Abstract
BMP-2 plays an essential role in osteoblast and chondrocyte differentiation, but its signaling mechanism has not been fully defined. In the present studies, we investigated the mechanism through which BMP-2 activates the Smad6 gene. A −2006/+45 Smad6 promoter-luciferase construct was generated along with deletions and Runx2 binding site mutations to examine the role of Smad1 and Runx2 signaling following BMP-2 stimulation in osteoblasts. Transfection of Runx2 or treatment with BMP-2-stimulated promoter activity of the −2006/+45 and −1191/+45 reporters but not the −829/+45 and −374/+45 reporters. No Smad1/5 binding site is present in the −1191/−829 region of the Smad6 promoter. Mutation of the OSE2-a site (−1036/−1031) completely abolished the stimulatory effect of Runx2 as well as BMP-2 on the −2006/+45 and −1191/+45 Smad6 reporters. Gel shift and chromatin immunoprecipitation (ChIP) assays showed that Runx2 binds the OSE2-a element. ChIP assays demonstrated that Smad1 also interacts with the OSE2-a site at the Smad6 promoter through Runx2. The protein degradation of Runx2 is mediated by the E3 ubiquitin ligase Smurf1. In the present studies, we found that Smurf1 binds the OSE2-a site through Runx2 and inhibits Smad6 gene transcription. Treatment with BMP-2 and transfection of Smad1 abolished Smurf1 binding to the OSE2 site. These results show that Smad1 binding excludes Smurf1 interaction with the OSE2 site and promotes Smad6 gene transcription.
Bone morphogenetic protein 2 (BMP-2)2 plays an important role in osteoblast and chondrocyte differentiation. BMP-2 signaling is mediated by its downstream molecules Smads 1 and 5. Although BMP-2 induces mRNA expression of many osteoblast and chondrocyte marker genes, the GC-rich Smad1/5 binding elements have only been identified in promoter regions of type X collagen and Smad6 genes (1, 2). Because Smad1/5 has relatively low DNA binding affinity, it has been proposed that Smad1/5 interacts with other sequence-specific DNA-binding proteins and forms stable DNA binding complexes to activate downstream target genes (3). It has been reported that Smad1/5 interacts with the bone-specific transcription factor Runt-related gene 2 (Runx2) (4, 5). The detailed mechanisms through which Smad1/5 and Runx2 activate downstream target genes remain undefined.
Runx2 is a bone-specific transcription factor that belongs to the Runx family. Runx2 acquires DNA binding activity by heterodimerizing with Cbfβ (6–8), and DNA binding sites for Runx2 have been identified in promoter regions of many osteoblast and chondrocyte-specific genes (9–16). Runx2 binds its specific responsive elements in the promoters of Runx2 target genes and regulates the transcription of these genes.
Runx2 plays a critical role in bone formation and chondrocyte maturation in vivo. Runx2−/− mice died just after birth and completely lack bone formation because of the absence of osteoblast differentiation, demonstrating that Runx2 is required for osteoblastogenesis (17, 18). Heterozygous mutant mice have skeletal abnormalities that mimick those observed in human cleidocranial dysplasia syndrome (19, 20) where delayed development of intramembranous bones occurs (17, 18). Further studies demonstrated that chondrocyte maturation is delayed in Runx2−/− mice (21, 22), suggesting that Runx2 promotes chondrocyte maturation. Consistent with this, dominant-negative Runx2 inhibited chondrocyte maturation (23, 24) while overexpression of Runx2 in chondrocytes restored chondrocyte maturation in Runx2−/− mice (25). Recent studies demonstrate that chondrocyte maturation was completely absent in Runx2 and Runx3 double knock-out mice (26), demonstrating that Runx2 and Runx3 are functionally redundant and play an essential role in chondrocyte maturation.
In the present studies, we investigated the molecular mechanism involved in Smad6 gene regulation downstream of BMP-2 signaling. We identified and characterized a Runx2 binding site at the Smad6 promoter and found that Smad1-mediated BMP-2 signaling on Smad6 gene transcription is through direct binding to the OSE2 site of the Smad6 promoter, while the E3 ubiquitin ligase Smurf1 binds the OSE2 site and suppresses Smad6 promoter activity. A BMP-2/Smad1 inhibits Smurf1 interaction with the promoter and permits Smad6 gene transcription.
MATERIALS AND METHODS
Cell Culture and Transfection
C2C12 cells were cultured in Dulbecco’s modified Eagle’s medium and 2T3 cells were cultured in α-minimal essential medium (α-MEM) supplemented with 10% fetal bovine serum. Deletion constructs of the Smad6 promoter were co-transfected with Runx2, Smad1, Smurf1, or control expression plasmids into C2C12 and 2T3 cells and treated with BMP-2 (100 ng/ml) or control medium in 6-cm culture dishes using the Lipofectamine 2000 reagents (Invitrogen). The total amount of DNA transfected was constant in the experiments. Luciferase assays were performed 24 h after transfection, and chromatin immunoprecipitation (ChIP) assays were performed 2 h after BMP-2 treatment.
Real-time RT-PCR Analysis
C2C12 and 2T3 cells were treated with different concentrations of BMP-2 (50, 100, and 200 ng/ml) or transfected with different amounts of Runx2 or Smurf1 expression plasmids (0.2, 0.4, and 0.8 µg/dish, 6-mm culture dish). Total RNA of cells was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. After reverse transcription reaction, real-time PCR was performed by an ABI PRISM 7000 sequence detection system according to the manufacturer’s instructions. The reaction mixture contained 1×EvaGreen dye, 0.5 pm of each primer, 2.5 mm MgCl2, and 0.5 µl of cDNA from 20 µl of reverse transcription reaction. The conditions of real-time PCR were as follows: 94 °C for 4 min followed by 40 cycles at 94 °C for 30 s, 60 °C for 1.5 min. There is no nonspecific amplification determined by dissolve curve. There are four samples in each group. The real-time RT-PCR results were reported as the fold induction of relative light units for the treatment over vehicle after normalization to GAPDH expression. The primers used for this analysis were: Smad6: 5′-TACCACTTCAGCCGGCTCTG-3′ (forward) and 5′-AGTACGCCACGCTGCACCAGT-3′ (reverse); GAPDH: 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse).
