Abstract
Cytotoxic T lymphocytes (CTL) control the replication of human cytomegalovirus (CMV). Previous studies assessed the clonotypic composition of CTL specific for individual immunodominant peptides within a certain HLA context. Such an approach has inherent limitations and may not assess the true clonal CTL response in vivo. Here, the clonotypic composition of CMV-specific CTL was determined in HLA-A2, CMV-seropositive kidney transplant recipients and healthy blood donors after stimulation of peripheral blood mononuclear cells with either a pp65 whole-peptide pool or a single immunodominant peptide. Even after stimulation with the whole peptide pool, CMV-specific CTL remained monoclonal or oligoclonal. Regarding intraindividual variation, the CDR3 motifs of the dominant clones were identical to those observed in CTL generated by the single immunodominant peptide. Sequencing of the CDR3 regions demonstrated significant interindividual variation; however, structural homology was observed for immunodominant clonotypes in three individuals. In conclusion, the highly focused T cell receptor repertoire found after stimulation with either a single immunodominant peptide or a peptide pool demonstrates a pivotal role for immunodominant epitopes in the generation of a clonal repertoire. These results provide new insights into the regulation of CMV clonal dominance and may contribute to the design and monitoring of adoptive immunotherapy.
Human cytomegalovirus (CMV) is a ubiquitous pathogen (seropositivity rate 60 to 90%) that can result in significant morbidity and mortality in immunocompromised hosts such as transplant recipients, HIV-positive patients, and congenitally infected newborns.1 Several lines of evidence, including clinical observations, laboratory studies, and animal models, indicate that the T cell immune response constitutes a key component in protection against CMV-related disease.2,3
Previously, CMV-specific cytotoxic CD8+ T cell (CTL) responses initiated through memory T cells in blood of seropositive healthy control subjects and immunosuppressed CMV carriers were analyzed, and CMV-specific T cells were quantified.4–6 Until recently, chromium-release assays and limiting-dilution T cell precursor analysis were the mostcommon experimental techniques used to measure specific T cell responses.7,8 Enzyme-linked immunospot assay analyzing antigen-induced cytokine secretion improved the detection.9 Furthermore, HLA peptide tetrameric complexes facilitate counting of antigen-specific T cells by flow cytometry10; however, use of such antigenic peptide-based reagents necessitates knowledge of both the immunodominant epitopes and their HLA restriction. In addition, responses to single immunodominant epitopes for which CMV tetramers are available may not be reflective of the response to whole antigen, whereas other epitopes dependent on other HLA-restriction elements may produce efficient T cell response in the same individual.1,11,12
Recent studies aiming at quantification of CMV-specific T cells performed by our and other groups used flow cytometric analysis of peptide-inducible intracellular IFN-γ production.13,14 Previously, we demonstrated that whole protein overlapping peptide libraries containing a full spectrum of T cell epitopes can be used to monitor frequencies of antigen-specific CD8+ and CD4+ lymphocytes12 as well as to generate CD4+ and CD8+ T cell lines capable of antigen-specific proliferation and effector function, such as killing.13,15 Such libraries consist of overlapping 15-mer peptides, each peptide 11–amino acid overlap and spanning the entire protein's amino acid sequence. Libraries designed this way should theoretically contain all possible 9- to 12-mer epitopes for all HLA types and do not require knowledge of individual epitopes/HLA restriction elements needed for T cell stimulation.
T cells recognize viral antigen via the T cell receptor (TCR) αβ heterodimer that is generated during thymic development by rearrangement of variable (V), diversity (D), and joining (J) gene segments. Additional TCR diversity is generated by imperfect joining of these segments, exonucleotide nibbling at the joins, and addition of non–germline-encoded N-region nucleotides. The regions spanning the V-D-J joins constitute the hypervariable CDR3 regions that interact with the middle of the bound peptide.4 Analysis of TCR diversity has been previously performed in many different diseases,16–20 but TCR repertoire of virus-specific T cells has been studied only for a few virus-derived peptides.4,17,21 These data are not reflective of conditions occurring in vivo, because other peptides than the known immunodominant one might play a role in clonal distribution of virus specific CTL.
We designed experiments to determine how an entire spectrum of virus derived peptides (peptide pool) may affect the clonal diversity of virus-specific CTL responses. We hypothesized that independent of the source of stimulation, identical immunodominant clone will be found within the same individual. To address the relationship between the type of antigenic stimulation and the clonal composition of CMV-specific effector memory T cells, we determined TCR repertoire in CMV pp65-specific cytotoxic T cells generated with pp65 peptide pool or NLVPMVATV (hereafter referred to as NLVP) single peptide (known to be immunodominant for HLA-A2) in HLA-A2, CMV-seropositive healthy, and immunosuppressed individuals.
RESULTS
Flow Cytometric Analysis of CMV-Reactive CD8+ T Cells
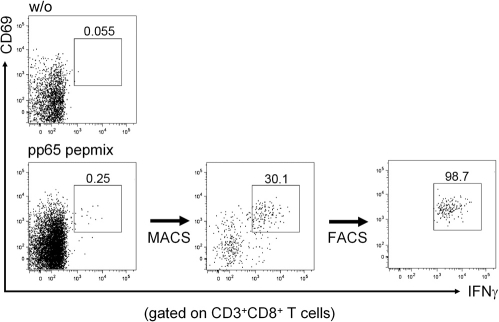
Functional characterization of CMV-reactive CD8+ T cells from seropositive individuals was performed after short time stimulation of PBMC with pp65 peptide pool or NLVP. CMV-specific CD3+CD8+ T cells were identified according to IFN-γ and CD69 coexpression. Purities of CMV-specific CD3+CD8+CD69+IFN-γ+ cells obtained directly after combined magnetic cell sorting (MACS) and FACS isolation ranged from 97 to 99.5% (Figure 1).
