Abstract
We detect internal water molecules in a membrane-embedded receptor-transducer complex and demonstrate water structure changes during formation of the signaling state. Time-resolved FTIR spectroscopy reveals stimulus-induced repositioning of one or more structurally active water molecules to a significantly more hydrophobic environment in the signaling state of the sensory rhodopsin II (SRII)-transducer (HtrII) complex. These waters, distinct from bound water molecules within the SRII receptor, appear to be in the middle of the transmembrane interface region near the Tyr199(SRII)-Asn74(HtrII) hydrogen bond. We conclude that water potentially plays an important role in the SRII→HtrII signal transfer mechanism in the membrane's hydrophobic core.
Recent studies indicate that structural changes of internal water molecules play a critical role in membrane protein function and in a soluble system in protein-protein interactions. A hydrogen bonded network of water molecules seen in the high resolution crystallographic structure of bacteriorhodopsin (BR) have been implicated in its proton pump mechanism (1, 2), and a hydrogen bonded network formed by nine water molecules located in an interhelical cavity of squid rhodopsin has been suggested to be involved in the cognate G-protein activation (3). Crystal structural analysis of interactions of the widely studied model protein-protein complex formed by barnase and barstar has revealed that water molecules in the interface region play a critical role in the stability of the complex and that a single mutation alters the structure and mobility of water at the interface (4). These findings raise the possibility that structural changes in water molecules located in the interface region of interacting membrane proteins such as membrane receptor-transducer complexes could also play an important role in signal transduction.
Here we report evidence that structural changes of water molecules located at the protein-protein interface do occur during the photoactivation of the membrane-embedded sensory rhodopsin II - transducer (SRII-HtrII) complex, one of the best characterized receptor-transducer signaling complexes to be studied (5). This system mediates blue-light repellent phototaxis in halophilic archaea, using a signaling pathway similar to that in bacterial chemotaxis (6). Much of the process of activation of SRII by light is understood at the atomic level (5, 7, 8). The next step, the process of signal transfer from the light-activated receptor SRII to HtrII, has become a focus of interest as an example of protein-protein communication between membrane proteins.
X-ray crystallography has provided an atomic structure of the membrane-embedded portion of the SRII-HtrII interface (9), and recently functional measurements including chimera and deletion mapping have localized the signal relay process to this membrane-embedded region of the complex (10, 11). This region is a four-helix bundle held together by predominantly non-polar interactions as well as hydrogen bonds between Tyr199 (SRII) and Asn74 (HtrII) located in the middle of the membrane and Thr189 (SRII) - Glu43/Ser62 (HtrII) near the extracellular surface (Figure 1).