Western Blot Analysis
C2C12 and 2T3 cells were treated with different concentration of BMP-2 (50, 100, and 200 ng/ml) or transfected with different amounts of Runx2 or Smurf1 expression plasmids (0.2, 0.4, and 0.8 µg/dish, 6-mm culture dish). Cells were lysed on ice for 30 min in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, and 0.1% SDS supplemented with protease inhibitors (10 µg/ml leupeptin, 10 µg/ml pepstatin A, and 10 µg/ml aprotinin) and phosphatase inhibitors (1 mm NaF and 1 mm Na3 VO4). Proteins were fractionated by SDS-PAGE, transferred to a nitrocellulose membrane, and detected using the anti-Smad6 (Imgenex, San Diego, CA) and anti-β-actin (Sigma) antibodies. Immunostaining was detected using an enhanced chemiluminescence (ECL) system (Amersham Biosciences).
Luciferase Assay
C2C12 and 2T3 cells were co-transfected with wild-type (WT) or mutant deletion constructs of the Smad6 promoter (−2006/+45, −1191/+45, −829/+45, and −374/+45) and Runx2, Smad1, Smurf1, mutant Smurf1 (C710A), or control expression plasmids and treated with BMP-2 (100 ng/ml) or control medium. Cell lysates were extracted 24 h after transfection, and luciferase activity was measured using a Promega dual luciferase reporter assay kit (Promega, Madison, WI).
PCR-based Site-directed Mutagenesis
Smad6 promoter constructs, −2006/+45 and −1191/+45 with mutations at either OSE2-a or OSE2-b site were constructed using Stratagene QuikChange Site-directed mutagenesis kit and cloned into pGL3 vector (Stratagene). The mutations were confirmed by sequencing the entire promoter fragment.
Electrophoresis Mobility Shift Assay
Nuclear extracts (5 µg) from C2C12 cells were incubated with radioactive end-labeled double-stranded oligonucleotide probes containing consensus binding sites for Runx2 in the mouse Smad6 promoter (5′-CCGGGTCGCGGTGGGTTCACC-3′, the binding site is highlighted). Incubation was performed in 10 µl of binding buffer (10 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm EDTA, 0.1% Triton-X 100, 10% glycerol, 1 µg of poly(dI-dC), 1 mm dithiothreitol), for 15–20 min at room temperature. For competition, the cold wild type or mutant oligonucleotides (5′-CCGGGTCGCGGTGTTTTCACC-3′ for the mutant oligo, nucleotide mutations indicated by underscore) were added in a 1:1 or 1:20 excess. For supershift assay, the nuclear extract was incubated with Runx-2 specific (Abcam, Cambridge, MA) or control antibodies for 30 min before addition of the labeled probe. After incubation with labeled probe for an additional 20–30 min, samples were fractionated on a 4% native polyacrylamide gel and visualized by exposing dry gel to Kodak Film.
Chromatin Immunoprecipitation
ChIP assay was performed using a kit from Upstate Technologies (Lake Placid, NY) with the following modifications. C2C12 or 2T3 cells were treated with 1% formaldehyde at 37 °C in the presence of 4% CO2 for 15 min. Cells were sonicated on ice for 6 times at 50 watts using a Sonic Dismembrator (Scientz, China) for 25 s with a 60-s interval between each sonication. The following antibodies (10 µg) were used: anti-Runx2 antibody (MBL), anti-Myc antibody (Santa Cruz Biotechnology), and anti-FLAG antibody (Sigma) with 100 µg of chromatin per ChIP. The amounts of each specific DNA fragment in immunoprecipitates were determined by PCR or quantitative PCR reactions. The primers used for this analysis were: A (−1108 to −937) 5′-GTGCTATGCGCGTTTGCAAGT-3′ (forward) and 5′-CTCAAGCTCACGTCGTGCCTT-3′ (reverse); B (−1968/−1839) 5′-CAGTCCAGAGCACAGGTATCCT-3′ (forward) and 5′-CCGAGCTCAATGAAAGAAATTCC-3′ (reverse); C (+281/+432) 5′-GCGCCAAAGGGTATCGTATG-3′ (forward) and 5′-AGGCTCAGCTCGGCTGCCCA-3′ (reverse). The conditions of PCR for the detection of each specific DNA fragment were as follows: 94 °C for 4 min followed by 28 cycles at 94 °C for 30 s, 62 °C for 30 s, 72 °C for 60 s.
Quantitative ChIP
For the quantitative ChIP (qChIP), a standard curve was generated using primer set A. Copy numbers for the DNA fragment −1108 to −937 in each anti-Runx2-and anti-Smurf1-immunoprecipitated samples were determined and compared with copy numbers of the DNA fragment without IP (input DNA). Anti-FLAG antibody was used as control for IP. The percentage of the input was then calculated. The final value was the percentage input obtained with specific antibody minus the percentage input obtained with the anti-FLAG control antibody. The dissociation curve was determined for each quantitative PCR reaction to assure that a single band was produced. Each data point represents four independent samples.
RESULTS
BMP-2 and Runx2 Stimulate and Smurf1 Inhibits Smad6 mRNA and Protein Expression
Smad6 is a downstream target gene of BMP signaling (2). BMP signaling protein Smad1 interacts with Runx2 (4, 5) and E3 ubiquitin ligase Smurf1 mediates Runx2 degradation (5, 27). In the present studies, we first examined effects of BMP-2, Runx2, and Smurf1 on Smad6 mRNA and protein expression in myoblast/osteoblast precursor C2C12 cells and osteoblast precursor 2T3 cells. We found that BMP-2 (50–200 ng/ml) as well as Runx2 (0.2–0.8 µg/dish, 6-mm culture dish) stimulated Smad6 mRNA and protein expression in a dose-dependent manner. In contrast, Smurf1 (0.2–0.8 µg/dish) inhibited Smad6 mRNA and protein expression in C2C12 (Fig. 1a) and 2T3 (data not shown) cells.