Figure 1.
Frequency of IFN-γ–positive T cells upon specific stimulation. PBMC were stimulated for 6 h with CMVpp65 peptide pool. Live antigen-specific cells were identified and magnetically enriched according to secreted IFN-γ (MACS). CD3+CD8+CD69+IFN-γ+ cells were further purified on a FACSAria cells sorter. Intracellular cytokine staining and flow cytometry analysis were performed for detection of the frequencies of IFN-γ–positive T cells. The figure shows frequency of IFN-γ–positive T cells after appropriate separation. The upper image (w/o) demonstrates an unstimulated negative control.
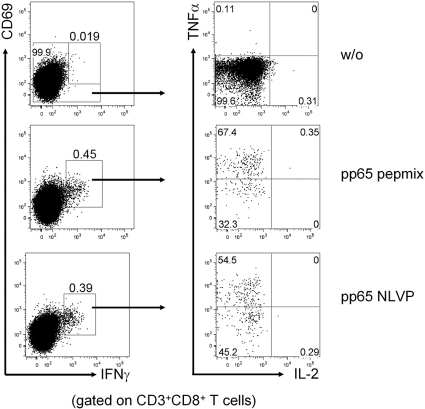
All generated cell lines contained mainly CD3+ lymphocytes (range 87 to 100%). The frequencies of expanded CD3− natural killer (NK) cells were <15% (range 0 to 13.9%). Distribution analysis of cytotoxic CD8+ lymphocytes and CD4+ helper T cells demonstrated that CTL constituted the predominant cell type in generated cell pools (Figure 2). Generated cell lines as well as the cells obtained directly after combined MACS and FACS isolation exhibited a typical effector/memory-like functional potential characterized by TNF-α but no IL-2 co-production (Figure 2).
Figure 2.
Dominance of effector memory CD8+ cells in generated T cells. PBMC were stimulated for 6 h with CMVpp65 single peptide/peptide pool in the presence of Brefeldin A. Antigen-specific CD3+CD8+ cells were identified according to coexpression of IFN-γ and CD69. Within this population, coexpression of TNF-α and IL-2 was studied. Quadrants were set according to total CD3+CD8+ T cells in the unstimulated sample. Examples for an unstimulated sample (DMSO, w/o) as well as for samples stimulated by pp65pepmix and pp65 NLVP peptide are shown. The cells exhibited a typical effector/memory phenotype characterized by IFN-γ and TNF-α but no IL-2 co-production.
Functional Characterization of CMV-Reactive T Cells
In the next set of experiments, we analyzed the specificity profile of cell lines generated through expansion in the presence of pp65-specific library. We tested in vitro killing efficiency for various target cells. Autologous targets loaded with pp65 peptide pools (that serve as a surrogate for infected cells), control peptides (that resemble uninfected cells), and allogeneic targets (as a model for a transplanted graft) were used. We observed dosage-dependent killing of specific target T cell lines generated from healthy and transplant donors. The mean killing of specific targets was 70% for transplant patients and 61% for healthy donors at an effector/target ratio of 20:1. The mean killing of autologous and allogeneic targets was <3% for healthy blood donors and transplant patients. NK cell activity was analyzed using K562 as an NK cell–sensitive target. Mean killing was 5 and 9% for healthy blood donors and transplant patients, respectively.
Monoclonal/Oligoclonal Composition of Effector Memory T Cells Obtained by pp65 Peptide-Pool Stimulation
To determine the clonal composition of pp65-specific effector memory T cells in asymptomatic healthy and immunosuppressed CMV carriers, we generated pp65-specific cytotoxic T cells using pp65 peptide pool or single NLVP peptide stimulation, respectively, MACS or combined MACS and FACS separation, and analyzed the TCR repertoire spectrum by sequencing the hypervariable V-D-J region of the TCR β chain. The number of different T cell clones contributing to the population of T cells specific for pp65 peptide pool or NLVP control peptide was determined, and the percentage of each clone within each individual cell population was calculated. We found mono- or oligoclonal composition of CTL in all study probands. In each of the nine individuals studied, the clonal composition of the effector memory T cells was highly focused: Six individuals demonstrated only one dominant CDR3-TCR β clonotype, in three other patients were detected two different CDR3 sequences with percentages of the immunodominant clonotypes within the whole clonotype repertoire ranging from 25 to 90% as demonstrated in Table 1. Comparable results were obtained from three independently performed sequencing analyses for each cell line (Table 1). To exclude any potential bias toward high-avidity clones resulted by culturing of MACS-sorted CMV-specific cells, we performed a parallel analysis of TCR repertoire of CMV pp65-specific T cells obtained directly after stimulation and separation by MACS combined with FACS in two blood donors. Identical immunodominant clones were found in each particular individual independent of the source of analyzed cells (MACS plus culture versus MACS plus FACS without culture), demonstrating, therefore, that the culture conditions did not alter the clonality spectrum of CTL generated by pp65 peptide pool.
Table 1.