Figure 1.
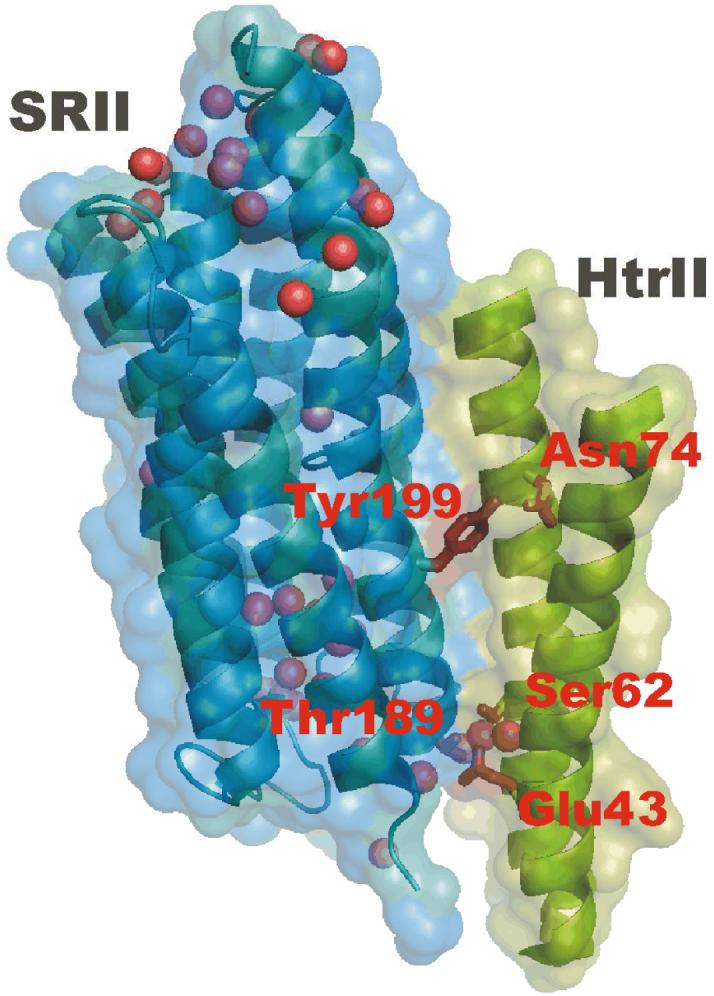
An X-ray crystallographic structure of the SRII-HtrII complex (Protein Data Bank (PDB) access code 1H2S) with the focus on the receptor-transducer interface region with intermolecular Tyr199-Asn74 and Thr189-Glu43/Ser62 hydrogen bonds shown. Internal water molecules are shown as spheres. The figure was prepared using the program PyMOL (http://www.pymol.org).
Fourier transform infrared (FTIR) difference spectroscopy can detect subtle changes even in the hydrogen-bonding strength of single water molecules by probing the frequency of their OH stretching vibrations (12). In proteins, these vibrations appear in a broad spectral region between 2900-3800 cm-1 with higher frequency vibrations corresponding to weaker hydrogen bonds (13, 14). However, the strong absorption of bulk solvent water, which is necessary to maintain physiological conditions, leads to higher noise between 3200-3600 cm-1 and often obscures many weaker peaks arising from individual water molecules in this region.
We have previously used millisecond time-resolved FTIR difference spectroscopy in combination with site-directed mutagenesis to identify protein conformational changes that occur at the SRII-HtrII interface in the receptor signaling state. For example, changes involving alterations in the intermolecular Tyr199-Asn74 hydrogen bond during the light-activation of the SRII-HtRII signaling complex were deduced on the basis of Tyr199→Phe (SRII) and Asn74→Asp (HtrII) substitutions which led to disappearance of several FTIR difference bands in the 800-1800 cm-1 region, consequently assigned to these residues (15). Here we present the results of FTIR difference spectra recorded in the 3600-4000 cm-1 water region. As described previously (15), the measurements were carried out on the SRII-HtrII120 complex that included full-length SRII connected via a flexible linker to the N-terminal 120 amino acids of the transducer comprising the membrane and membrane-proximal regions of HtrII.
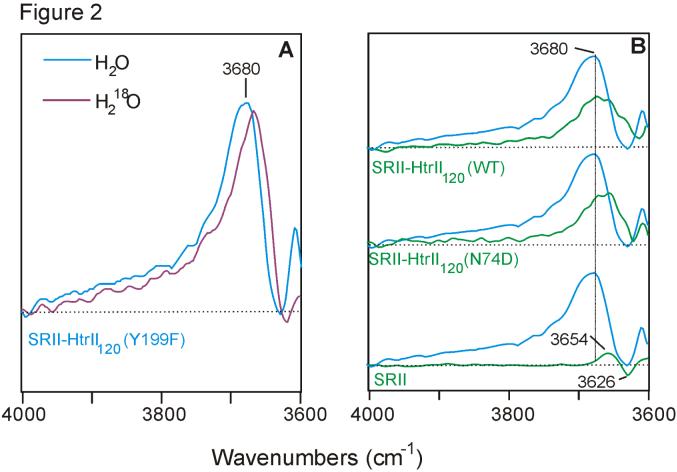
The most prominent spectral feature in this water region is found in the SRII-HtrII Y199F mutant, where an unusually strong positive band with a maximum near 3680 cm-1 and a broad shoulder extending above 3800 cm-1 (Figure 2A) appears within the first 10 ms of the SRII photocycle. It is noted that 3680 cm-1 is at a much higher frequency than previously reported for water molecules in the SRII receptor alone or other related microbial rhodopsins. The downshift of this peak by ~12 cm-1 in the spectra recorded in H218O confirms its assignment to one or more water molecules undergoing a structural change upon light-activation. A band with a similar frequency at 3680 cm-1, although with a lower intensity, is also observed in the spectra of wild-type and N74D mutant SRII-HtrII (Figure 2B). In contrast, spectra of the SRII receptor devoid of the transducer do not exhibit such a strong, high frequency and broad peak (Figure 2B, lower trace). The latter spectrum exhibits a pair of positive/negative bands at 3654/3626 cm-1, which was previously assigned to water located in the hydrophilic protein core of SRII near its retinal chromophore (16).
