Abstract
Discrepancy between the abundance of cognate protein and RNA molecules is frequently observed. A theoretical understanding of this discrepancy remains elusive, and it is frequently described as surprises and/or technical difficulties in the literature. Protein and RNA represent different steps of the multi-stepped cellular genetic information flow process, in which they are dynamically produced and degraded. This paper explores a comparison with a similar process in computers - multi-step information flow from storage level to the execution level. Functional similarities can be found in almost every facet of the retrieval process. Firstly, common architecture is shared, as the ribonome (RNA space) and the proteome (protein space) are functionally similar to the computer primary memory and the computer cache memory respectively. Secondly, the retrieval process functions, in both systems, to support the operation of dynamic networks – biochemical regulatory networks in cells and, in computers, the virtual networks (of CPU instructions) that the CPU travels through while executing computer programs. Moreover, many regulatory techniques are implemented in computers at each step of the information retrieval process, with a goal of optimizing system performance. Cellular counterparts can be easily identified for these regulatory techniques. In other words, this comparative study attempted to utilize theoretical insight from computer system design principles as catalysis to sketch an integrative view of the gene expression process, that is, how it functions to ensure efficient operation of the overall cellular regulatory network. In context of this bird’s-eye view, discrepancy between protein and RNA abundance became a logical observation one would expect. It was suggested that this discrepancy, when interpreted in the context of system operation, serves as a potential source of information to decipher regulatory logics underneath biochemical network operation.
1. Introduction
The genomic sequences are readily available for an increasing number of species. These sequences, like English literature, represent static strings of symbols/alphabets (A, T, C, and G). Hence, genomic sequences are often termed as the “book” of life. The “reader” of the book is usually a cell. Reading English literature requires sufficient knowledge of English grammar and the meaning of the words; the literature would represent meaningless one-dimensional string of alphabets to people who do not have any basic knowledge of the language. Likewise, an urgent task in biology is to elucidate how cells “read” the “book” (Searls, 2001; Wang, 2005), i.e., the mechanisms by which cells utilize embedded information to specify systems operations. A key step is dynamic information retrieval from the genome so that a specific set of genes is expressed at specific physiological conditions. Major steps in this process include transcription, splicing in case of higher eukaryote, and translation into proteins, which execute encoded actions upon transportation to their cellular destination. An integral component of cellular regulatory machinery, this multi-step process is tremendously complex and tightly regulated. A lack of bird’s-eye views of this dynamic process, in my view, represents a major impediment in systems biology research.
Application of genomic and proteomic technologies has generated large amounts of gene expression data, primarily in the form of mRNA and protein abundance. These data are frequently used in statistical inference of biochemical network models (Lee et al., 2004; Lu et al., 2005), however, with very limited success in generating high-quality predictive models. Theoretical interpretations of these data that would better guide such network model inference efforts, thus, remain elusive. Indeed, many observations still puzzle us. In particular, mRNA abundance correlates too weakly with protein abundance for it to be a reliable predictor of protein abundance. This discrepancy has long been observed (Anderson and Seilhamer, 1997; Gygi et al., 1999). The discrepancy is further confirmed by more recent studies using high-throughput proteomic techniques (Flory et al., 2006; Ghaemmaghami et al., 2003; Griffin et al., 2002; Ideker et al., 2001; Le Roch et al., 2004; Tian et al., 2004; Washburn et al., 2003). Such prevalent observations are unlikely to be merely noises; these discrepancies might in stead prove to be informative (Greenbaum et al., 2003). A plausible explanation from the perspective of cellular system operation, on the other hand, has yet to be devised.
The scheme of utilizing one-dimensional simplistic codes to manage complex system operations is, on the other hand, not unique to the cells. A computer stores all necessary information in the hard disk/drive and dynamically retrieves specific sections under specific conditions, analogous to the permanent storage of genetic information in the genome and the dynamic gene expression (information retrieval) process respectively. Moreover, functional similarity has long been discussed between proteins and computational elements (Bray, 1995), as well as between cellular processes and computational processes (Bray, 1990; Brent and Bruck, 2006). Biological materials have been used to assemble computing machineries for challenging issues (Adleman, 1994; Unger and Moult, 2006). It was further suggested that a cell can be studied as a DNA-based molecular computer (Ji, 1999). A comparative examination of system architectures suggested that a computer, even though a much simpler system, shares common functional components with a cell (Wang and Gribskov, 2005). As they are engineered by us, we have a complete understanding of computers. It was therefore suggested that systems biologists look into computer system design for theoretical insights in analyzing cellular systems, as simpler model systems have historically been used in biology, such as the use of yeast as a model organism for higher eukaryotes. This is also consistent with an underlying notion in systems biology, that is, to explore similarities between biological and engineered complex systems (Csete and Doyle, 2002; Zheng, 2006).