Immunodominant TCR repertoire of analyzed CTL lines generated by pp65 peptide pool mix and single pp65 (NLVP) peptidea
| Patient | AA Sequence of ID Clone | Vβ Family | Jβ Family | % of All Clones |
|---|---|---|---|---|
| pp65 peptide pool mixb | ||||
| 1 | CASSLEQHTEAFF | 14S1 | 1–1 | 62 |
| 2 | CASSSQTGTIYGYTF | 13S1 | 1–2 | 52 |
| 4a | CASSSRQGADTGELFF | 6S4 | 2–2 | 25 |
| 4b | CASRGLSGLTEAFF | 13S1 | 1–1 | 33 |
| 5 | CASQGTAATNTGELFF | 17S1 | 2–2 | 56 |
| 9 | CASTWTGPDQPQHF | 17S1 | 1–5 | 38 |
| 14a | CAANQREPYEQYF | 13S1 | 2–7 | 48 |
| 14b | CASSAWTSGLNEQFF | 17S1 | 2–1 | 33 |
| 19 | CASSYQTGAAYGYTF | 13S1 | 1–2 | 90 |
| 28a | CASKWGPANPEAFF | 14S1 | 1–1 | 32 |
| 28b | CASKQGAGGNTEAFF | 14S1 | 1–1 | 41 |
| 30 | CASSPQTGVGYGYTF | 13S1 | 1–2 | 70 |
| Single pp65 (NLVP) peptide | ||||
| 1 | CASSLEQHTEAFF | 1–1 | 88 | |
| 4a | CASSSRQGADTGELFF | 2–2 | 35 | |
| 4b | CASRGLSGLTEAFF | 1–1 | 30 | |
| 5 | CASQGTAATNTGELFF | 2–2 | 86 | |
| 19 | CASSYQTGAAYGYTF | 13S1 | 1–2 | 79 |
| 28a | CASKWGPANPEAFF | 14S1 | 1–1 | 36 |
| 28b | CASKQGAGGNTEAFF | 14S1 | 1–1 | 38 |
The results were obtained from 3 independently performed sequencing analyses for each cell line/CTL separation. The mean size/percentage of the immunodominant clone is shown.
Cell lines obtained from patients 2, 19, and 30 showed the same Vβ and Jβ usage (italics), length of AA sequence, and central TCR motif (QTG, underline). For proving that the found TCR repertoire is not altered by in vitro expansion, TCR of pp65 peptide pool–specific cells obtained directly after their FACS separation were additionally analyzed in patients 1, 4, 5, 19, and 28. The found sequences of immunodominant clones were identical within the same patient.
Comparison of T Cell Repertoire in T Cell Lines Generated by Stimulation with a Single pp65 Immunodominant Peptide and the Whole pp65 Peptide Pool
Repertoire of T cells generated by stimulation with a single immunodominant peptide has been shown in previous studies4–6; however, the whole protein peptide library-stimulated cell pools may resemble more in vivo situation. Consequently, we compared clonotypic repertoire of T cells generated by single peptide and peptide pool, respectively. Using pp65 NLVP single peptide, known to be immunodominant for HLA-2–positive individuals, and pp65 peptide pool as a source for T cell stimulation, we obtained four different T cell lines in two healthy blood donors. In three other blood donors, highly purified antigen-specific T cells were obtained directly after appropriate stimulation (single peptide/peptide pool) and a combination of MACS and FACS isolation without culture. After sequencing of subcloned CDR3 region of TCR β chain, the amino acid motif of CDR3 was deduced and the frequencies of the found T cell clones assessed for each individual. The same immunodominant clone (identical clonotype) was found to be shared in each particular individual independent of the source of stimulation (peptide pool versus single immunodominant peptide; Table 1).
Analysis of pp65-Specific T Cells in CTL Lines and Peripheral Blood Using Clonotype-Specific PCR
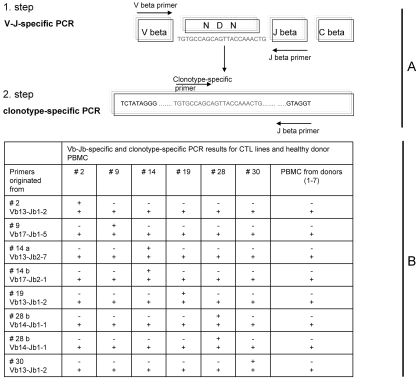
To establish that the TCR sequence found in each individual is unique and does not occur in other HLA-A2, CMV-seropositive patients, we performed PCR assay using clonotype-specific primers. To increase the sensitivity of the assay, we performed two-step (nested) PCR using Vβ forward/Jβ reverse (one-step) and clonotype-specific CDR3 forward/Jβ reverse (two-step) primer panels (Figure 3). All PCR amplification products were subsequently sequenced and PCR fidelity was confirmed. PBMC and/or CTL lines of all study individuals as well as PBMC of seven other HLA-2, CMV-seropositive healthy blood donors were analyzed by PCR using eight clonotype-specific primer panels. The created primer panels showed a very high specificity: Whereas a very bright PCR band was detected using DNA obtained from the patient whose CDR3 sequence was used to design this particular primer panel, no amplifications were seen in other HLA-2, CMV-seropositive individuals using the same primer panel (Figure 3).
Figure 3.
