF hBE cell preparations with lentiviral vectors expressing Bmi-1 and hTERT. All of the CF cells were homozygous for the common ΔF508 CFTR mutation. Bmi-1/hTERT expression extended cell life span, although not as profoundly as viral oncogenes. However, the cells at passage 15 recapitulated normal structure and function more closely than prior cell lines. Bmi-1/hTERT cell lines are a resource for diverse basic studies and will fill key gaps currently hindering CF research and therapeutic development.
MATERIALS AND METHODS
Cell culture.
Human lung tissue was procured under an Institutional Review Board-approved protocol, and hBE cell harvest and culture was performed using established procedures previously described in detail (11). Unless specified otherwise, reagents were obtained from Sigma-Aldrich (St. Louis, MO). Samples for cell line creation originated from three non-CF and three CF lungs, as described in Table 1. Cryopreserved passage 1 (P1) cells were cultured in bronchial epithelial growth medium (BEGM) on collagen type I/III-coated plastic dishes at a seeding density of 1 × 106 viable cells per 100-mm dish. Collagen coating of plastic dishes is used when plating freshly harvested P0 cells or cells from cryopreservation but is not needed after routine passaging (11). Beginning 48 h after plating, the cells were infected with human immunodeficiency virus (HIV)-1-based lentiviral vectors (described below) expressing Bmi-1 or hTERT or with a control vector expressing GFP or were sham infected as indicated, all in the presence of 2 μg/ml polybrene. To create the novel cell lines characterized in detail, P1 or P2 cells were infected with a 50:50 mixture of lentiviral vectors expressing Bmi-1 and hTERT for 3 h each on 2 consecutive days beginning 24–48 h after plating. After lentiviral or sham infection, the cells did not undergo selection and were repeatedly passaged on plastic to obtain growth curves. Between 70 and 90% confluence, cells were split 1:4 using trypsin-EDTA and conventional tissue culture procedures. Population doublings were calculated as the log base 2 of the ending cell number divided by the starting cell number, and growth curves were created by plotting the cumulative population doublings as a function of time.
Table 1.
Demographics of tissue donors
| Cell Line | Age, yr | Sex | Race | Smoking | Cause of Death or Diagnosis |
|---|---|---|---|---|---|
| UNCN1T | 47 | M | Cau | NS | Head injury |
| UNCN2T | 39 | F | Cau | 18 pack years | Cerebral hemorrhage |
| UNCN3T | 16 | F | Cau | NS | Head injury |
| UNCCF1T | 33 | M | Cau | NS | CF, ΔF508/ΔF508 |
| UNCCF2T | 18 | F | Cau | NS | CF, ΔF508/ΔF508 |
| UNCCF3T | 54 | F | Cau | NS | CF, ΔF508/ΔF508 |
M, male; F, female; Cau, Caucasian; NS, nonsmoker. Non-cystic fibrosis (CF) lungs were obtained from organ donors whose lungs were unsuitable for transplant due to acute injury or lack of a matching recipient. CF lung tissues were obtained at transplantation; the CF patient genotype was obtained from the medical record and was confirmed by a second round of PCR and sequencing of resulting CF cell lines.
ALI cultures.
At 70–90% confluence, uninfected P1 or P2 parental primary cells and each of the novel cell lines at P14–P16 were trypsinized, counted, and transferred to human placental type IV collagen-coated (no. C7521; Sigma, St. Louis, MO) 0.4-μm pore size Snapwell (Corning Costar, Cambridge, MA), T-Clear (Corning Costar), or Millicell CM membranes (Millipore, Bedford, MA) in ALI medium. Cells were seeded at a density of 1 × 105 cells/cm2, ∼125,000 cells for a 10- to 12-mm support and 1 × 106 cells for a 24- to 30-mm support. Upon confluence, usually beginning at days 5–7, cells were maintained at an ALI. The apical surface was washed with PBS, and medium was replaced only in the basal compartment three times per week. Ussing chamber studies (described below) were performed using Snapwell inserts. The histology of cells grown on Millicell CM membranes at various times after plating was studied using formalin fixation and paraffin embedding. Cytokine production was assessed using cells grown on Millicell CM membranes. For all experiments, the minimum sample size was three replicate culture wells.
We note that Bmi-1/hTERT growth-enhanced cell lines described in this report must be seeded at >1 million viable cells per 100-mm plastic dish and passaged at no greater than 1:4 on plastic before creating ALI cultures. Furthermore, the passaged cells must be seeded at the specified high densities (or greater) on porous supports, especially at later passages, to create successful ALI cultures. The ability of the cells to maintain long-lasting ALI cultures is decreased in large-format wells (24–30 mm vs. 10–12 mm in diameter), since the cells ultimately form islands and retract, creating holes. For unknown reasons, the tendency to retract and form holes is more pronounced in the larger format cultures.
Viral vectors.
Plasmids allowing creation of HIV-1-based lentiviral vectors expressing mouse Bmi-1, hTERT, or green fluorescent protein (GFP) have been described previously (34). Vectors were produced by triple transfection of 293T cells with appropriate plasmids encoding the gene of interest (Bmi-1, hTERT, or GFP), HIV gag and pol, and the VSV-G env gene. Supernatants were filtered and titered by transducing 293T cells, followed by PCR to quantitate the copy number of the WPR cis-acting element per genome compared with reference cells. For lentiviral infection, hBE cells on plastic were directly exposed to virus-containing 293T cell-conditioned medium in the presence of 2 μg/ml polybrene, as described above.
Western blots and CFTR immunoprecipitation-Westerns.
Antibody against p16Ink4a was obtained from BD Pharmingen (San Diego, CA), anti Bmi-1 was obtained from Cell Signaling Technology (Danvers, MA), and anti-actin was obtained from Sigma. Further details of the Western blot procedure are given in the Supplemental Data. (Supplemental data for this article are available online at the American Journal of Physiology-Lung Cellular and Molecular Physiology website.) CFTR immunoprecipitation (IP)-Westerns were performed as previously described (8). Briefly, 12 × 12-mm well-differentiated ALI Millicell CM cultures were lysed in 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris·HCl, pH 7.4, and 10 mM NaMoO4 with protease inhibitors (1 μg/ml leupeptin, 2 μg/ml aprotinin, 50 μg/ml Pefabloc, 121 μg/ml benzamidine, and 3.5 μg/ml E64). CFTR was immobilized using mouse anti-CFTR monoclonal antibody (MAb) 596 cross-linked to protein G Dynabeads (Invitrogen, Carlsbad, CA). Beads were washed three times with lysis buffer, and proteins were eluted, separated by SDS-PAGE, and probed by Western blotting, again using MAb 596.