This paper explored a detailed comparison of cellular gene expression process with computer information retrieval process. Remarkable similarities were discovered. It became obvious that the ribonome (RNA space) can be treated as cellular equivalent to computer primary memory, and the proteome (protein space) to computer cache memory. The computer memory management principles, which are vital for computer process management and system optimization, provide useful insights for an integrative understanding of cellular gene expression regulation. In particular, they provide a starting framework for an interpretation, in the context of cellular process management, of the discrepancy between mRNA and protein abundance.
2. Analysis and Discussion
2.1: Information and System Operation
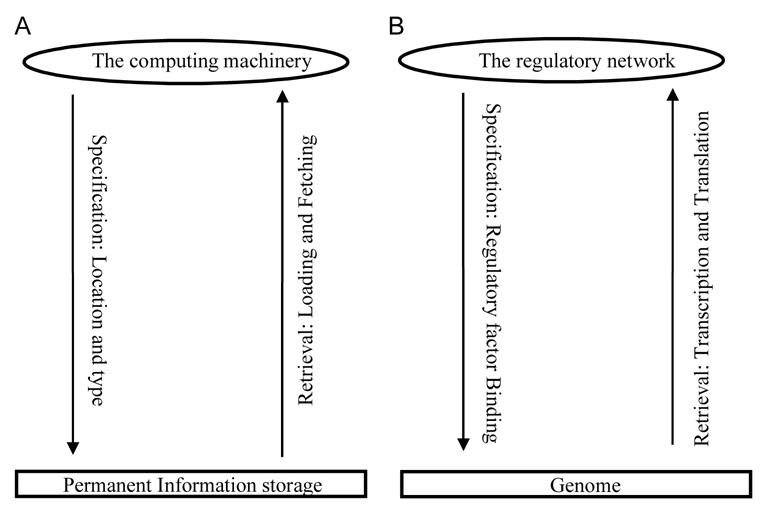
A striking analogy between a computer and a cell appears to exist in that they both use seemingly one-dimensional codes to regulate the operation of a multi-layered dynamic system (Wang and Gribskov, 2005) ( Fig. 1). In a cell, it is the quadruple genomic code. In a computer, it is the binary code carried by the information storage devices, primarily the hard drive. The code provides instruction for an operation such as adaptation to an environmental signal in cells or initiation of a computing process upon a user keyboard input in computers. The state of the system, on the other hand, determines how the codes will be interpreted and/or used. In the cells, the state of the cellular machinery determines which genes should be expressed or shutdown. In computers, this means where in the memory the CPU will go to fetch data or instructions (Wang, 2005).
Figure 1. The information utilization cycle in a computer (A) and in a cell (B).
A computer retrieves information through memory loading, and CPU instruction fetching (See Figure 2A). A cell retrieves genetic information through transcription, splicing in case of higher eukaryote, and translation (See Figure 2B).