FIGURE 1. BMP-2 and Runx2 stimulate and Smurf1 inhibits Smad6 mRNA and protein expression.
C2C12 cells were treated with different concentrations of BMP-2 (50, 100, and 200 ng/ml) or transfected with different amounts of Runx2 or Smurf1 expression plasmids (0.2, 0.4, and 0.8 µg/dish, 6-mm culture dish). Total RNA and cell lysates were extracted 24 h after transfection or BMP-2 treatment. Changes in Smad6 mRNA and protein levels were analyzed by real-time PCR (a) and Western blotting (b). BMP-2 and Runx2 stimulated and Smurf1 inhibited Smad6 mRNA and protein expression in a dose-dependent manner.
BMP-2 Stimulates Smad6 Gene Transcription
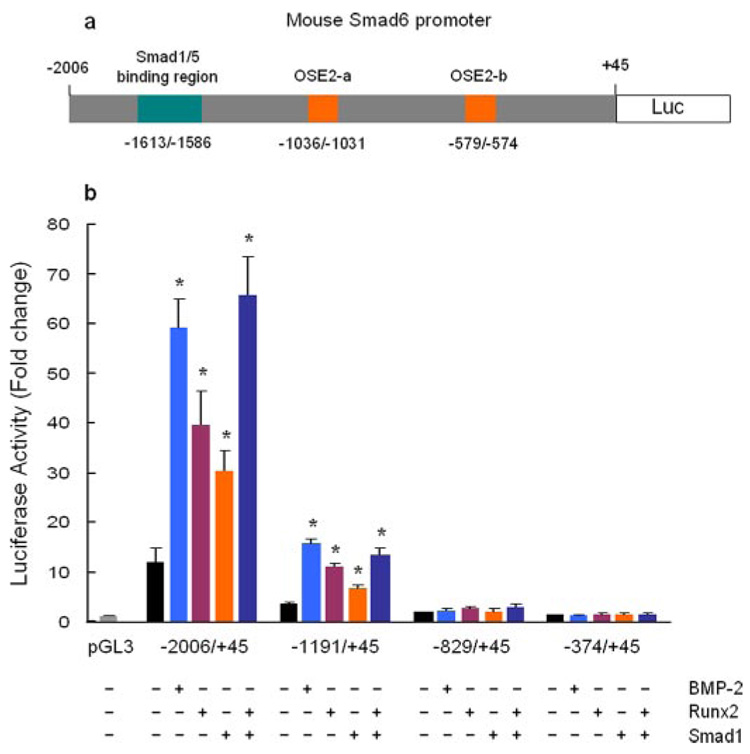
To investigate the mechanism of BMP-2 on Smad6 gene transcription, we amplified the mouse Smad6 promoter −2006/+45 by PCR (GenBank™ accession number: AF010133) using mouse genomic DNA as a template (Fig. 2a) and generated three deletion constructs, −1191/+45, −829/+45, and −374/+45. The various Smad6 promoter fragments were cloned into pGL3 vector. A significant reduction in the promoter activity was observed in −1191/+45 reporter compared with −2006/+45 reporter (Fig. 2b), suggesting the positive transcription units exist in the −2006/−1191 region of the Smad6 promoter. In −1906/−1878 region of the Smad6 promoter, three overlapping GC-rich sequences have been identified and Smad1/5 binds this region of the Smad6 promoter (2). To determine the effect of BMP-2 on Smad6 promoter, we transfected the deletion constructs of the Smad6 promoter and treated cells with BMP-2 (100 ng/ml) in C2C12 cells. The luciferase ctivity was measured 24 h after transfection. BMP-2 stimulated the promoter activity of the −2006/+45 and −1191/+45 reporters but not the −829/+45 and −374/+45 reporters (Fig. 2b). BMP-2 stimulated the −1191/+45 reporter over 5-fold although no Smad1/5 binding site has been found in the −1191/+45 region of the Smad6 promoter by sequence analysis. The results suggest that BMP-2 may activate the −1191/+45 Smad6 promoter indirectly or through other transcription factor binding sites, such as Runx2.
FIGURE 2. BMP-2 and Runx2 activate the Smad6 promoter.
a, diagram shows the structure of the mouse Smad6 promoter −2006/+45 construct. The putative Runx2 binding sites (OSE2-a, OSE2-b), and Smad1/5 binding region (three overlapping GC-rich sequences) are indicated. b, C2C12 cells were co-transfected with the Smad6 promoter construct (−2006/+45) or three deletion constructs (−1191/+45, −829/+45, and −374/+45) with Runx2 or Smad1 expression plasmid or treated with BMP-2 (100 ng/ml). Cell lysates were collected 24 h after transfection, and a luciferase assay was performed. BMP-2 and Runx2 stimulated the promoter activity of −2006/+45 and −1191/+45 constructs with similar potency but had no effect on −829/+45 and −374/+45 reporters. Transfection of Smad1 further enhanced Runx2-induced promoter activity of −2006/+45 and −1191/+45 constructs compared with the transfection of each plasmid separately. *, p < 0.05, unpaired Student’s t test, compared with vector transfection alone.