Karyotyping.
P14 and P15 Bmi-1/hTERT cell lines on plastic at 30–90% confluence were treated with Karyomax Colcemid solution (Invitrogen) at 0.15 mg/ml for 16–26 h. Parallel experiments were performed using AALEB cells, a previously described SV40ER/hTERT cell line (23), except that this rapidly growing cell was only incubated in colchicine solution for 3.5 h. Karyotyping was performed as described in the Supplemental Data. At least 20 chromosome spreads were analyzed using a ×100 oil-immersion objective on a Nikon Microphot microscope, and two representative images were captured for each cell type.
Bioelectric properties of parental cells and cell lines.
Between 18 and 24 days after plating, P2 parental cells and P14–P16 derivative cell lines grown at an ALI on collagen type IV-coated Snapwell inserts were mounted in Ussing chambers (Physiologic Instruments, San Diego, CA). The epithelium was voltage clamped, and short-circuit current (Isc) and Rt were measured (Physiologic Instruments). Data were analyzed using Acquire and Analysis (version 1.2) software (Physiologic Instruments). The solution on both sides of the membrane was 5 ml of 37°C Krebs-bicarbonate-Ringer containing (in mM): 140 Na+, 120 Cl−, 5.2 K+, 1.2 Ca2+, 1.2 Mg2+, 2.4 HPO42−, 0.4 H2PO4−, 25 HCO3−, and 5 glucose, circulated with 95% O2-5% CO2 gas, pH 7.4. Compounds were added, in order, as indicated: amiloride, 100 μM, mucosal; forskolin, 25 μM, mucosal and serosal; CFTR(inh)-172, 10 μM, mucosal; and UTP, 100 μM, mucosal.
DNA fingerprinting, confirmation of CFTR genotype, and mycoplasma testing by PCR.
Cellular genomic DNA was extracted using SDS-spermidine-proteinase K digestion and potassium acetate precipitation and was quantitated using spectrophotometry. To determine that cell lines had not become cross-contaminated with each other or by another cell type during viral infection, expansion, and passage, we performed DNA fingerprinting on P14 and P15 cells using the PowerPlex 1.2 system (Promega, Madison, WI), which enables detection of eight polymorphic tetranucleotide repeat loci, plus amelogenin, as described in the Supplemental Data.
We performed PCR to confirm the CF tissue donor genotype. Briefly, exon 10 of CFTR in genomic DNA prepared as described above was amplified using the following primers: forward 5′-GCA GAG TAC CTG AAA CAG GA-3′ and reverse 5′-CAT TCA CAG TAG CTT ACC CA-3′. PCR products were purified using QIAquick PCR purification columns (Qiagen,Valencia, CA), and sequencing was performed using the primer 5′-TAA TGG CGA GGC AAG TGA ATC CTG AG-3′ in conjunction with the BigDye Terminator version 1.1 cycle sequencing kit (Applied Biosystems). Samples were run on an ABI Prism 310 genetic analyzer, and the sequence was deduced from electrophoretograms.
To evaluate whether cell lines were contaminated with mycoplasma, DNA was extracted from cell lines at P12–P15, using a QiaAmp mini DNA kit (Qiagen), and analyzed using the MycoFind version 1.0 mycoplasma detection PCR reaction kit (Clongen, Germantown, MD) according to the manufacturer's instructions.
Cell stimulation and IL-8 measurement.
Cell lines at P14 and P15 were grown at an ALI in low-endotoxin medium (3) on collagen type IV-coated Millicell CM membranes for 21–24 days. The apical surface was washed twice with PBS, and the inserts were transferred to individual wells of a 12-well plate with 0.8 ml of medium on the basolateral side. Twenty-four hours later, the apical side was washed again, basolateral medium was changed, and the cultures were challenged both apically (in 50 μl of ALI medium) and basolaterally (in 800 μl of ALI medium) with IL-1β (5 ng/ml; R&D Systems, Minneapolis, MN), TNF-α (20 ng/ml; R&D Systems), and the Toll-like receptor 2 agonist Pam3Cys-Ser-(Lys)4 (Pam3Cys; 25 μg/ml; EMD Microcollections, Tubingen, Germany). Twenty-four hours later, the basolateral conditioned media was sampled and the apical surface was washed twice with 200 μl of PBS, pooled, and centrifuged, and the supernatant was collected. Aliquots of apical and basolateral media were stored frozen at −20°C. The IL-8 concentration in apical and basolateral fluids from each well was assayed in duplicate using ELISA (R&D Systems; sensitivity of 30 pg/ml). When IL-8 standard was spiked into representative samples, the results were additive (data not shown).
RESULTS AND DISCUSSION
SV40ER/hTERT immortalized hBE cells create poor ALI cultures.
Great need and limited supply of primary hBE cells coupled with high variability, expense, and shortcomings of methods for experimental manipulation of primary cultures has stimulated the creation of cognate cell lines. We initially used HPVE6/E7 or SV40ER in combination with hTERT to create airway epithelial cell lines (Refs. 23, 27, and unpublished data). In our experience, cells immortalized with HPVE6/E7 grew rapidly but were relatively poor at forming tight hydrostatic and electrical resistance barriers in ALI cultures compared with SV40ER cells (data not shown). Thus we discontinued using HPVE6/E7 for cells intended for ALI cultures. Many features of SV40ER/hTERT immortalized airway epithelial cells have been previously reported (23), and a non-CF hBE cell line, designated AALEB, when grown on plastic, has proved very useful for studies of innate immunity (42). We also created a corresponding ΔF508 homozygous CF hBE cell line, designated KKLEB. We examined the long-term growth of AALEB and KKLEB on plastic as well as their morphological and electrophysiological properties in ALI culture at early and late passage (Supplemental Data, Fig. S1). AALEB and KKLEB grew continuously and robustly when cultured on plastic (Fig. S1A). When grown at an ALI, the cells formed a multilayered hyperplastic, abnormal epithelium that resembled carcinoma in situ. Cuboidal, apically oriented cells appeared to polarize but did not form mucous secretory or ciliated cells (Fig. S1B). When tested in Ussing chambers at low passage, the cells had Rt values >150 Ω·cm2 and had small but measurable Isc and PD values but minimal amiloride responses (Fig. S1C). At later passages, cultures of both cell types displayed progressively variable and abnormal morphology and irreproducible electrophysiological behavior, failing to consistently create high-resistance membranes. Despite a considerable investment in their creation and testing, we realized that AALEB and KKLEB cells were unsuitable as in vitro model for studying many differentiated hBE cell functions.