2.2: Multi-tiered Memory Hierarchies
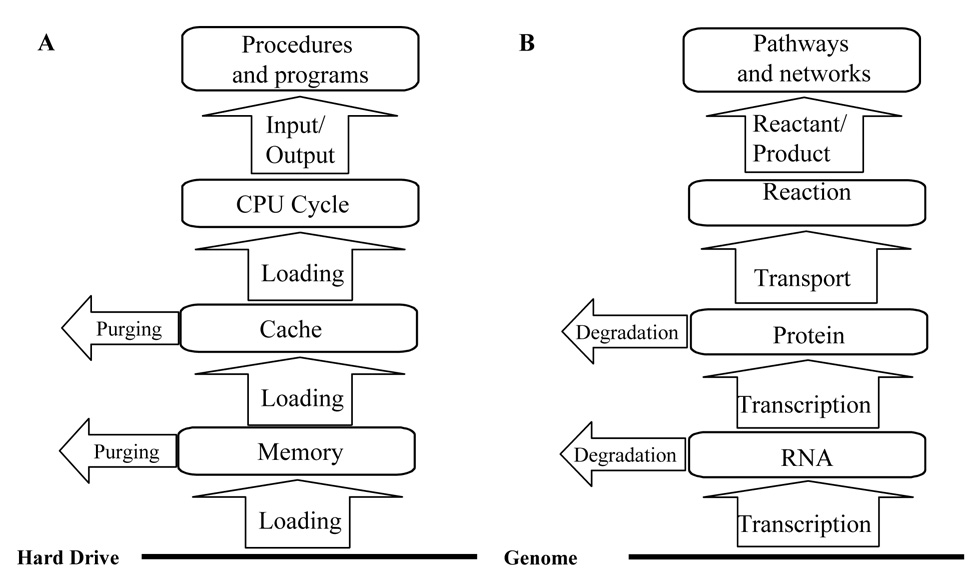
Integral to both systems is the information utilization cycle, which consists of their dynamic retrieval out of storage, execution of actions they encode, and their dispersal (Fig. 1). The information, before reaching execution stage, flows through multiple steps in the two systems. This retrieval process is termed gene expression in biology, with protein and RNA representing different steps (Figure 2B). In computers, this multi-step process flows through a structure termed multi-tiered memory hierarchy. This structure consists of storage/secondary memory (usually hard drive), primary memory, and cache (Fig. 2A), in parallel to the genome, the RNA, and the protein levels in cells as discussed below.
Figure 2. The multi-tiered memory architecture.
A comparative examination of the information retrieval process in a computer (A) and the genetic information flow process in a cell (B) is delineated.
The hard drive and the genome are used, in computers and cells respectively, as permanent information storages with the stablest information content among the memory hierarchy. A computer usually functions for a long time without change in its hard drive content; and the genomic sequences of an organism remain mostly constant throughout a whole life cycle. This level has the largest storage capacity as well, but only specific portions of the content are used at any instant. Portions to be used are dynamically specified in a temporal fashion. In other words, information stored at this level serve as preparatory measures for a wide array of functionalities. In computers, some hard drive content codes for specific programs. Some, such as the operating systems, functions to maintain a stable execution environment for them. Collectively, they ensure that user requested tasks can be completed without further programming requirement. Likewise, the genomes are utilized by the evolution process as a depository to prepare the cells for dynamic environmental conditions - to ensure their integrity and functionality.
At the execution level, the system, in adjusting to signals, executes actions encoded in specific sections of the storage. In a computer, this leads to the corresponding program being run – the CPU sequentially executes a set of instructions in a defined order. In a cell, activities of specific proteins are augmented, enabling specific chemical reactions through catalysis. Such reactions can be metabolic transformations, protein modification events such as phosphorylationh, protein and RNA synthesis, et al.
As discussed above, the information is not retrieved to the execution level directly from storage. Instead, it goes through intermediate steps. In computers, it is retrieved to primary memory, then to cache, and finally fed into the CPU to dictate CPU cycles. In cells, genomic information is transcribed into RNA molecules, and is then translated into proteins. Before realizing their biochemical activities, many proteins then go through intracellular transportation and posttranslational modification, a process similar to computer CPU instruction fetching from the cache.
The functions of these intermediate steps in computers are to meet the needs of executing programs - to always contain sections of the storage expected by the CPU and to provide program execution environments such as storage of intermediate results. Their content therefore changes dynamically. The content at the information storage level, on the other hand, remains stable once all programs have been installed, as this level functions as repository for all programs. When new programs are added to the storage, the operating system would automatically manage their retrieval to memory in order to incorporate them into system functionality. In other words, these intermediate steps decouple dynamic execution of programs from their creation and storage. This enables end-users add new programs into a computer without venturing into its internal working, which is expensive, technically demanding, and therefore not feasible in the market place. System functionality can thus be expanded at reduced efforts and costs, a major factor for this design to dominate as computer architecture evolves through competition in the market place. We don’t know yet why, and how, the gene expression process evolved into this multi-step format during biological evolution. But this should remain an interesting point of investigation, since the advantages of such design in computers are clear.
2.3: Organization of retrieved actions into networks/processes
Each execution action represents a step of transformation action and is encoded by an atomic information unit, a CPU instruction in computers and a gene in the cells. A computer instruction dictates one CPU cycle, by controlling conductivity of its transistors, to transform binary input signals into output signals in specified manner. Similarly, a protein, proposed as basic computational elements in living cells (Bray, 1995), is often involved in controlling a step of biochemical reaction through catalysis.