Runx2 Activates Smad6 Gene Transcription
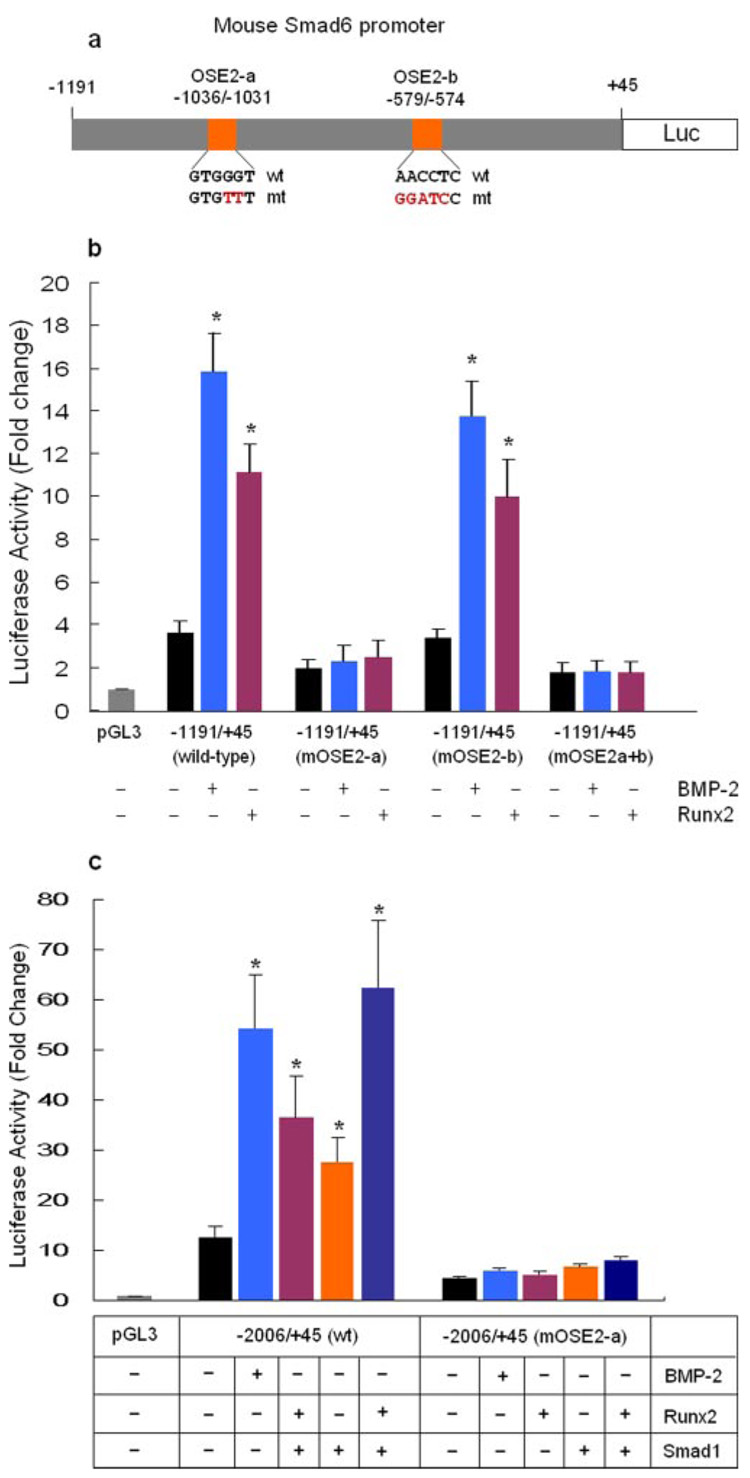
In the present studies, we identified two putative Runx2 binding sites located at −1036/−1031 (OSE2-a) and −579/−574 (OSE2-b) regions of the Smad6 promoter by sequence analysis (Fig. 2a). Transfection of Runx2 stimulated the promoter activity of −2006/+45 and −1191/+45 reporters but not −829/+45 and the −374/+45 reporters (Fig. 2b). Co-transfection of Smad1 with Runx2 further enhanced Runx2-induced promoter activity of the −2006/+45 and −1191/+45 reporters (Fig. 2b). To determine if these OSE2 sites are functional, we performed site-direct mutagenesis. The sequences of the wild type and mutant OSE2-a and OSE2-b are shown in Fig. 3a. Mutation of the OSE2-b at −579/−574 did not affect basal promoter activity and had no significant effect on the responsiveness of the −1191/+45 reporter to BMP-2 (100 ng/ml) treatment or Runx2 transfection (Fig. 3b). In contrast, mutation of OSE2-a at −1036/−1031 caused a significant reduction in the basal promoter activity and completely abolished BMP-2 (100 ng/ml) and Runx2-induced promoter activity of the −1191/+45 and −2006/+45 reporters (Fig. 3, b and c). These results indicate that OSE2-a is the major functional site for Runx2 at the Smad6 promoter and suggest that Runx2 is required for BMP-2 to activate Smad6 promoter. Runx2 and BMP-2 retain their stimulatory activity when the multiple copies of OSE2-a elements (6xOSE2-a) was cloned in front of the TK basal promoter (data not shown), suggesting that OSE2-a is a true Runx2 responsive element.
FIGURE 3. BMP-2 and Runx2 activate Smad6 promoter through the OSE2-a site.
a, diagram shows the construct harboring wild-type (wt) or mutant (mt) OSE2-a and OSE2-b sites derived from −1191/+45 reporter. The wt and mt sequences of OSE2-a and OSE2-b sites are indicated. b, C2C12 cells were transfected with wt and mt −1191/+45 Smad6 reporter with Runx2 expression plasmid or treated with BMP-2 (100 ng/ml). Cell lysates were collected 24 h after transfection, and a luciferase assay was performed. Mutation of OSE2-a caused a significant reduction in basal promoter activity and completely abolished BMP-2- or Runx2-induced promoter activity of the −1191/+45 reporter. Mutation of OSE2-b had no significant effect on the basal promoter activity or the responsiveness to Runx2 or BMP-2. *, p < 0.05, unpaired Student’s t test, compared with vector alone. c, C2C12 cells were transfected with wt and mt −2006/+45 Smad6 reporter with Runx2 or Smad1 expression plasmid or treated with BMP-2 (100 ng/ml). Mutation of OSE2-a in −2006/+45 reporter significantly reduced basal promoter activity and inhibited BMP-2, Runx2, or Smad1-induced promoter activity. *, p < 0.05, unpaired Student’s t test, compared with vector alone.