Decreased Bmi-1 and elevated p16Ink4a protein content precedes senescence in primary hBE cells.
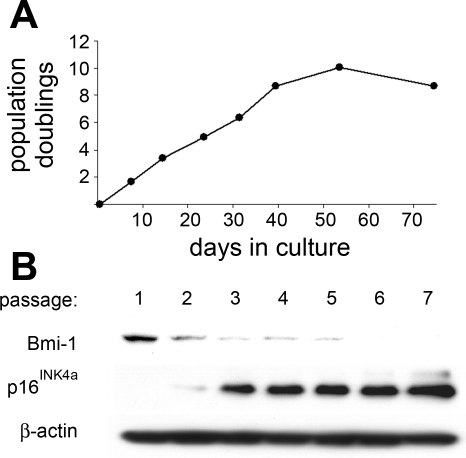
Dysregulation of the cyclin-dependent kinase inhibitor p16Ink4a is a common event in lung cancer. Despite the well-known role of p16Ink4a in growth arrest of normal cells, its expression during the life span of normal hBE cells under various growth conditions is not well documented. Since p16Ink4a expression is reported to be dependent on culture conditions (29) and also suppressed by the Bmi-1 protooncogene (16), we performed Western blots for p16Ink4a and Bmi-1 protein during progression of the hBE cell growth curve under our conditions by using cells from three lung specimens. We found that Bmi-1 protein expression was highest in very early passage cells and that enhanced p16Ink4a expression preceded the development of hBE cellular senescence, defined as no increase in cell number within 21 days (Fig. 1). These results agree with those of Haga et al. (16) using different media and suggest that suppression or bypass of p16Ink4a is necessary to extend the life span of hBE cells as grown under the conditions used in our laboratory.
Fig. 1.
Decreased Bmi-1 and elevated p16Ink4a protein content precedes senescence in primary human bronchial epithelial (hBE) cells. A: growth curve from a representative hBE preparation. B: corresponding Western blots for Bmi-1, p16Ink4a, and actin. The actin blot represents reprobing of the p16Ink4a blot. Similar results were obtained in 2 additional specimens.
Bmi-1 alone extends the life span of hBE cells, but not indefinitely in most specimens.
The protooncogene Bmi-1 suppresses p16Ink4a expression and extends the life span of many cell types. We infected three non-CF and three ΔF508 homozygous CF hBE cell preparations, derived from the tissue donors described in Table 1, with Bmi-1- or GFP-expressing HIV-1-based lentiviral vectors or sham controls and performed growth curves (Supplemental Data, Fig. S2). All control cell types (sham or GFP vector) senesced between P2 and P16, before 25 population doublings. Bmi-1 vector alone significantly enhanced the growth of all six of the cell lines, but four of six senesced between P21 and P24. Consistent with the observation that cell lines created with Bmi-1 did not express significant telomerase activity (data not shown), we hypothesized that the senescence of cells whose growth was enhanced by Bmi-1 alone was due to telomere shortening. To test this hypothesis, we subjected Bmi-1 cell lines to sham infection or hTERT-expressing HIV-1-based lentiviral vector approximately five passages before their predicted senescence. As shown in the Supplemental Data, Fig. S3, hTERT further enhanced the growth of Bmi-1 cell lines. Thus, although addition of Bmi-1 alone enables a significant expansion in cell number, a proportion of the new cell lines become senescent when repeatedly passaged on plastic in the absence of hTERT. Previous reports found that Bmi-1 activated hTERT transcription and induced telomerase activity in mammary epithelial cells (10), but a subsequent study found only weak telomerase activation by Bmi-1 in mammary cells and no activation in human dermal keratinocytes or human small airway epithelial cells (16). The most straightforward interpretation of our results is that hBE cells expressing Bmi-1 in the absence of hTERT were growth enhanced, but extended culture variably decreased telomere length to the point of triggering senescence. Since addition of hTERT to late-passage cells may enable propagation of prior genetic damage, we infected low-passage hBE cells with both Bmi-1 and hTERT as described below.
Bmi-1 plus hTERT extends the life span of hBE cells, but not as potently as viral oncogenes.
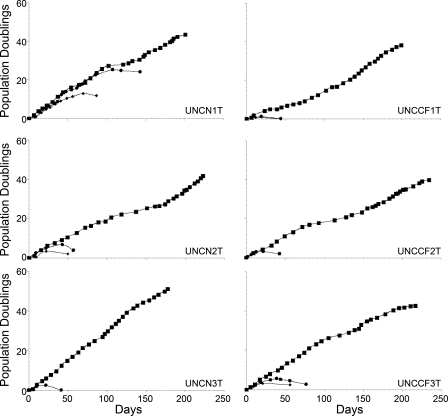
We thawed primary cell aliquots of all six hBE specimens described in Table 1 and infected P1 or P2 cells with both Bmi-1 and hTERT. For increased efficiency, we infected the cells twice on consecutive days with both lentiviruses. The three non-CF and three CF cell lines were named UNCN1T, UNCN2T, UNCN3T and UNCCF1T, UNCCF2T, UNCCF3T, respectively. We chose viral constructs that did not express selection markers so that all resulting cell lines would remain pan-sensitive to selection agents, facilitating later studies. All cells infected with control vectors (sham infection or GFP vector) senesced before 25 population doublings, but Bmi-1- and hTERT-transduced cells grew for at least 38 population doublings, and all cells displayed no signs of senescence at P30 (Fig. 2). In our experience, the Bmi-1 plus hTERT cells do not grow as rapidly as viral oncogene/hTERT immortalized cells (compare with Supplemental Data, Fig. S1). They grew slowly if passaged more than 1:4 and were sensitive to the growth factor content of BEGM (data not shown). There are subtle differences between the different Bmi-1 plus hTERT cell lines; for example, the growth curve of UNCN3T is shifted somewhat upward and to the left compared with that of the others, indicative of slightly faster growth. There are inflections in some of the growth curves, which may represent husbandry changes over the extended study period, but the cells apparently did not go through a crisis period. These results are consistent with prior reports of extension of replicative life span of several human epithelial cell types by Bmi-1 plus hTERT (9, 16, 34).