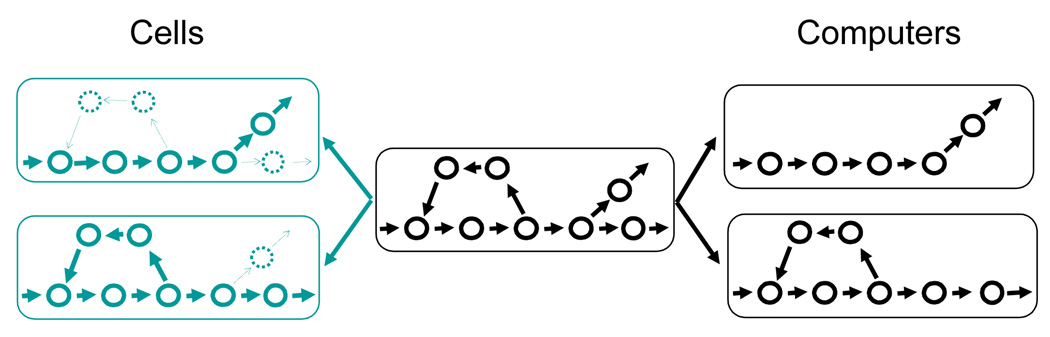
Actions encoded in individual information units are organized into modular functional units and the notion of network unifies this organization in the two systems. The term network is used here synonymously with program and process, the two terms commonly used in computer community to describe such functional units and their execution in computers. A computer program is essentially a collection of CPU instructions in predefined combinatorial order; individual instructions are chained together through their input-output relationship into procedures, which in turn interconnect to form the program. A computer program thus represents a virtual network, each node of which represents a CPU instruction and through which the CPU travels each time the program is executed. If one were to visualize it, each snapshot would display CPU executing one instruction. But a time series will visualize this traveling process (Figure 2A and 3).
Figure 3. A network model for the organization of individual actions.
The network (center) assumes dynamic configuration through on-off procedure switch in computers (black, right), and through managing pathway throughput in the cells (green, left, with less active pathway denoted by thin broken circles and arrows).
Similarly, proteins, through their mutual interactions or substrate/reactant-product relationships, form pathways, which in turn join together to form biochemical networks (Figure 2B and 3; See Wang and Gribskov, 2005 for further details). Among the best such examples are the observations that protein kinases form signaling cascades and that these cascades join together to form signaling networks. Computer procedures and programs are thus analogous to biochemical pathways and networks, respectively.
Such networks underlie functionalities of both systems. As the system adjusts to environmental signals, the networks assume dynamic configuration - in term of how active individual paths are – in order to perform specific tasks (Figure 3). In computers, a procedure is either on or off. Depending on inputs the program gets and its dynamic execution environment, the CPU might travel through many possible routes through the network – a procedure used in one execution cycle might not be used in another. In other words, binary logic is used. For biochemical networks, actions at each node of the network proceed in parallel, and such mathematical framework for describing process management has yet to be established. In general, throughput of a pathway in a biochemical network displays, as oppose to the on/off switch control in computers, a value range. As biochemical reactions rely on catalysis - usually proteins - to proceed, the throughput is controlled through two major mechanisms, catalysis abundance and how active the catalysis is. Gene expression and protein degradation processes control the abundance of proteins, thus regulating availability of catalysis for a biochemical pathway. Posttranslational regulation, such as phosphorylation and allosterical interactions, controls the activity of proteins already produced. Thus, cellular process management can be qualitatively described as regulation of pathway throughput, which is similar to the term bandwidth1 used in telecommunication. If one were to consider binary (on/off) logic as a special case of it, this scheme would unify network management in the two systems. It is noteworthy that protein phosphorylation often acts in a binary fashion by turning on/off its targets.
Thus, the information retrieval process functions to support the operation of regulatory networks, which in turn operate in support of functions manifested at system level. Efficient network operations are vital to both systems. Fine-tuning the multi-step information retrieval process, as discussed below, becomes integral to optimizing operation of this network.