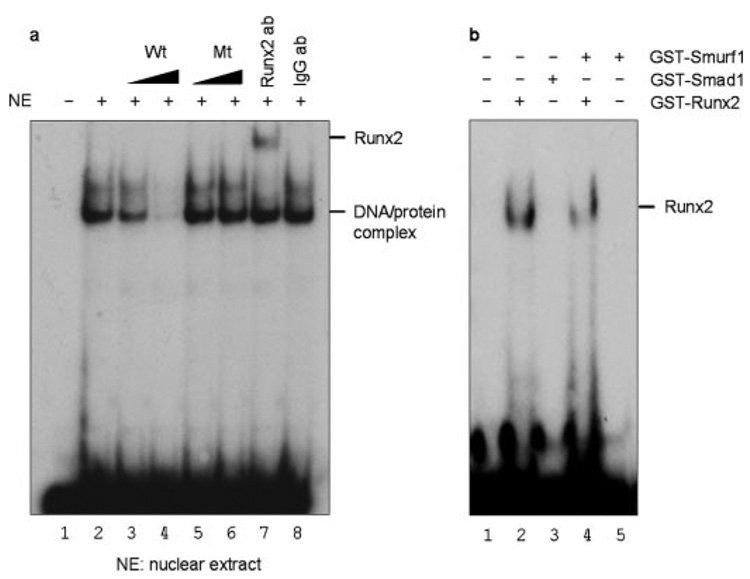
To demonstrate the direct binding of Runx2 to the Smad6 promoter in vitro, electrophoresis mobility shift assay (EMSA) was performed using nuclear protein extracts from C2C12 cells. The oligonucleotide sequence used was derived from the Runx2 consensus site in the mouse Smad6 promoter (−1046/−1026). Following the incubation, binding complexes were detected. The specificity of the complex was suggested by the competition and supershift assays. An unlabeled consensus binding sequence competed with the binding complex in a dose-dependent manner (Fig. 4a, lanes 3 and 4), while oligonucleotides mutated in Runx2 binding site had no effect (Fig. 4a, lanes 5 and 6), confirming the specificity of the complex for the Runx2 binding. Moreover, antibody specific for Runx2 but not control antibody resulted in supershift of the Runx2 binding complex (Fig. 4a, lane 7), indicating the presence of Runx2 in the DNA-protein complex. Altogether, these results demonstrate that Runx2 binds to the OSE2-a site in the Smad 6 promoter. To determine if Smad1 or Smurf1 binds directly to the OSE2-a site, we performed gel shift assay using GST-Smad1 and GST-Smurf1. The results demonstrated that neither Smad1 nor Smurf1 directly binds the OSE2-a site of the Smad6 promoter (Fig. 4b, lanes 3 and 5). To investigate if Smurf1 interferes with Runx2 binding to the OSE2-a site, we examined Runx2 binding in the absence or presence of Smurf1 in gel shift assay using GST-Runx2 and GST-Smurf1. The results showed that Smurf1 slightly reduced Runx2 binding to the OSE2-a site (Fig. 4b, lane 4), suggesting that Smurf1 does interfere with Runx2 binding to the Smad6 promoter.
FIGURE 4. Runx2 binds OSE2-a site gel shift assay.
a, nuclear extracts from C2C12 cells were assayed by gel shift using 32P end-labeled oligonucleotide probes derived from the mouse Smad6 promoter (−1046/−1026). Lane 1, free probe control (without nuclear extracts); lane 2, addition of nuclear extracts only. The binding of OSE2-a was competed with excess amounts of cold wild-type (lanes 3 and 4) or mutant (lanes 5 and 6) oligonucleotide (1:10 and 1:100 ratio). The same nuclear extracts were also examined in supershift assays using antibody for Runx2 (lane 7) or IgG control antibody (lane 8). DNA protein binding complexes and the Runx2-specific supershifted band were observed (lane 7). b, binding of Runx2, Smad1, or Smurf1 was further determined by gel shift assay using in vitro translated GST-Runx2, GST-Smad1, and GST-Smurf1 proteins. Smad1 or Smurf1 did not bind the OSE2-a site (lanes 3 and 5). The binding of Runx2 was reduced in the presence of Smurf1 (lane 4).
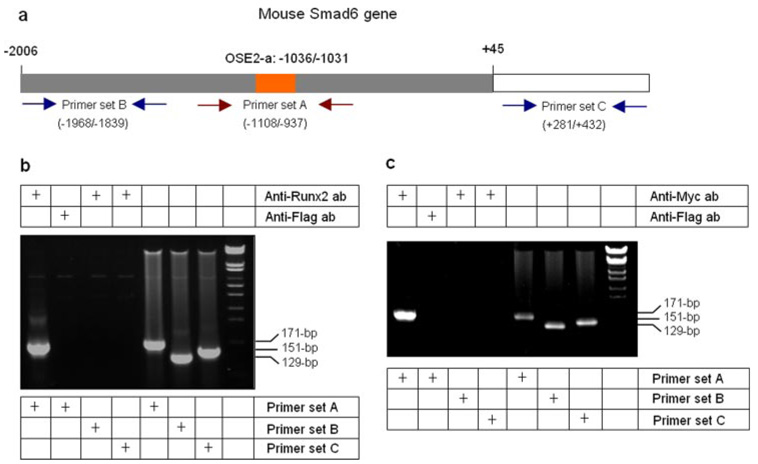
To further determine the interactions of Runx2, Smad1 and Smurf1 with the OSE2-a site, we performed ChIP assays. Immunoprecipitation (IP) was performed using the anti-Runx2 antibody. Anti-FLAG antibody was used as a control in IP experiment. A segment of the Smad6 promoter containing the OSE2-a (−1108/−937) site was amplified by PCR using the primer set A, and DNA templates were extracted from protein/DNA cross-linked samples. As negative controls, primer sets recognizing 5′- and 3′-distal regions (primer sets B and C) of the Smad6 gene were used in the PCR assays (Fig. 5a). ChIP assay showed that Runx2 binds specifically to −1108/−937 region (containing OSE2-a) of the Smad6 promoter (Fig. 5b). These findings further confirmed the binding of Runx2 to the OSE2-a site of the Smad6 promoter.