Clonotype-specific PCR confirmed individually unique structure of TCRβ chain CDR3. Genomic DNA from all generated CTL lines as well as from PBMC of seven HLA-2, CMV-seropositive healthy blood donors was amplified using clonotype-specific primer panels. Clonotypic forward primers were designed from immunodominant clonotypic sequences derived from each individual T cell line to span the CDR3 region. Jβ family region was used to design reverse primers. For increasing sensitivity of the assay, a two-step (nested) clonotypic PCR was performed. In the first step, Vβ family–specific forward primers with Jβ family–specific reverse primers were used to amplify an individual's V-D-J region. PCR products (including blank) were diluted 1:100 in water, and 6 μl was used in a second-step (clonotype-specific) PCR (for sequences, see Table 2). Whereas all samples demonstrated amplification bands in the first-step PCR (+), clonotype-specific PCR products were detected only in individuals from which clonotypes were originated (+). No amplifications were seen in other HLA-2, CMV-seropositive individuals using the same primer panel (−). (A) Design of clonotype-specific nested PCR. The specificity of the product was confirmed by sequencing of the amplicon. (B) Results on clonotype-specific PCR for generated CMV-CTL lines and PBMC using all designed individual clonotype-specific primer panels
DISCUSSION
The immune response mediated by antigen-specific T cells plays a pivotal role in the course and the outcome of CMV infection or reactivation. The introduction of MHC-peptide tetramers made possible the individual analysis of the peptide-responsive T cell clones on the basis of the specificity of their TCR. Molecular analysis of TCR repertoire enabled investigation of the clonotypic TCR repertoire and quantification of individual T cell clones without the knowledge of their TCR specificity, as we previously described for aplastic anemia, myelodysplastic syndrome, and large granular lymphocyte.18–20 Clonotypic composition of CMV-specific CTL has been investigated in various experimental systems. With the use of a single immunodominant CMV pp65-derived peptide for CTL stimulation, mono/oligoclonal T cell response was demonstrated in previous studies4–6,22; however, this type of T cell stimulation may not always accurately reflect in vivo conditions. Consequently, we applied the whole-protein peptide library for stimulation of pp65-specific T cells and separated the cells by IFN-γ secretion assay MACS with or without combination of FACS.
Our study revealed that although a very high degree of interindividual clonal diversity exists within CMV T cell recognition spectrum, a marked clonal restriction among the effector memory T cells is generated from one donor using a CMV pp65 peptide library and/or a single immunodominant pp65 peptide. Within individual donors, a remarkable clonal sharing was observed; identical immunodominant clones were isolated in CTL generated by stimulation with pp65 single immunodominant peptide and pp65 whole-protein peptide pool, respectively. This finding suggests the importance of the immunodominant peptide in the intrinsically polyclonal CTL immune response. Our results are in general agreement with previous reports showing mono/oligoclonal composition of T cells obtained after stimulation with viral immunodominant peptides.4,5,11,22 Although the strong intraindividual TCR focusing was demonstrated in studies with CMV and other antigenic targets,4,11 the most important difference between our study and previous results is that the previous studies used T cell generated by single peptide stimulation. In contrast, we obtained CMV-responsive T cell populations using stimulation with all potential pp65 peptide epitopes, which could at least theoretically lead to a very large clonal diversity. The identical sequences of the immunodominant T cell clones obtained after stimulation with single peptide and peptide pool suggest the importance of the dominant peptide in the evolution of antiviral immune response.
The TCR clonotypes derived from CTL of different donors that recognize the same peptide-MHC complex were not identical but often may show some conservation of gene Vβ segment usage, although they differ in hypervariable sequence.4 Consequently, in CTL clones derived from different donors, individual Vβ family and β chain CDR3 motifs were conserved between TCR that recognized an HLA-A11 restricted Epstein-Barr virus peptide.17 Similar observation was made for two different influenza virus peptides.23,24 In our study, CMV-specific T cells displayed an even higher degree of TCR conservation in three unrelated individuals. The conserved CTL clonotypes were the only one shared between individuals that showed very high levels of expansion despite that these T cell lines were generated by stimulation not by a single permissive peptide but by a whole protein peptide pool. Similar CDR3 Vβ clonotypes were found in two additional HLA-2, CMV-seropositive individuals reported by another group.5 This finding strongly suggests that immunodominant clonal responses are likely to include identical/similar targets and consequently, if identified, can be used as marker clonotypes for PCR-based monitoring.
Although the high conservation of TCR motif in CTL clones obtained by stimulation with a single immunodominant peptide has been already shown by other groups, our data demonstrate to our knowledge for the first time the highly focused TCR repertoire in dominant clones reactive for the entire pp65 peptide pool. This observation might affect the design of adoptive T cell therapy. Further functional studies should be performed to prove the role of CDR3 conservation in epitope recognition.
Except for the similarity of the CDR3 motif in three patients, the found genotypes seem to be highly unique. Using clonotype-specific primer designed from the obtained TCR sequencing, we analyzed the presence of the identical T cell clones in different HLA-2, CMV-seropositive transplant patients and healthy blood donors. In agreement with other authors,4,6 public TCR motifs were not found. Moreover, comparative analysis of the found immunodominant TCR sequences with those of the database containing more than 5000 TCR sequences identified by sequencing in our laboratory did not reveal identical amino acid/nucleotide sequences (data not shown).