2.4: Information retrieval and network regulation/optimization scheme
This retrieval process creates issues in system design. The sequential feature of the retrieval process results in a delay – execution of encoded actions has to wait until it is available. A determining factor of the delay is the bandwidth of the retrieval process – the higher the bandwidth, the shorter the delay will be. In computers, the delay is termed latency and is determined by the speed of the bus (clock-speed and number of parallel wires) and capacity at each step (memory and cache). This is paralleled in a cell by a delay from gene activation to protein production; RNA synthesis by RNA polymerase II takes ~30 seconds/kb and synthesizing an average protein of ~400 amino acids takes about 30 seconds. The delay is also determined by the capacity of the gene expression machinery, such as transcription and translation apparatuses, as well as the speed of the transportation process. Increasing the bandwidth, however, carries economical overhead. In the cells, the bandwidth represents the speed in which RNA and protein are produced. Increasing RNA and protein synthesis would elevate the rate at which nucleotide, amino acid, as well as energy (in the form of ATP molecules) are consumed, increasing the load on the metabolic network. Furthermore, aberrant production of RNA and proteins, when they are not needed, is often troublesome. As for computers, any improvement of bus clock-speed and the number of parallel bus wires is technologically and financially challenging. Optimal computer system design represents a balance between enhanced speed of the bus and economical factors. Therefore, the bandwidth of the retrieval process is limited. Optimal utilization of the limited bandwidth becomes imperative.
For better performance, a computer implements various mechanisms to fine-tune the multi-step information retrieval process, often decoupling these steps, in order to maximally reduce delay incurrence and to optimize usage of the bandwidth. It was examined whether these mechanisms have biology counterparts and therefore help interpret the observed discrepancy in cells. These mechanisms regulate information organization, loading, or purging. All of the three topics are vital at each retrieval step and will be discussed one-by-one below.
The capacity of each retrieval cycle is much bigger than an information unit – each cycle retrieves a collection of information units. The body of information thus needs to be organized into retrieval units, each containing a collection of functionally related information units, for retrieval to the next level (Table 1). Consequently, when the CPU finishes one retrieved instruction, expected instructions for future steps are more likely retrieved in the same cycle already, minimizing the number of retrieval cycles required to complete a computer program. The strategy in computers is straightforward. A page and a cache line represent retrieval unit to the memory and to the cache level, respectively (Table 1). A computer program tends to occupy, when possible, a contiguous stretch of information units. Instructions belong to the same segment of a program, and therefore functionally related, reside in close proximity (in the same retrieval unit or in units next to each other). Similarly, pro-karyote cells often organize many genes of a biochemical pathway, which is analogous to a segment of a computer program, into one operon (Table 1). Even though such contiguous distribution pattern is not preserved in eukaryote as each gene represents an independent transcription unit, similarity does exist – functionally related genes share common indexes (transcription factor binding sites), a phenomenon termed regulon. Operon/regulon can be regarded as retrieval unit at transcription level in the cells. Although translation regulation is not as well studied, we do have a glimpse of the sketches of regulatory schemes - functionally related genes have been shown co-regulated at this level (Le Roch et al., 2004; MacKay et al., 2004). Binding of regulatory factors, such as RNA binding proteins, to cognate cis-elements in un-translated region on individual mRNA molecules plays important roles (Fan and Steitz, 1998; Keene, 1999; Morris, 1997; Tenenbaum et al., 2002). The Post-transcriptional RNA regulon (PTRO) concept has been postulated to describe this phenomenon (Keene and Lager, 2005; Tenenbaum et al., 2002). It is tempting to treat a PTRO as a retrieval unit at translation level.
Table 1.
Comparative view of tiers in the information flow process. Physical materials and the unit of information organization in each tier are presented.
| Tiers | Computer | Cell | ||
|---|---|---|---|---|
| Hardware | Organization | Material | Organization | |
| Storage | Hard-drive | Paging | Genome | Operon; Regulon |
| Intermediate | Memory | Cache-line | mRNA | Post-transcriptional RNA Operon (PTRO) |
| Cache | CPU Cycle | Protein | A reaction | |
| Execution | CPU | Binary transformation | Catalysis | A reaction |
Loading and purging dynamically change the content of memory and cache in order to always optimize the partition of limited capacity at each step among parallel executing processes. The goal is, as in the case of information organization scheme described above, to stay steps ahead of the computing machinery so that expected data and instructions are already available by the time computing machinery requests them, minimizing chances of delay.