Fig. 2.
Bmi-1 plus human telomerase reverse transcriptase (hTERT) extends the growth capacity of hBE cells. Growth curves of UNCN1T, UNCN2T, UNCN3T, UNCCF1T, UNCCF2T, and UNCCF3T cell lines on plastic were recorded in sham-infected cells (•) or cells infected with green fluorescent protein (GFP) lentivirus (⧫) or the combination of Bmi-1 and hTERT lentivirus (▪) as described in materials and methods. The addition of Bmi-1 plus hTERT enabled at least 38 population doublings, whereas all controls senesced before a maximum of 25 doublings. All Bmi-1 plus hTERT cell lines have grown for at least 30 passages.
To verify that the new cell lines remained uncontaminated with each other or by another cell line during viral infection, expansion, and passage, we performed DNA fingerprinting on the cell lines at P14 and P15. The cell type-specific genotypes of eight polymorphic tetranucleotide repeat loci plus amelogenin were unique in each cell type (Supplemental Data, Table S4). The CFTR genotype was confirmed to be ΔF508 homozygous in all three of the CF cell lines (data not shown). Mycoplasma DNA was undetectable in the cells between P12 and P16 (Supplemental Data, Fig. S5).
Unlike viral oncogene immortalized cells, Bmi-1 plus hTERT hBE cells are diploid at P14 and P15.
Many cell lines are aneuploid and genetically unstable, but Bmi-1 immortalized cells are reported to behave more like normal cells (9). We analyzed the karyotype of the SV40ER/hTERT immortalized cell line AALEB (23) at P13 and of the six novel cell hBE lines created with Bmi-1/hTERT at P14 and P15 (Table 2). In AALEB, no two metaphase spreads had the same chromosome complement, varying from 85 to 92 in number and displaying rearrangements, telomeric associations, and dicentrics: chromosomes 4, 8, 11, 14, and 20 were most frequently affected. UNCN1T, UNCN3T, UNCCF2T, and UNCCF3T were all found to have normal chromosome complements in the majority of cells analyzed, a result similar to what would be expected in primary cell cultures. UNCN2T and UNCCF1T had chromosome abnormalities in 32 and 75% of the cells analyzed, respectively. There were several different abnormalities in UNCN2T, whereas UNCCF1T had only one aberration, a consistent translocation involving chromosomes 2 and 6. It is possible that higher percentages of early passage UNCN2T and UNCCF1T cells have a normal chromosome complement. In summary, although AALEB was created by sequentially adding SV40ER and hTERT at early passage, by P13 the cells were aneuploid and presumably unstable and progressively variable, as indicated by dicentric chromosomes. Bmi-1 plus hTERT hBE cells were diploid, and although there were sporadic chromosomal abnormalities that were propagated in two of six cell lines, the cells appeared to be relatively stable genetically. On the basis of these results, we predict that Bmi-1 plus hTERT hBE cells will be less susceptible than viral oncogene immortalized cells to spontaneous genetic change over time and will be more suitable for purposeful genetic manipulation and subsequent experimental analysis of sublines. The combination of genetic stability, extended growth capacity, and selection agent sensitivity in Bmi-1 plus hTERT hBE cell lines will facilitate experimental approaches including conditional overexpression of normal or mutant proteins and/or short-hairpin RNA gene silencing. These cells can be infected with viral gene transfer vectors and selected, yet will still retain adequate growth capacity for subpassage to conditions enabling differentiation and testing of cell type-specific functions.
Table 2.
Karyotype of AALEB and Bmi-1/hTERT immortalized cell lines
| Cell Line | Karyotype |
|---|---|
| AALEB (P13) | 46, X or XX with additions/deletions of parts of chromosomes 4, 8, 14, and 20, dicentrics and telomeric associations in 9 spreads |
| Counts of 85–92 with rearrangements, telomeric associations and dicentrics in 7 spreads | |
| 44, XX, −4, −8, add(20q) in 1 spread | |
| 45, X with add(20q) and telomeric association plus other rearrangements in 1 spread | |
| 45, XX, −14, del(10q), del(11q), add(16p), add(20q) in 1 spread | |
| 47, XX, +6 with other rearrangements in 1 spread | |
| UNCN1T (P14) | 46, XY normal male karyotype in 16 spreads |
| 45, X in 2 spreads | |
| 45, XY with random losses in 2 spreads | |
| UNCN2T (P15) | 46, XX normal female karyotype in 15 spreads |
| 46, XX, t(2p;10q) in 2 spreads | |
| 47, XX,+20 in 2 spreads | |
| 47, XX, +7 in 2 spreads | |
| 46, XX, del(Xq) in 1 spread | |
| UNCN3T (P14) | 46, XX normal female karyotype in 19 spreads |
| 45, XX, −19 in 1 spread | |
| UNCCF1T (P14) | 46, XY, t(2p;6p) in 15 spreads |
| 46, XY normal male karyotype in 5 spreads | |
| UNCCF2T (P14) | 46, XX normal female karyotype in 18 spreads |
| 46, XX, del(13p) in 1 spread | |
| 47, XX, +9 in 1 spread | |
| UNCCF3T (P14) | 46, XX normal female karyotype in 18 spreads |
| 46, XX, del(11q) in 1 spread | |
| 45, X in 1 spread |
Karyotypes of a non-CF cell line immortalized with the early region from simian virus 40 (SV40ER) and the human telomerase reverse transcriptase (hTERT) (AALEB cell line; P13) and non-CF and CF cell lines (P14 and P15) created with Bmi-1/hTERT are indicated. P, passage.