One important limitation of our experiments was the purity of the T cell in one part of the population studied. Unlike other studies, in which T cells used for CDR3 sequencing were separated by flow cytometric sorting, T cells in some blood donors were obtained by immunomagnetic selection of activated IFN-secreting cells and expansion in vitro; however, despite the possible presence of unspecific “background” cells within the analyzed T cell population, we were able to identify clearly immunodominant clones with a high percentage of clone-related colonies (in five patients >50% and in one patient 38% of all colonies belonged to one clone).
Another important limitation is a potential bias in results that can be introduced at multiple steps of molecular TCR analysis. Preferential amplification of the immunodominant clone family over background clones during multiplex PCR can be one of the most important limitations; however, this mechanism may constitute an advantage in that irrelevant low copy number clones are experimentally “diluted.” Clonal size could also be affected during selection of colonies for further amplification. Because the large number of colonies was obtained for the sequencing and the colonies were picked up randomly from the whole plate, the bias in the results by this step seems to be unlikely. In any event, while detection of immunodominance is facilitated, the precise clonal sizes may be over- or underestimated. Of note is that all results were confirmed by three independently performed cloning/sequencing analyses for each cell line.
Furthermore, it could be assumed that the found TCR repertoire of CMV pp65-specific T cell lines obtained after MACS separation with culture is altered by in vitro expansion. To examine/exclude this, we additionally analyzed the TCR repertoire of pp65-specific cells obtained directly after combined MACS and FACS separation in the same blood donors. The found sequences of immunodominant clones were identical to those obtained after single peptide/peptide pool stimulation and subsequent culture, confirming, therefore, that the culture conditions did not bring bias toward high-avidity clones.
What can be the reason for the high degree of intraindividual clonal restriction observed in individuals studied? One of the possible explanations is an affinity/avidity-driven selection process.5 Preferential expansion of high-affinity/avidity clonotypes could explain the overrepresentation of the certain clonotypes among the clones found in our patients. As already discussed by several authors, the role of TCR affinity/avidity in peripheral T cell clonal selection remains unclear.5 Selective loss of clonotypes expressing low-affinity TCR after primary antigenic stimulation was found to occur without further selection above this affinity threshold of clones expressing TCR with the highest affinity or half-life.5 The possibility of further clonal focusing on secondary/chronic antigenic challenges and subsequently the mechanisms underlying this phenomenon remain a subject of speculation. It is important to note that another possible explanation for the high degree of clonal focusing is repeated exposure to the viral antigen with preferential selection of T cells expressing high-affinity TCR during maturation into long-term memory. Our results suggest that obviously only one peptide is responsible for the generation of one immunodominant T cell clone.
By combining peptide pool–based IFN-secretion MACS/FACS assay and subsequent TCR sequencing, we demonstrated a new approach for the clonotypic analysis of pp65-specific T cells independent of knowledge of donor's HLA type or immunodominant epitopes. Major disadvantages of peptide-based protocols reported in other studies arise from the restricted knowledge of immunodominant epitopes. In most viral diseases, relevant epitopes are known for certain HLA types, such as HLA-A1, -A2, or -B7, but knowledge is limited for others. Moreover, in some patients, a peptide known to be immunodominant within certain HLA context can induce a very low response or no response at all, whereas another peptide that would not be expected on the basis of the HLA background induces a very high reactivity.5 In our protocol, specific stimulation was provided by peptide libraries loaded on autologous PBMC. Peptide libraries are designed and synthesized solely on the basis of the protein's amino acid sequence and, theoretically, contain all T cell epitopes for all existing HLA types. Thus, in contrast to most peptide-based approaches, the protocol applied in our study does not depend on previously identified epitopes.
In summary, we demonstrate a new effective, HLA-independent method for the analysis of TCR motif and CTL clonotype using CMV pp65 overlapping peptide pool spanning the entire protein amino acid sequence. Generation of clonal repertoire seems to be influenced mainly by the immunodominant peptides, because identical TCR repertoire was found within certain individuals after stimulation with both a single immunodominant peptide and a peptide pool. This finding could be predicted but has not been shown so far. Assuming the leading role of the immunodominant CTL clones for the protective CMV immunity, our data may have important implication for the design and application of the adoptive immunotherapy as well as creation of molecular tools for monitoring strategies.
CONCISE METHODS
Patients
After informed consent was obtained, blood samples were collected from randomly selected kidney transplant patients and healthy control subjects on the basis of the presence of HLA-A2 and seropositivity for CMV IgG and according to the protocols approved by the institutional review board of the Cleveland Clinic Foundation and Charité University Medicine, Berlin. Healthy blood donors and transplant patients showed no signs of active viral replication (no measurable CMV load, negative pp65-antigenemia test). Kidney transplant blood donors were recruited from the Transplant Outpatients Unit, Charité, Campus Virchow, University Medicine, Berlin. The mean transplant age of the patients was 6.3 yr, the patients were on a triple immunosuppressive therapy (tacrolimus, mycophenolate mofetil, steroid) with a current stable graft function and no other clinical complications.
Isolation of CMV-Specific T Cells
CMV-specific T cells were obtained after appropriate stimulation (peptide pool or single peptide) and IFN-γ secretion assay using MACS (six donors) or MACS in combination with FACS (three donors) technology. Because the number and purity of MACS-separated T cells were not high enough for direct TCR analysis, the cells were cultured as described in the next section (generation of CTL lines). FACS-generated T cells were directly used for TCR analysis.