FIGURE 5. Interaction of Smad1 with Runx2 at the OSE2-a site-ChIP assay.
a, diagram shows the structure of the mouse Smad6 gene. The putative Runx2 binding site (OSE2-a) and priming sites for ChIP assay are indicated. b, IP was performed using anti-Runx2 antibody (lanes 1, 3, and 4) or anti-FLAG control antibody (lane 2). The PCR was performed using primer set A (lanes 1, 2, and 5) to detect Runx2 binding and the PCR product (171 bp) of OSE2-a region (−1108/−937, lane 1) was amplified using the samples of cross-linked IP products as templates. Primer set B (−1968/−1839, lanes 3 and 6) and set C (+281/+432, lanes 4 and 7) were used as negative controls for PCR. Results showed that Runx2 binds specifically to the OSE2-a region (lane 1). c, C2C12 cells were transfected with Myc-Smad1 plasmid and treated with or without BMP-2 for 2 h before the ChIP assay. The IP was performed using the anti-Myc (lanes 1, 3, and 4) antibody and the anti-FLAG control antibody (lane 2). The PCR was performed using primer set A (lanes 1, 2, and 5) for detecting Smad1 interaction with the OSE2-a region (−1108/−937) and primer set B (−1968/−1839) and C (+281/+432) as negative controls. Results showed that Smad1 interacts with the OSE2-a region (lane 1).
Smad1 Interacts with the OSE2 Site at the Smad6 Promoter
Although BMP-2 stimulated −1191/+45 reporter, sequence analysis does not demonstrate any GC-rich Smad1/5 binding sites (GCCGnCGC) in the −1191/−829 region of the Smad6 promoter. Our previous results showed that the Runx2 is necessary for BMP-2 activation of its target gene reporter (5). Furthermore, previous reports from ours and other laboratories showed that Smad1 interacts with Runx2 in COS and C2C12 cells (4, 5). To determine if Smad1 binds the OSE2-a site at the Smad6 promoter, we performed ChIP assays. Myc-Smad1 expression plasmid was transfected into C2C12 cells and IP was performed using an anti-Myc antibody. The same DNA segment of the Smad6 promoter containing the OSE2-a site (−1108/−937) was amplified by PCR from IP samples (Fig. 5c). The results suggest that Smad1 interacts indirectly with the OSE2-a site at the Smad6 promoter possibly through Runx2 and BMP-2 may activate Smad6 promoter through Smad1 interaction with the OSE2-a site in addition to its direct binding to the Smad1/5 responsive elements.
Smurf1 Interacts with the Smad6 Promoter and Inhibits Smad6 Gene Transcription
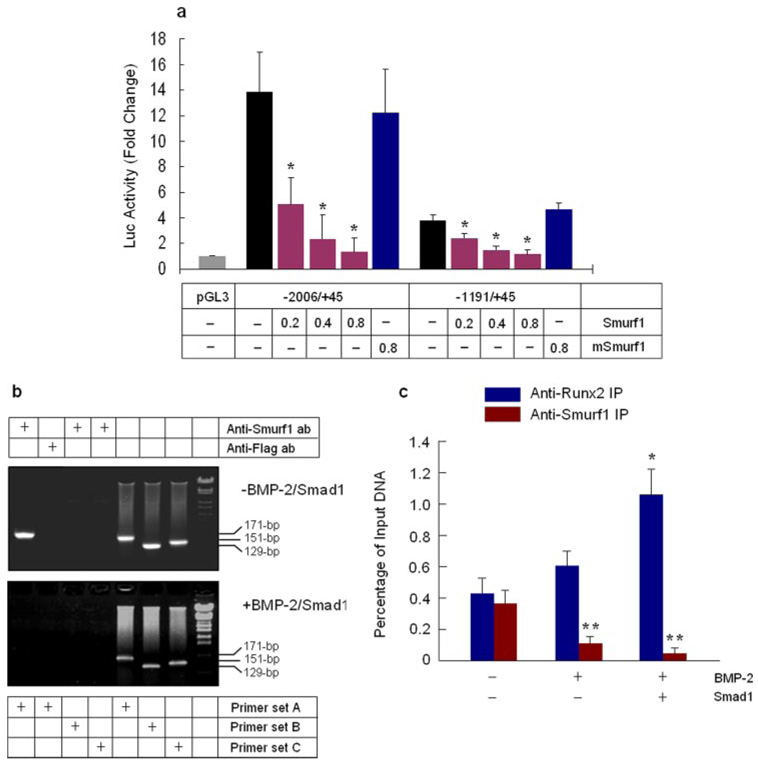
Smurf1 is a Hect domain E3 ubiquitin ligase and was first identified by its interaction with Smads 1 and 5 and induces the degradation of these Smad proteins (28). In previous studies, we found that Smurf1 binds Runx2 and induces Runx2 degradation (5). To determine if Smurf1 regulates Smad6 gene transcription, different amounts of Smurf1 plasmid were co-transfected with Smad6 promoter (−2006/+45 and −1191/+45), and changes in Smad6 promoter activity were examined. Transfection of Smurf1 inhibited Smad6 promoter activity in a dose-dependent manner. In contrast, mutant Smurf1 (C710A) had no significant effect on Smad6 promoter activity (Fig. 6a). To determine if Smurf1 interacts with Runx2 at the Smad6 promoter, we performed ChIP assays. IP was performed using an anti-Smurf1 antibody and the anti-FLAG antibody was used as a negative control in the IP assay. The DNA segment of the Smad6 promoter containing the OSE2-a site (−1108/−937) was amplified by PCR using the primer set A. The primer sets B and C recognizing 5′- and 3′-distal regions of the Smad6 gene were used as negative controls in PCR assays. The results showed that Smurf1 interacts specifically with the OSE2-a region (−1108/−937) of the Smad6 promoter (Fig. 6b, upper panel).