We made no attempt to derive clones of Bmi-1/hTERT hBE cells. Because of vector efficiency and lack of selection, the cells at early passage are very likely polyclonal populations of growth-enhanced cells. We speculate that the populations may become more highly represented by faster growing cells and potentially become oligoclonal at later passages.
Cell lines created with Bmi-1 plus hTERT formed ALI cultures suitable for Ussing chamber studies at P14–P16.
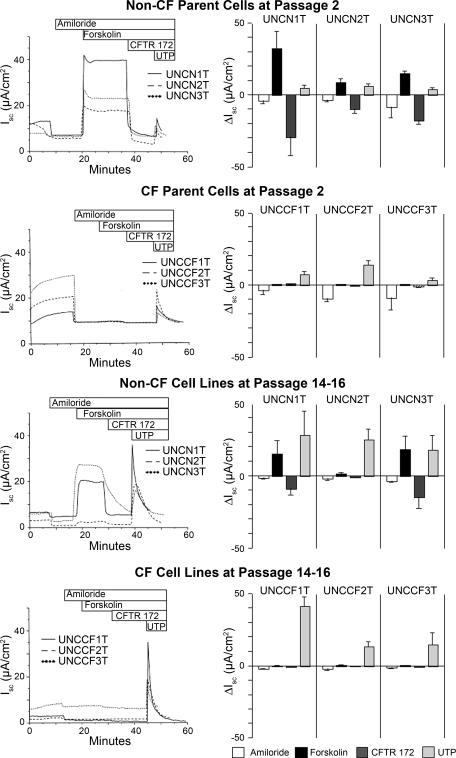
When studied in Ussing chambers, CF airway epithelial cells do not efflux anions across the apical plasma membrane in response to increased cellular cAMP. The restoration of cAMP-stimulated anion currents is thus a key functional test of therapeutic strategies to treat CF. We studied P2 primary cells of all six parental cell lines and their Bmi-1 plus hTERT growth-extended derivatives at P14–P16 in Ussing chambers. All cells were seeded on human collagen type IV-coated Snapwell inserts at designated seeding densities, and medium was changed regularly. We noted in later passage cultures, or if low seeding densities were used on larger porous supports, that the cells would form islands and would take an extended period to become confluent. However, all cells that were maintained and seeded under the conditions specified grew to confluence and maintained a patent ALI. Mean baseline resistance (R), PD, and Isc values are given in Table 3. Representative Ussing chamber traces and changes in Isc in response to amiloride, forskolin, CFTR(inh)-172, and UTP are shown in Fig. 3. The growth-enhanced cells exhibited variable Rt values ranging from ∼200 to 1,200 Ω·cm2. Isc responses stimulated by forskolin and inhibited by CFTR(inh)-172 were true to the cell's genotype, with negligible “residual” forskolin-stimulated currents in CF cells, although there was a small CFTR(inh)-172 response, as in the parent cells. Forskolin responses were comparable to early passage primary cells, except for one non-CF cell line (UNCN2T) with relatively low CFTR Isc values at P15. Interestingly, the parent of UNCN2T had the lowest forskolin-stimulated current among the three non-CF tissue donors. The reason that UNCN2T displays low CFTR currents is unknown but may relate to the smoking history (Table 1) or chromosomal changes (described above) or morphologic appearance (described below). Ethical concerns prohibit us from genotyping CFTR in cells from people not carrying a CF diagnosis, so we cannot determine whether the UNCN2T parental cells were originally heterozygous for CFTR mutations or whether the cell lines have acquired one. Amiloride-sensitive currents were not different between CF and non-CF cells but tended to be lower in the growth-enhanced cells compared with the parental cells. UTP-stimulated Isc of variable magnitude were present and were not different between CF and non-CF cells but were somewhat higher in growth-enhanced cells compared with the parental cells.
Table 3.
Basal electrophysiological characteristics of non-CF and CF parent cells at P2 and derivative cell lines at P14–P16 as measured in Ussing chambers
| Cell | Isc, μA/cm2 | PD, mV | Rt, Ω·cm2 |
|---|---|---|---|
| UNCN1T-parent | 11.0±4.7 | −5.5±3.3 | 401±134 |
| UNCN2T-parent | 7.9±3.8 | −3.6±4.0 | 283±262 |
| UNCN3T-parent | 9.9±2.0 | −7.1±3.9 | 598±271 |
| UNCCF1T-parent | 8.0±3.7 | −6.8±6.6 | 1075±803 |
| UNCCF2T-parent | 15.6±1.8 | −13.2±5.3 | 2583±932 |
| UNCCF3T-parent | 13.2±9.3 | −8.1±7.8 | 1187±776 |
| UNCN1T | 5.4±1.3 | −2.0±1.3 | 522±578 |
| UNCN2T | 4.3±1.8 | −3.5±1.7 | 748±274 |
| UNCN3T | 3.0±5.3 | −0.7±1.1 | 195±72 |
| UNCCF1T | 2.6±0.7 | −0.6±0.2 | 317±96 |
| UNCCF2T | 2.8±3.0 | −0.9±0.5 | 511±429 |
| UNCCF3T | 7.9±0.6 | −10.4±4.0 | 1210±330 |
Isc, short-circuit current; PD, potential difference; Rt = transepithelial resistance. Data are means ± SD; n = 3–14 wells per cell type.
Fig. 3.
Ussing chamber studies of parental primary cells and each of the novel cell lines. Representative Ussing chamber traces and changes in short-circuit current (Isc) are shown in response to amiloride, forskolin, CFTR(inh)-172, and UTP in passage 2 (P2) primary cells of all 6 parental cell lines and their Bmi-1 plus hTERT growth-extended derivatives at P14–P16. Each data point represents the mean ± SD of between 3 and 14 individual 12-mm Snapwell inserts per cell type, cultured for 18–24 days. CF, cystic fibrosis.