Figure 2.
(A): Comparison of FTIR difference spectra of Y199F mutant of SRII-HtrII recorded in H2O (blue trace) and H218O (purple trace). (B): Comparison of Y199F mutant SRII-HtrII complex (blue traces) with the wild-type (green trace; top) and N74D mutant complex (green trace; middle) and wild-type SRII receptor (green trace; bottom) spectra. The baseline is shown as dotted line. All spectra were recorded at 8 cm-1 spectral resolution corresponding to the data acquisition window of 9.5 ms.
Our results indicate that in addition to structurally active water previously identified within the SRII receptor (16), water molecules are perturbed in the SRII-HtrII complex during receptor activation. As noted above, the current experimental limitations allowed reliable detection of difference bands only above 3600 cm-1, which are associated with weakly hydrogen-bonded or free OH groups. In this region, the positive 3680 cm-1 band not present in the receptor-only difference spectrum dominates the SRII-HtrII spectra and is particularly strong in the Y199F mutant. Because the FTIR difference spectra are recorded as [light state] minus [dark state], the positive sign of this peak indicates that one or more water molecules undergo a transition to a more weakly hydrogen-bonded and possibly more hydrophobic environment upon photoactivation. Furthermore, the effects of the Y199F substitution suggest that the affected water molecules are located at the receptor-transducer interface, most likely near the Tyr199-Asn74 pair. The appearance of a broad shoulder extending beyond 3800 cm-1 in the Y199F SRII-HtrII spectra compared to the wild-type and N74D mutant can be explained by the introduction of a significantly more hydrophobic phenylalanine residue instead of tyrosine in this mutant, which leads to increased disorder in the water in this region. The replacement of Asn74 with an Asp does not have a large effect as expected since the N74D mutation in full length HtrII preserves phototaxis signaling by the mutant SRII-HtrII complex (E.N.S. and J.L.S., unpublished data). Similarly Y199F mutation preserves phototaxis suggesting that neither mutation causes large perturbations in the protein structure outside of their immediate vicinity.
The detection of structural changes of water in the transmembrane region of SRII-HtrII is somewhat unexpected since the x-ray crystallographic structure of the resting (inactive) state of the complex does not reveal water molecules in this region, although they are present near the Thr189 - Glu43/Ser62 hydrogen bond and also near the cytoplasmic surface (Figure 1). Also, no water molecules in the vicinity of Tyr199-Asn74 were detected in the late M (M2) photointermediate state (17). One possibility is that the molecules observed by FTIR are disordered and therefore not easily detected in the x-ray structure. Alternatively, lattice constraints may have prevented the full structural change of the activated state to occur in the crystal (5). The spectral changes likely reflect water molecules located initially at a different site in the protein or in the extramembrane aqueous milieu, which are displaced during the photocycle in response to the protein structural changes. In fact, one of the main features hypothesized in the signal transfer mechanism is a shift of the transducer helix TM2 in response to the receptor activation (5, 17). This movement could potentially affect the position of some of the water molecules present at the SRII-HtrII interface near the cytoplasmic and extracellular surface regions in the resting state of the protein complex (Figure 1). The fact that no negative band was observed in the FTIR difference spectra supports the latter hypothesis since the altered water molecules likely exist in a more hydrophilic environment in the protein initial state. In that case, the corresponding peak is expected to appear below 3600 cm-1 and would not be detected under the current experimental setup.