FIGURE 6. Smad1 competes with Smurf1 on Runx2 interaction at the OSE2-a site.
a, Smad6 promoter −2006/+45 and −1191/+45 were co-transfected with different amounts of Smurf1 (0.2, 0.4, and 0.8µg/dish, 6-mm culture dish) or mSmurf1 (0.8 µg/dish) expression plasmids into C2C12 cells. Cell lysates were collected 24 h after transfection, and a luciferase assay was performed. Smurf1 but not mSmurf1 inhibited the promoter activity of −2006/+45 and −1191/+45 constructs in a dose-dependent manner. *, p < 0.05, one-way analysis of variance followed by Dunnett’s test, compared with vector alone. b, Smurf1 ChIP assay. C2C12 cells were transfected with Smad1 and treated with BMP-2 (100 ng/ml). Cell lysates were collected 2 h after BMP-2 treatment. The IP was performed using the anti-Smurf1 (lanes 1, 3, and 4) or anti-FLAG (lane 2, negative control) antibodies. The PCR was performed using primer set A (lanes 1, 2, and 5) (OSE2-a region, −1108/−937), B (−1968/−1839), and C (+281/+432) (negative control). Results showed that Smurf1 interacts with the OSE2-a region in the absence of BMP-2 and Smad1 (upper panel). With BMP-2 treatment and Smad1 transfection, Smurf1 was no longer able to interact with the OSE2-a region of the Smad6 promoter. c, qChIP assay. Copy numbers of the DNA fragment containing OSE2-a site (−1108/−937) in anti-Runx2 and anti-Smurf1 immunoprecipitated samples before (Input DNA) and after IP were quantified by real-time PCR using primer set A. The bars indicate final values of the percentage of the DNA input obtained from specific antibody-immunoprecipitated samples subtracting the percentage of DNA input obtained from anti-FLAG control antibody-immunoprecipitated samples. The results showed that BMP-2/Smad1 enhanced Runx2 binding and inhibited Smurf1 binding to the OSE2-a site. *, p < 0.05, (n = 4), unpaired Student’s t test, Runx2 binding was compared with untreated group. **, p < 0.05 (n=4), unpaired Student’s t test, Smurf1 binding was compared with untreated group.
Smad1 binds the C-terminal region of Runx2 (4) and Smurf1 presumably binds the PY motif of Runx2 (located at C terminus) through its WW domain. To determine if Smad1 activates Smad6 promoter through binding to Runx2 and competing with Smurf1, we performed ChIP assays to examine changes in Smurf1 binding to OSE2-a site in the presence or absence of BMP-2 and Smad1. We found that Smurf1 was no longer able to bind the OSE2-a site of the Smad6 promoter in the presence of BMP-2 and Smad1 (Fig. 6b, lower panel). To further evaluate the effects of BMP-2 and Smad1 on the binding of Runx2 and Smurf1 to the OSE2-a site, we performed quantitative ChIP assays using cells transfected with Smad1 and treated with BMP-2. Copy numbers of the DNA fragment containing the OSE2-a site were determined by quantitative PCR using primer set A before (Input DNA) and after IP. The anti-FLAG antibody was used as a negative control for IP. The specific percentages of input DNA for anti-Runx2 and anti-Smurf1-immunoprecipitated samples were determined. The results showed that the binding of Runx2 to the OSE2-a site was enhanced by BMP-2/Smad1. Smurf1 interacts specifically with the OSE2-a site in the absence of BMP-2 and Smad1. The interaction of Smurf1 with the OSE2-a site was completely abolished when cells were transfected with Smad1 and treated with BMP-2 (Fig. 6c). These results suggest that Smad1 activates the Smad6 promoter by displacing Smurf1 and Smad1/Runx2 co-activate Smad6 gene transcription. Similar ChIP assay results for the interactions of Runx2, Smad1, and Smurf1 with the OSE2-a site were also obtained in 2T3 cells (data not shown).
DISCUSSION
In the present studies, we investigated mechanisms of BMP-2 signaling and Smad6 gene regulation. We identified and characterized a Runx2 binding site at the Smad6 promoter. Following BMP-2 stimulation, Smad1 interacts with Runx2 at the OSE2 site and stimulates Smad6 gene expression. In addition, we also discovered that the E3 ubiquitin ligase Smurf1 interacts with Runx2 at the OSE2 site and suppresses Smad6 gene transcription. More importantly, Smurf1 interacts with Runx2 and hinders Runx2 to activate Smad6 gene transcription. Smad1 displaces Smurf1 binding at the OSE2 site. Thus, Smad1 and Smurf1 appear to compete for their interaction with Runx2 at the OSE2 site and Smad6 gene expression appears related to their relative interaction. These results provide novel evidence about BMP signaling and regulation of Smad6 gene transcription.
BMP-2 plays an important role in osteoblast and chondrocyte differentiation but signaling mechanism remains unclear. Many osteoblast marker genes are activated by BMP-2 but lack GC-rich Smad1/5 binding elements in their 5′-promoter regions. Smads 1 and 5 have weak DNA binding activity, and they require interaction with other transcription factors to activate downstream target genes (3). Our previous results show that BMP-2 stimulated reporter activity (9xSBE-Luc or 12xSBE-Luc) only when multiple Smad1/5 binding elements (SBE) were cloned in front of an osteocalcin basal promoter containing an OSE2 site (29, 5). In contrast, BMP-2 activity is completely abolished when the OSE2 site was mutated in the osteocalcin basal promoter (5), suggesting a critical role of Runx2 in BMP-2 signaling. Other laboratories also demonstrate that mutations of Runx2 binding sites in type X collagen and COX-2 promoters inhibit effects of BMP-2 on activation of these promoters (15, 30). These findings demonstrate that interaction of Smad1 with Runx2 at the OSE2 site may be the major mechanism for BMP-2 to activate its downstream target genes in bone cells. Recently it has been reported that BMP-2 completely lost its stimulatory effects on expression of osteoblast marker genes in immortalized calvarial cells derived from Runx2−/− mice (31), demonstrating the requirement of Runx2 for BMP signaling in bone cells.
Smurf1 is a Hect domain E3 ubiquitin ligase and first identified by its interaction with Smads 1 and 5 and induces the degradation of these Smad proteins (28). We recently demonstrated that Smurf1 interacts with Runx2 and induces Runx2 ubiquitination and proteasome degradation (5, 27). Smurf1 interacts with the PY motif of its substrate proteins through its WW domain (28). A conserved PY motif has been identified in the C terminus of Runx family members by sequence analysis (27) and Smurf1 may induce Runx2 degradation through direct binding to Runx2 at the PY motif. We recently demonstrated an alternative mechanism of Runx2 degradation that was dependent upon Smurf1/Smad6/Runx2 interaction (27). In the present studies, we demonstrate that Smurf1 interacts with Runx2 at the OSE2 site of the Smad6 promoter and inhibits Smad6 promoter activity, suggesting that interaction of Smurf1 with Runx2 at the OSE2 site will hinder Runx2 to activate its downstream target gene.