There was substantial variability in electrophysiological properties (e.g., Rt) of the growth-enhanced cell lines. Similar variability was present in the parental cultures and was also found previously in P2 primary hBE cells under similar conditions (3). The data in this report are completely nonselected, and unless there was an obvious technical failure, no samples were excluded from the Ussing chamber analysis. Resistances tended to be higher in parent CF cells, which is consistent with a missing pathway for ion transport. Beyond the effects of CF vs. non-CF genotype, the variability is likely a function of person-to-person genetic and epigenetic differences, tissue specimen handling, growth capacity of the primary cells when first harvested, culture stage, and other subtle factors, which may be amplified in the cell lines, especially at later passages. Absence of functioning CFTR is reported to upregulate the epithelial sodium channel (ENaC). For unknown reasons, and as reported previously (3), ENaC upregulation is not consistently preserved in CF hBE cells in the robust BEGM-ALI system. The lack of relative ENaC hyperactivity in CF primary, and especially Bmi-1/hTERT hBE, cell lines may relate to the relatively low levels of amiloride-sensitive current. Because primary cells can typically only form ALI cultures suitable for Ussing chambers at low passages, growth enhancement with Bmi-1 plus hTERT will very significantly increase the number of hBE cells suitable for electrophysiological studies of CFTR, especially for critical tests of strategies that enhance ΔF508 CFTR transit to the plasma membrane and/or Cl− transport function.
Western blots and CFTR IP-Westerns.
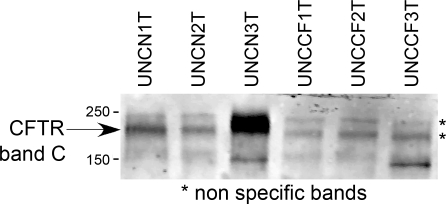
To determine whether differences in CFTR protein expression correlated with differences in forskolin-stimulated currents seen in the Bmi-1/hTERT hBE cell lines, we performed CFTR IP-Westerns. Based on previous experience with primary hBE cell cultures, we anticipated that detection of endogenous levels of CFTR would be difficult and variable compared with high-expressing cell lines or even freshly excised tissues. We harvested cellular proteins from 12 × 12-mm well-differentiated Millicell CM cultures of each cell line, performed IP as described in materials and methods, and applied the entire eluate to a single lane on a Western blot. As shown in Fig. 4, the strongest visible bands, putatively correlating with CFTR band C, were present in lines UNCN1T and UNCN3T, and a very faint band was present in line UNCN2T, which agrees with the electrophysiological results in Fig. 3. Presumably due to the large amount of input protein required, nonspecific bands with slightly different mobilities from CFTR band C were present in all lanes. Although there were suggestive bands that may have correlated with CFTR band B, the results were not consistent among different samples, and it was not possible to draw conclusions related to precursor forms of the CFTR protein. Although there was reasonable correlation between CFTR band C protein expression and function, these results illustrate the difficulty of CFTR detection at endogenous levels of expression and highlight the relatively greater sensitivity of the electrophysiological techniques.
Fig. 4.
CFTR protein detection in Bmi-1 plus hTERT cell lines. Immunoprecipitation (IP)-Western blot analysis was performed using lysates of 12 × 12-mm Millicell CM inserts per lane, beads coupled to monoclonal antibody (MAb) 596, and probing of the Western blot with MAb 596. The strongest putative CFTR band C signals originated from UNCN1T and UNCN3T cells, the same 2 cell lines with the greatest forskolin-stimulated and CFTR(inh)-172-inhibitable currents in Ussing chambers. We assume that the bands marked with asterisks are nonspecific and are due to the necessarily high levels of input protein.
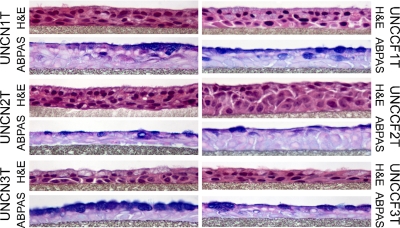
Cell lines created with Bmi-1 plus hTERT exhibit a pseudostratified morphology at P14–P16.
We performed routine histological analysis of parental cell lines (data not shown) and growth-enhanced cell lines in ALI culture. The cell lines underwent time-dependent changes in differentiation similarly to primary cells. Early cultures were a homogeneous population of proliferative, squamoid cells (data not shown) that ultimately polarized and differentiated. Representative histology examples after 28 days of culture on porous supports are shown in Fig. 5. All of the cell lines formed a pseudostratified columnar epithelium with distinct basal cells and abundant mucous secretory (goblet) cells but few ciliated cells. Apical polarization of luminal cells was clearly visible. The cells exhibited a more normal nuclear morphology and fewer apoptotic bodies than similar preparations of cells immortalized using viral oncogenes (for example, compare with Fig. S1). In some of the cell lines, for example, UNCN3T, almost all of the apically oriented cells in day 28 ALI cultures were mucous secretory cells. We anticipate that these cell lines will be very useful for understanding events controlling mucous secretory cell differentiation and function. The UNCN2T cultures appeared thicker in cross section and less systematically polarized, which may contribute to the relatively lower CFTR Isc values noted in Ussing chambers. The subtle differences in cell morphology between the different cell lines may be influenced by the spectrum of growth and differentiation potentials of the polyclonal, and possibly oligoclonal, progenitor cell populations.
Fig. 5.
Bmi-1 plus hTERT cell lines at P14–P16 exhibit a pseudostratified morphology. Representative photomicrographs are shown of histology sections from day 28 air-liquid interface (ALI) cell cultures of all 6 novel Bmi-1/hTERT growth-enhanced hBE cell lines. Note the sparse ciliated cells and abundant mucous secretory cells, routine formalin-fixed paraffin sections, hematoxylin and eosin (H&E) and alcian blue-periodic acid-Schiff (ABPAS) stain (original magnification, ×400). Sections are representative of at least 3 individual cultures for each cell line.
Ciliated cells were infrequent but increased somewhat in cultures maintained for longer periods, up to 35 days (data not shown). The precise reason why ciliated cell abundance is lower in Bmi-1/hTERT hBE cells than in corresponding P2 primary cells requires further study but may relate to the effects of Bmi-1 on the complex process of ciliated cell differentiation. Since loxP sites flank the Bmi-1 and hTERT coding sequences (34), it is possible to excise introduced genes using Cre recombinase. In an effort to increase ciliated cell number, we exposed several of the new cell lines on plastic to an adenovirus expressing Cre recombinase (Ad Cre) with the intent to subpassage these cells to ALI. Unfortunately, Ad Cre infection of cells whose growth capacity was already extended beyond their normal life expectancy resulted in widespread cell death, precluding this approach toward increasing ciliated cell abundance.