The results of this study also help further elucidate the mechanism of structural changes involving the Tyr199-Asn74 hydrogen bond. Our earlier FTIR measurements revealed substantial changes in the hydrogen-bonding strength of both Tyr199 and Asn74 suggesting that these residues form hydrogen bonds with other groups during photoactivation(15). It is therefore possible that the water molecules we observe in this study function as hydrogen-bonding partners to these residues when their interaction is disrupted in the light-activated state. In the case of the N74D mutant of HtrII, water may also serve as a proton acceptor for Asp74, which was shown to undergo at least a partial deprotonation during the photocycle (15).
It is well established that in the case of bacteriorhodopsin, a closely related microbial rhodopsin that functions as a light-driven proton pump, water molecules including those with weakly bonded hydroxyl groups play an important role in the proton transport (2). The detection of water structural changes in the SRII-HtrII phototaxis complex indicates that water molecules also play a significant role in the signal transduction mechanism employed by sensory rhodopsins, in this case in the SRII-HtrII membrane-embedded interface where signal relay takes place. In particular, it is possible that hydrogen-bonding changes involving the Tyr199-Asn74 pair and water molecules help stabilize the receptor-transducer complex in its light-activated state thus increasing the signal relay efficiency of the SRII receptor.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01GM069969 (to K.J.R.) and R37GM27750 (to J.L.S.) and US Department of Energy grant DE-FG02-07ER15867 and the Robert A. Welch foundation (to J.L.S.). We thank Jason Amsden and Joel Kralj for assistance and helpful discussions.
Footnotes
SUPPORTING INFORMATION AVAILABLE Experimental details of sample preparation and FTIR spectroscopic measurements. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mathias G, Marx D. Proc. Natl. Acad. Sci. U S A. 2007;104:6980–6985. doi: 10.1073/pnas.0609229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garczarek F, Gerwert K. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 3.Murakami M, Kouyama T. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 4.Urakubo Y, Ikura T, Ito N. Protein Sci. 2008;17:1055–1065. doi: 10.1110/ps.073322508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spudich JL. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Hoff WD, Jung KH, Spudich JL. Annu. Rev. Biophys. Biomol. Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 7.Klare JP, Chizhov I, Engelhard M. Results Probl. Cell Differ. 2008;45:73–122. doi: 10.1007/400_2007_041. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki J, Spudich JL. Photochem. Photobiol. 2008;84:863–868. doi: 10.1111/j.1751-1097.2008.00314.x. [DOI] [PubMed] [Google Scholar]
- 9.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 10.Sudo Y, Spudich JL. Proc. Natl. Acad. Sci. U S A. 2006;103:16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki J, Nara T, Spudich EN, Spudich JL. Mol. Microbiol. 2007;66:1321–1330. doi: 10.1111/j.1365-2958.2007.05983.x. [DOI] [PubMed] [Google Scholar]
- 12.Kandori H. Biochimica Et Biophysica Acta-Bioenergetics. 2000;1460:177–191. doi: 10.1016/s0005-2728(00)00138-9. [DOI] [PubMed] [Google Scholar]
- 13.Mohr SC, Wilk WD, Barrow GM. J. Am. Chem. Soc. 1965;87:3048–3052. [Google Scholar]
- 14.Glew DN, Rath NS. Can. J. Chem. 1971;49:837–856. [Google Scholar]
- 15.Bergo VB, Spudich EN, Rothschild KJ, Spudich JL. J. Biol. Chem. 2005;280:28365–28369. doi: 10.1074/jbc.M505555200. [DOI] [PubMed] [Google Scholar]
- 16.Kandori H, Furutani Y, Shimono K, Shichida Y, Kamo N. Biochemistry. 2001;40:15693–15698. doi: 10.1021/bi011621n. [DOI] [PubMed] [Google Scholar]
- 17.Moukhametzianov R, Klare JP, Efremov R, Baeken C, Goppner A, Labahn J, Engelhard M, Buldt G, Gordeliy VI. Nature. 2006;440:115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.