Combined MACS and FACS Approach for Isolation of Live CMV-Specific CD3+CD8+ T Cells According to Secreted IFN-γ
PBMC of CMV-positive donors were isolated from 30 ml of blood by Ficoll gradient centrifugation (Amersham Bioscience, Munich, Germany). PBMC were stimulated at a density of 107/ml for 6 h with 1 μg/ml CMVpp65 peptide pool (15-mers, 11–amino acid overlap) or with 1 μg/ml CMVpp65 derived A2 restricted NLVP peptide (Jerini Peptide Technologies [JPT], Berlin, Germany) in the presence of 1 μg/ml anti-CD28 (BD Biosciences, San Jose, CA). After stimulation, cells were stained for secreted IFN-γ by cytokine secretion assay technology according to the manufacturer's protocol (Miltenyi Biotec, Bergisch-Gladbach, Germany). In addition, CD3, CD8, and CD69 were co-stained (BD Biosciences). After pre-enrichment of IFN-γ+ T cells over an LS+ MACS-column (Miltenyi Biotec), live antigen-specific CD3+CD8+PI+CD69+IFN-γ+ T cells were sorted on a FACS Aria (BD Biosciences) and lysed using Qiagen RLT Buffer (Qiagen, Hilden, Germany). Purity was checked on a FACS LSRII (BD Biosciences).
Generation of pp65- and NLVP-Specific CTL Lines
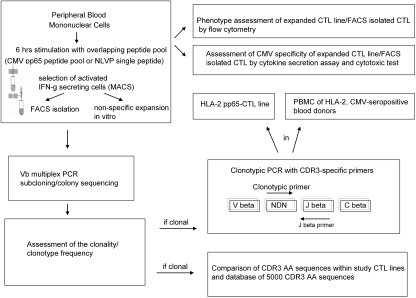
CMV pp65- or NLVP-specific CTL were generated as recently described by our group.25 Briefly, PBMC were isolated from 30 ml of blood by Ficoll gradient centrifugation (Amersham Bioscience, Munich, Germany). PBMC were stimulated for 6 h with 1 μg/ml for each peptide using an overlapping pp65 peptide pool (JPT). Thereafter, IFN-γ–secreting cells were labeled and isolated using the immunobead-based IFN-γ secretion assay according to the manufacturer's instructions (Miltenyi Biotec). Cells selected on the basis of IFN-γ secretion profile were cultured in 24-well plates in the presence of irradiated autologous feeder cells in complete medium (RPMI + 10% FCS + 1% penicillin/streptomycin; Biochrom, Berlin, Germany) and 100 U/ml rIL-2 (Chiron, Munich, Germany). The medium was changed on day 4 and then every 2 d for the next 20 to 30 d. After formation of confluent cell layers, cells were divided 1:1. Figure 4 illustrates the procedure for generation of the T cell lines and the study design.
Figure 4.
Experimental study design.
Identification and Functional Characterization of CMV-Specific CD8+ T Cells
PBMC of CMV-positive donors were stimulated at a density of 107/ml for 6 h with 1 μg/ml CMVpp65 peptide pool (15-mers, 11–amino acid overlap) or with 1 μg/ml CMVpp65 derived A2 restricted NLVP peptide (JPT) or DMSO (negative control) in the presence of 1 μg/ml anti-CD28 (BD Biosciences). Brefeldin A was added for the last 4 h to inhibit secretion of cytokines. After stimulation, cells were fixed (FACSLysing solution; BD Biosciences); permeabilized (FACSPerm solution; BD Biosciences); and stained for CD3, CD8, CD69, IFN-γ, TNF-α, and IL-2. Cells were analyzed on a FACS LSRII (BD Biosciences; Figure 1 and 2). Data were evaluated using FlowJo software.
Cytotoxicity Assay
A modified Calcein-AM release assay was used for cytotoxicity testing. Target cells were labeled with Calcein-AM (15 mM) according to the manufacturer's instructions (MoBiTec, Gottingen, Germany). Target cells (1 × 104) were co-cultured with T cell lines in a 96-well plate for 4 h at decreasing effector/target ratios. For spontaneous and maximum release, targets were incubated with complete media or 0.9% Triton X100 (Sigma-Aldrich, Hamburg, Germany). Samples were analyzed in quadruplicate using a fluorescence plate reader (Tecan, Zurich, Switzerland). Lytic activity was calculated as follows: Specific cytotoxicity = (sample release − spontaneous release)/(maximum release − spontaneous release) × 100%.
Outliers were excluded by the Nalimow's test using the formula
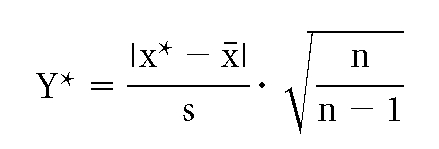 |
where Y* is sensitivity, x* is possible outliner, x is mean value, s is standard variation, and n is number of measurement. The sensitivity was set at 1.645, yielding a 95% probability that x* was not an outlier if the result was <1.645.
Multiplex TCR Vβ PCR
The hypervariable regions of TCR Vβ chain were amplified by a two-tube multiplex PCR with primer mix that covers all Vβ TCR gene rearrangements on genomic DNA or cDNA samples as described previously.18 RNA was isolated from separated cells by Absolutely RNA Miniprep Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The RNA was quantified by the Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA) and used for cDNA synthesis as described elsewhere.26 DNA was isolated using a commercially available DNA extraction kit (QIAamp Blood Kit; Qiagen) according to the manufacturer's instructions.