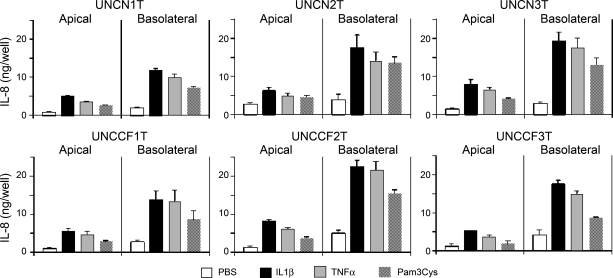
No systematic differences in IL-8 production between non-CF and CF cell lines at baseline or following proinflammatory stimulation.
We studied IL-8 secretion, as measured by ELISA, into the apical and basolateral culture compartments by the six cell lines created with Bmi-1 plus hTERT (Fig. 6). The range of IL-8 production at baseline or after stimulation with IL-1β, TNF-α, or Pam3Cys (Toll-like receptor 2 ligand) varied among the cell lines and was typical of human-to-human variability documented previously (3). There was no systematic difference in IL-8 production between CF and non-CF cells. There is a great deal of controversy regarding the presence of an “intrinsic hyperinflammatory” state in CF cells (reviewed in Ref. 24). The reasons for the discrepant results in the literature are unclear, but our data suggest that absence of functioning CFTR does not necessarily cause activation of NF-κB, a key transcriptional regulator of IL-8, and enhanced production of proinflammatory cytokines. However, the induction of IL-8 secretion by diverse proinflammatory mediators indicates that relevant signal transduction pathways are intact and predicts usefulness of the cell lines for studies related to airway inflammation.
Fig. 6.
No systematic differences in IL-8 production between non-CF and CF cell lines. IL-8 content in apical washings and basolateral conditioned media was measured 24 h after IL-1β, TNF-α, or Pam3Cys (Toll-like receptor 2 ligand) challenge of well-differentiated ALI cultures of all 6 Bmi-1/hTERT novel hBE cell lines. Results are expressed as the total amount of IL-8 secreted into the apical and basolateral culture compartments over a 24-h period and are means ± SE of quadruplicate wells.
In conclusion, we created three CF (homozygous for the ΔF508 CFTR mutation) and three non-CF novel hBE cell lines by HIV-1-based lentiviral expression of the protooncogene Bmi-1 and hTERT. The cells grew for at least 38 population doublings under the conditions described. At P14 to P15, the cells remained diploid and were genetically stable compared with cells immortalized with viral oncogenes. At P14 to P15, when propagated and grown on porous supports under the specified conditions, the cells established a confluent cell layer whose electrophysiological properties in Ussing chambers were mainly true to the parent cell genotype. The ALI cultures at days 28–35 displayed a pseudostratified morphology with abundant mucous secretory cells. Preservation of relatively normal structure and function is paralleled by relatively slow and limited growth compared with typical cell lines created with viral oncogenes. The novel cells are similar to primary hBE cells regarding sensitivity to adequate culture seeding densities, substrate and media quality, and husbandry requirements, especially at later passages. Nevertheless, Bmi-1/hTERT growth-enhanced hBE cells will be useful for many lines of research relevant to airway diseases and will specifically help fill gaps currently hindering CF therapeutic development.
GRANTS
This work was supported by Cystic Fibrosis Foundation Grant Randel04G0 and National Heart, Lung, and Blood Institute Grant HL080322.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Hirsh for early Ussing chamber studies of AA and KKLEB cells; Dr. William Hahn for checking telomerase activity in “Bmi-1 only” cells; Dr. Didier Trono for generous access to lentiviral vector technology and advice; Jeffrey Sailus and Dr. Jessica Booker for DNA fingerprinting; Rhonda Pace and Dr. Michael Knowles for confirmatory genotyping of CF cells; Dr. Jack Riordan for anti-CFTR antibodies and advice regarding IP-Westerns; Jennifer Becker, Anna Krause, Victoria Sefcik, and Tarra Wasilchen for technical assistance; the University of North Carolina Cystic Fibrosis Center Histology Core for embedding, sectioning, and staining of cultures; and Lisa Brown for editing assistance.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Alkema MJ, van der Lugt NM, Bobeldijk RC, Berns A, van Lohuizen M. Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature 374: 724–727, 1995. [DOI] [PubMed] [Google Scholar]
- 2.Bea S, Tort F, Pinyol M, Puig X, Hernandez L, Hernandez S, Fernandez PL, van Lohuizen M, Colomer D, Campo E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res 61: 2409–2412, 2001. [PubMed] [Google Scholar]
- 3.Becker MN, Sauer MS, Muhlebach MS, Hirsh AJ, Wu Q, Verghese MW, Randell SH. Cytokine secretion by cystic fibrosis airway epithelial cells. Am J Respir Crit Care Med 169: 645–653, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Benali R, Tournier JM, Chevillard M, Zahm JM, Klossek JM, Hinnrasky J, Gaillard D, Maquart FX, Puchelle E. Tubule formation by human surface respiratory epithelial cells cultured in a three-dimensional collagen lattice. Am J Physiol Lung Cell Mol Physiol 264: L183–L192, 1993. [DOI] [PubMed] [Google Scholar]
- 5.Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am J Respir Crit Care Med 157: 2000–2006, 1998. [DOI] [PubMed] [Google Scholar]
- 6.Breuer RH, Snijders PJ, Smit EF, Sutedja TG, Sewalt RG, Otte AP, van Kemenade FJ, Postmus PE, Meijer CJ, Raaphorst FM. Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia 6: 736–743, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breuer RHJ, Snijders PJF, Sutedja GT, Sewalt RGAB, Otte AP, Postmus PE, Meijer CJLM, Raaphorst FM, Smit EF. Expression of the p16INK4a gene product, methylation of the p16INK4a promoter region and expression of the polycomb-group gene BMI-1 in squamous cell lung carcinoma and premalignant endobronchial lesions. Lung Cancer 48: 299–306, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Chang XB, Mengos A, Hou YX, Cui L, Jensen TJ, Aleksandrov A, Riordan JR, Gentzsch M. Role of N-linked oligosaccharides in the biosynthetic processing of the cystic fibrosis membrane conductance regulator. J Cell Sci 121: 2814–2823, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cudre-Mauroux C, Occhiodoro T, Konig S, Salmon P, Bernheim L, Trono D. Lentivector-mediated transfer of Bmi-1 and telomerase in muscle satellite cells yields a Duchenne myoblast cell line with long-term genotypic and phenotypic stability. Hum Gene Ther 14: 1525–1533, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, van Lohuizen M, Campisi J, Wazer DE, Band V. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res 62: 4736–4745, 2002. [PubMed] [Google Scholar]