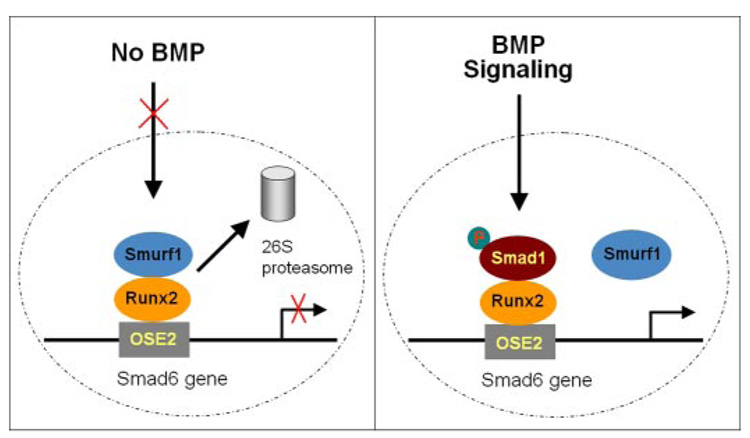
Based on our findings, we propose a model in which BMP-2 activates its downstream target gene Smad6 through Smad1 interaction with Runx2 at Runx2 DNA binding site and prevents Runx2 interaction with Smurf1 at the OSE2 site (Fig. 7).
FIGURE 7. A proposed model for BMP-2 activation of Smad6 gene transcription.
In the absence of BMP-2, Smurf1 binds Runx2 and induces Runx2 degradation. In the presence of BMP-2, Smad1 replaces Smurf1 and binds to the OSE2 site.
Footnotes
This work was supported in part by Grant 05YFJMJC01800 (to T. Z.) from the Natural Sciences Foundation, Tianjin, China and Grants R01 AR051189 and K02 AR052411 (to D. C.) from the National Institutes of Health.
The abbreviations used are: BMP-2, bone morphogenetic protein 2; Runx2, runt-related gene 2; Smurf1, Smad ubiquitin regulatory factor 1; IP, immunoprecipitation; ChIP, chromatin immunoprecipitation; wt, wild-type; mt, mutant; EMSA, electrophoretic mobility shift assay; GST, glutathione S-transferase.
REFERENCES
- 1.Kusanagi K, Inoue H, Ishidou Y, Mishima HK, Kawabata M, Miyazono K. Mol. Biol. Cell. 2000;11:555–565. doi: 10.1091/mbc.11.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishida W, Hamamoto T, Kusanagi K, Yagi K, Kawabata M, Takehara K, Sampath TK, Kato M, Miyazono K. J. Biol. Chem. 2000;275:6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- 3.Derynck R, Zhang Y, Feng XH. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. (Review) [DOI] [PubMed] [Google Scholar]
- 4.Hanai J, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, Stavnezer J, Kawabata M, Miyazono K, Ito Y. J. Biol. Chem. 1999;274:31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 5.Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. J. Biol. Chem. 2003;278:27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- 6.Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, Stein GS, Liu PP. Nat. Genet. 2002;32:639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 7.Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, Speck NA. Nat. Genet. 2002;32:645–649. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Nat. Genet. 2002;32:633–638. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 9.Ducy P, Karsenty G. Mol. Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geoffroy V, Ducy P, Karsenty G. J. Biol. Chem. 1995;270:30973–30979. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 11.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, Shimizu H, Yasui T, Ogihara H, Yasui N, Ochi T, Kitamura Y, Ito Y, Nomura S. Oncogene. 1998;17:1517–1525. doi: 10.1038/sj.onc.1202064. [DOI] [PubMed] [Google Scholar]
- 13.Kern B, Shen J, Starbuck M, Karsenty G. J. Biol. Chem. 2001;276:7101–7107. doi: 10.1074/jbc.M006215200. [DOI] [PubMed] [Google Scholar]
- 14.Javed A, Barnes GL, Jasanya BO, Stein JL, Gerstenfeld L, Lian JB, Stein GS. Mol. Cell. Biol. 2001;21:2891–2905. doi: 10.1128/MCB.21.8.2891-2905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leboy P, Grasso-Knight G, D’Angelo M, Volk SW, Lian JV, Drissi H, Stein GS, Adams SL. J. Bone Joint Surg. Am. 2001;83:S15–S22. [PubMed] [Google Scholar]
- 16.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. J. Cell Biol. 2003;162:833–842. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 18.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 19.Mundlos S, Mulliken JB, Abramson DL, Warman ML, Knoll JH, Olsen BR. Hum. Mol. Genet. 1995;4:71–75. doi: 10.1093/hmg/4.1.71. [DOI] [PubMed] [Google Scholar]
- 20.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JHM, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 21.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, Ochi T, Endo N, Kitamura Y, Kishimoto T, Komori T. Dev. Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 22.Kim IS, Otto F, Zabel B, Mundlos S. Mech. Dev. 1999;80:159–170. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- 23.Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, Kurisu K, Komori T. J. Cell Biol. 2001;153:87–100. doi: 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stricker S, Fundele R, Vortkamp A, Mundlos S. Dev. Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 25.Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O’Keefe RJ, Chen D. J. Biol. Chem. 2006;281:3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 29.Zhao M, Harris SE, Horn D, Geng Z, Nishimura R, Mundy GR, Chen D. J. Cell Biol. 2002;157:1049–1060. doi: 10.1083/jcb.200109012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chikazu D, Li X, Kawaguchi H, Sakuma Y, Voznesensky OS, Adams DJ, Xu M, Hoshio K, Katavic V, Herschman HR, Raisz LG, Pilbeam CC. J. Bone Miner. Res. 2002;17:1430–1440. doi: 10.1359/jbmr.2002.17.8.1430. [DOI] [PubMed] [Google Scholar]
- 31.Bae J-S, Gutierrez S, Narla R, Pratap J, Devados R, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Javed A. J. Cell. Biochem. 2007;100:434–449. doi: 10.1002/jcb.21039. [DOI] [PubMed] [Google Scholar]