CDR3 Cloning and Sequencing
PCR products were separated on a 1.7% agarose gel, excised, and purified using the Gel Extraction Kit (Eppendorf, Hamburg, Germany) following the manufacturer's instructions. Six microliters of the purified PCR product was ligated into the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, CA) followed by overnight incubation at 14°C. Ligations were heat shock–transformed into TOP10F Escherichia coli and plated on agarose plate containing X-gal and ampicillin (100 μg/ml) followed by overnight incubation. For each individual, 32 to 40 bacterial colonies were picked up and analyzed. Colony PCR and subsequent sequencing of positive colonies were performed as described previously.18 Vβ CDR3 region sequences were analyzed using the ImMunoGeneTics information system TCR alignment tool (http://imgt.cines.fr) as described previously.18 An expanded immunodominant clonotype was defined according to its frequency among sequenced Vβ CDR3 regions. The frequency of the clones was defined as percentage of the particular unique clonotype out of total number of sequenced colonies as described previously.18,20 In this study, we focused on comparison of CDR3 regions that include invariant portions of Vβ and Jβ chains (in accordance with the CDR3 numbering proposed on the ImMunoGeneTics web site: http://imgt.cines.fr). To validate fidelity of the results, we performed all steps including multiplex PCR, subcloning, and sequencing analysis in triplicate, for each cell line/CTL separation independently.
Clonotype-Specific PCR in CTL and PBMC
Clonotypic forward primers were designed from immunodominant clonotypic sequences derived from each individual T cell line to span the CDR3 region and were used with Jβ family–specific reverse primers (for sequences, see Table 2). To increase sensitivity of the assay, a two-step clonotypic PCR was performed. In the first-step, Vβ family–specific forward primers with Jβ family–specific reverse primers were used to amplify individual's V-D-J region. PCR products (including blank) were diluted 1:100 in water, and 6 μl was used in a second-step PCR (for sequences, see Table 2). PCR products were analyzed on a 2.5% agarose gel. The fidelity of the clonotypic PCR was verified by direct sequencing of CDR3 amplicons.
Table 2.
Primer sequences used for PCR assays
| Patient | Vβ Family | Jβ Family | Forward Primer |
|---|---|---|---|
| Clonotype-specific primer (CDR3 region) | |||
| 2 | 13S1 | 1–2 | 5′-TGTGCCAGCAGTTCCCAAACAG |
| 9 | 17S1 | 1–5 | 5′-TGTGCCAGTACCTGGACAGGAC |
| 14 | 13S1 | 2–7 | 5′-TGTGCTGCCAATCAGCGGGAA |
| 17S1 | 2–1 | 5′-TGTGCCAGTAGTGCCTGGACTA | |
| 19 | 13S1 | 1–2 | 5′-TGTGCCAGCAGTTACCAAACTG |
| 28 | 14S1 | 1–1 | 5′-TGTGCCAGCAAGTGGGGGC |
| 14S1 | 1–1 | 5′-TGTGCCAGCAAACAGGGG | |
| 30 | 13S1 | 1–2 | 5′-TGTGCCAGCAGTCCACAGACAG |
| Jβ family primer | |||
| 1–1 | 5′-CTTACCTACAACTGTGAGTCTGGTG | ||
| 1–2 | 5′-CTTACCTACAACGGTTAACCTGGTC | ||
| 1–5 | 5′-CTTACCTAGGATGGAGAGTCGAGTC | ||
| 2–1 | 5′-CCTTCTTACCTAGCACGGTGA | ||
| 2–7 | 5′-CTTACCTGTGACCGTGAGCCTG | ||
| Vβ family primer | |||
| 13S1 | 5′-ATACATGTACTGGTATCGACAAGAC | ||
| 14S1 | 5′-GTATATGTCCTGGTATCGACAAGA | ||
| 17S1 | 5′-GGCCATGTACTGGTACCGACA |
DISCLOSURES
None.
Acknowledgments
The study was supported in part by the Rahel-Hirsch-Stiftung and Elsa-Kroener-Stiftung, Germany, to N.B.; National Institutes of Health grants RO1 HL073429-02, RO1 CA113972-02, K24 HL077522-01A1, and U54 RR019391-01 to J.P.M.; and BMBF Germany (BCRT) to P.R.
We gratefully acknowledge Dr. Andrew Schade for providing CMV serology data on healthy blood donors. We also thank Zachary Nearman for editing the manuscript.
Published online ahead of print. Publication date available at www.jasn.org.