- 11.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107: 183–206, 2004. [DOI] [PubMed] [Google Scholar]
- 12.Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J Cell Biochem Suppl 24: 1–11, 1996. [DOI] [PubMed] [Google Scholar]
- 13.Gruenert DC, Basbaum CB, Welsh MJ, Li M, Finkbeiner WE, Nadel JA. Characterization of human tracheal epithelial cells transformed by an origin-defective simian virus 40. Proc Natl Acad Sci USA 85: 5951–5955, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruenert DC, Finkbeiner WE, Widdicombe JH. Culture and transformation of human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 268: L347–L360, 1995. [DOI] [PubMed] [Google Scholar]
- 15.Gruenert DC, Willems M, Cassiman JJ, Frizzell RA. Established cell lines used in cystic fibrosis research. J Cyst Fibros 3, Suppl 2: 191–196, 2004. [DOI] [PubMed] [Google Scholar]
- 16.Haga K, Ohno Si Yugawa T, Narisawa-Saito M, Fujita M, Sakamoto M, Galloway DA, Kiyono T. Efficient immortalization of primary human cells by p16INK4a-specific short hairpin RNA or Bmi-1, combined with introduction of hTERT. Cancer Sci 98: 147–154, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in Eμ-myc transgenic mice. Cell 65: 753–763, 1991. [DOI] [PubMed] [Google Scholar]
- 18.Heckman CA, Marchok AC, Nettesheim P. Respiratory tract epithelium in primary culture: concurrent growth and differentiation during establishment. J Cell Sci 32: 269–291, 1978. [DOI] [PubMed] [Google Scholar]
- 19.Jorissen M, Van Der Schueren B, van den Berghe H, Cassiman JJ. The preservation and regeneration of cilia on human nasal epithelial cells cultured in vitro. Arch Otorhinolaryngol 246: 308–314, 1989. [DOI] [PubMed] [Google Scholar]
- 20.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396: 84–88, 1998. [DOI] [PubMed] [Google Scholar]
- 21.Lechner JF, Haugen A, Autrup H, McClendon IA, Trump BF, Harris CC. Clonal growth of epithelial cells from normal adult human bronchus. Cancer Res 41: 2294–2304, 1981. [PubMed] [Google Scholar]
- 22.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255–260, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, Fleming MD, Olsen JC, Miller SW, Weinberg RA, Hahn WC. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene 21: 4577–4586, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Machen TE Innate immune response in CF airway epithelia: hyperinflammatory? Am J Physiol Cell Physiol 291: C218–C230, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Masui T, Lechner JF, Yoakum GH, Willey JC, Harris CC. Growth and differentiation of normal and transformed human bronchial epithelial cells. J Cell Physiol Suppl 4: 73–81, 1986. [DOI] [PubMed] [Google Scholar]
- 26.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev 19: 1432–1437, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nissim-Rafinia M, Aviram M, Randell SH, Shushi L, Ozeri E, Chiba-Falek O, Eidelman O, Pollard HB, Yankaskas JR, Kerem B. Restoration of the cystic fibrosis transmembrane conductance regulator function by splicing modulation. EMBO Rep 5: 1071–1077, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol 70: 177–185, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez RD, Morales CP, Herbert BS, Rohde JM, Passons C, Shay JW, Wright WE. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev 15: 398–403, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, DiMaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 64: 9027–9034, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Randell SH, Walstad L, Schwab UE, Grubb BR, Yankaskas JR. Isolation and culture of airway epithelial cells from chronically infected human lungs. In Vitro Cell Dev Biol Anim 37: 480–489, 2001. [DOI] [PubMed] [Google Scholar]
- 32.Rheinwald JG, Hahn WC, Ramsey MR, Wu JY, Guo Z, Tsao H, De Luca M, Catricala C, O'Toole KM. A two-stage, p16INK4A- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol Cell Biol 22: 5157–5172, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizo A, Dontje B, Vellenga E, de Haan G, Schuringa JJ. Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood 111: 2621–2630, 2008. [DOI] [PubMed] [Google Scholar]
- 34.Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J, Trono D. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol Ther 2: 404–414, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoner GD, Katoh Y, Foidart JM, Myers GA, Harris CC. Identification and culture of human bronchial epithelial cells. Methods Cell Biol 21A: 15–35, 1980. [DOI] [PubMed] [Google Scholar]
- 37.Terzaghi M, Nettesheim P, Williams ML. Repopulation of denuded tracheal grafts with normal, preneoplastic, and neoplastic epithelial cell populations. Cancer Res 38: 4546–4553, 1978. [PubMed] [Google Scholar]
- 38.Van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der GH, Berns A. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell 65: 737–752, 1991. [DOI] [PubMed] [Google Scholar]
- 39.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 10: 789–799, 2004. [DOI] [PubMed] [Google Scholar]
- 40.Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, van Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer 84: 1372–1376, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitcutt MJ, Adler K, Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell Dev Biol 24: 420–428, 1988. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q, Lu Z, Verghese MW, Randell SH. Airway epithelial cell tolerance to Pseudomonas aeruginosa. Respir Res 6: 26, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zabner J, Karp P, Seiler M, Phillips SL, Mitchell CJ, Saavedra M, Welsh M, Klingelhutz AJ. Development of cystic fibrosis and noncystic fibrosis airway cell lines. Am J Physiol Lung Cell Mol Physiol 284: L844–L854, 2003. [DOI] [PubMed] [Google Scholar]
- 44.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, Hess JL, Slany RK. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol 24: 617–628, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.