REFERENCES
- 1.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA: Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol 169: 1984–1992, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Diamond DJ, York J, Sun JY, Wright CL, Forman SJ: Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood 90: 1751–1767, 1997 [PubMed] [Google Scholar]
- 3.Straten PT, Guldberg P, Seremet T, Reisfeld RA, Zeuthen J, Becker JC: Activation of preexisting T cell clones by targeted interleukin-2 therapy. Proc Natl Acad Sci U S A 95: 8785–8790, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weekes MP, Wills MR, Mynard K, Carmichael AJ, Sissons JG: The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J Virol 73: 2099–2108, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M: Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J Immunol 175: 6123–6132, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Peggs K, Verfuerth S, Pizzey A, Ainsworth J, Moss P, Mackinnon S: Characterization of human cytomegalovirus peptide-specific CD8+ T-cell repertoire diversity following in vitro restimulation by antigen-pulsed dendritic cells. Blood 99: 213–223, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Iglesias E, Samri A, Kamkamidze G, Decoville T, Carcelain G, Autran B: A systematic comparison of methods to measure HIV-1 specific CD8 T cells. J Immunol Methods 272: 23–34, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Waldman WJ, Adams PW, Orosz CG, Sedmak DD: T lymphocyte activation by cytomegalovirus-infected, allogeneic cultured human endothelial cells. Transplantation 54: 887–896, 1992 [DOI] [PubMed] [Google Scholar]
- 9.Heeger PS, Forsthuber T, Shive C, Biekert E, Genain C, Hofstetter HH, Karulin A, Lehmann PV: Revisiting tolerance induced by autoantigen in incomplete Freund's adjuvant. J Immunol 164: 5771–5781, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R: Counting antigen-specific CD8 T cells: A re-evaluation of bystander activation during viral infection. Immunity 8: 177–187, 1998 [DOI] [PubMed] [Google Scholar]
- 11.Betts MR, Casazza JP, Patterson BA, Waldrop S, Trigona W, Fu TM, Kern F, Picker LJ, Koup RA: Putative immunodominant human immunodeficiency virus-specific CD8(+) T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J Virol 74: 9144–9151, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kern F, Faulhaber N, Frömmel C, Khatamzas E, Prösch S, Schönemann C, Kretzschmar I, Volkmer-Engert R, Volk HD, Reinke P: Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur J Immunol 30: 1676–1682, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Ogg G, McMichael A: HLA-peptide tetrameric complexes. Curr Opin Immunol 10: 393–396, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Kern F, Surel IP, Brock C, Freistedt B, Radtke H, Scheffold A, Blasczyk R, Reinke P, Schneider-Mergener J, Radbruch A, Walden P, Volk HD: T cell epitope mapping by flow cytometry. Nat Med 4: 975–978, 1998 [DOI] [PubMed] [Google Scholar]
- 15.Hammer MH, Meyer S, Brestrich G, Moosmann A, Kern F, Tesfa L, Babel N, Mittenzweig A, Rooney CM, Hammerschmidt W, Volk HD, Reinke P: HLA type-independent generation of antigen-specific T cells for adoptive immunotherapy. Eur J Immunol 35: 2250–2258, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Melenhorst JJ, Sorbara L, Kirby M, Hensel NF, Barrett AJ: Large granular lymphocyte leukaemia is characterized by a clonal T-cell receptor rearrangement in both memory and effector CD8(+) lymphocyte populations. Br J Haematol 112: 189–194, 2001 [DOI] [PubMed] [Google Scholar]
- 17.Campos-Lima PO, Levitsky V, Imreh MP, Gavioli R, Masucci MG: Epitope-dependent selection of highly restricted or diverse T cell receptor repertoires in response to persistent infection by Epstein-Barr virus. J Exp Med 186: 83–89, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wlodarski M, O'Keefe CL, Washawski I, Loughran TP, Rodriguez A, Maciejewski JP: Pathologic clonal cytotoxic T-cell responses: Nonrandom nature of the T-cell-receptor restriction in large granular lymphocytic leukemia. Blood 106: 2769–2780, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Risitano AM, Maciejewski JP, Muranski P, Wlodarski M, O'Keefe C, Sloand EM, Young NS: Large granular lymphocyte (LGL)-like clonal expansions in paroxysmal nocturnal hemoglobinuria (PNH) patients. Leukemia 19: 217–222, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Wlodarski MW, Gondek LP, Nearman ZP, Plasilova M, Kalaycio M, Hsi ED, Maciejewski JP: Molecular strategies for detection and quantitation of clonal cytotoxic T-cell responses in aplastic anemia and myelodysplastic syndrome. Blood 108: 2632–2641, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weekes MP, Wills MR, Mynard K, Hicks R, Sissons JG, Carmichael AJ: Large clonal expansions of human virus-specific memory cytotoxic T lymphocytes within the CD57+ CD28- CD8+ T-cell population. Immunology 98: 443–449, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan N, Best D, Bruton R, Nayak L, Rickinson AB, Moss PA: T cell recognition patterns of immunodominant cytomegalovirus antigens in primary and persistent infection. J Immunol 178: 4455–4465, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Bowness P, Moss PA, Rowland Jones S, Bell JI, McMichael AJ: Conservation of T cell receptor usage by HLA B27-restricted influenza-specific cytotoxic T lymphocytes suggests a general pattern for antigen-specific major histocompatibility complex class I-restricted responses. Eur J Immunol 23: 1417–1421, 1993 [DOI] [PubMed] [Google Scholar]
- 24.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK: Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J Exp Med 181: 89–91, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammer MH, Brestrich G, Andree H, Engelmann E, Rosenberger C, Tillmann H, Zwinger S, Babel N, Nickel P, Volk HD, Reinke P: HLA type-independent method to monitor polyoma BK virus-specific CD4 and CD8 T-cell immunity. Am J Transplant 6: 625–631, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Kotsch K, Mashreghi MF, Bold G, Tretow P, Beyer J, Matz M, Hoerstrup J, Pratschke J, Ding R, Suthanthiran M, Volk HD, Reinke P: Enhanced granulysin mRNA expression in urinary sediment in early and delayed acute renal allograft rejection. Transplantation 77: 1866–1875, 2004 [DOI] [PubMed] [Google Scholar]