In order to achieve that, the loading process speculates the need of the computing machinery and pre-loads multiple retrieval units accordingly. Spatial locality principle is frequently used based on observation that memory/cache locations in close proximity to one recently accessed are likely to be accessed in the near future. Loading process exploits this property by bringing in adjacent retrieval units, with the expectation that future requests can be anticipated. This corresponds to co-loading of multiple pages (a working set) for each executing program at the memory level, and multiple cache-lines at the cache level. Similarly, production of RNA proceeds in parallel at multiple operons (or regulons) in the cells. At the translation level, this corresponds to parallel protein production from mRNAs of distinct translational PTRO unit (Table 2).
Table 2.
Many features of computer information retrieval process, designed to minimize latency incurrence, are also found in cells. These features, in conjunction with decoupling of steps of the retrieval process, help explain discrepancy in RNA and protein abundance.
| Computer | Cell | Potential Discrepancy | |||
|---|---|---|---|---|---|
| Step | Action | Feature | Action | Feature | |
| Storage To Memory | Load | By page | Transcription | Operon; Regulon | Higher RNA level, steady protein level |
| Spatial locality: working set | Parallel signaling pathways | ||||
| Purge | By page | RNA degradation | Degradation PTRO | Lower RNA level, steady protein level | |
| Temporal locality | Parallel degradation pathways | ||||
| Memory To Cache | Load | By cache-line | Translation | Translation PTRO | Higher protein level, steady RNA level |
| Spatial locality | Parallel signaling pathways | ||||
| Purge | By cache-line | Protein degradation | Co-degradation (shared mechanism) | Lower protein level, steady RNA level | |
| Temporal locality | Parallel degradation pathways | ||||
In order to load new information, portion of existing information will have to be purged first in order to free up adequate space. Erroneous purging, when the CPU requests for information just purged, will cause execution delay associated with retrieving them back. The goal of the purging process is therefore to identify information with the least reuse likelihood for deletion. Temporal locality principle is exploited by cache replacement algorithms, as low frequency of recent usage is the primary criteria in speculating what to discard. Retrieval unit (page or cache-line) serves as units for purging as well. As the loading process speculatively loads multiple retrieval units in batches, multiple units are simultaneously purged in order to free up space for them (Table 2). Similarly, the cell dynamically discard un-necessary mRNA and protein molecules to free up gene expression machinery for genes that need to be expressed, as well as to avoid detrimental effects these un-necessary RNA and protein molecules might otherwise cause. Multiple genes in a prokaryote poly-cistronic operon, analogous to multiple information units in a computer retrieval unit, are always retrieved to and purged from mRNA level simultaneously. In the eukaryote, the PTRO concept applies to RNA degradation as well, as mRNA decay in the yeast S. cerevisiae is orchestrated in functional groups (Wang et al., 2002). Such functional groups can be conveniently termed as degradation PTRO units (Table 2). When operons, or degradation PTRO units, are functionally related, it is conceivable that their degradation likely proceeds in coordinated fashion. At the protein level, a reliable high-throughput degradation measurement technique remains elusive. But it has been documented that protein half life, and thus its stability, is meticulously regulated in the cells.
2.5: A framework for interpreting discrepancy between RNA and protein abundance
Thus, commonalities can be found between computers and cells in almost every facet of their information retrieval process; not only common multi-layered architecture, but also shared regulatory mechanisms at each step of the process (Table 1 and 2). These regulatory mechanisms control loading information to, and purging information from, intermediate steps, thus determining content at each of these steps. In computers, the dynamically changing content of the memory and the cache results from a combination of loading and purging (Figure 2A). Likewise, observed mRNA and protein abundance in cells represents steady state levels, which are determined by production and degradation (Figure 2B).
RNA and protein levels, therefore, dynamically change. When changes in mRNA and protein levels differentiate, discrepancy between mRNA and protein abundance would occur. The regulatory mechanisms, which control information loading and purging at these intermediate steps, would help understand observed discrepancies. Firstly, decoupling actions in multiple steps of the process makes this discrepancy conceivable – without this decoupling, RNA and protein abundance should be perfectly correlated except a delay (the time it takes to produce the cognate proteins). Secondly, capability for efficient batch information management is acquired through organizing multiple information units into management (loading/purging) unit. This capability is further enhanced by parallel loading/purging of multiple management units, which in computers is achieved by exploiting locality principle. Batch information management, in turn, would result in various types of discrepancy between RNA and protein abundance. As discussed below, four scenarios of differential changes in mRNA and protein levels can be expected: a. Steady RNA, lower protein; b. Steady RNA, higher protein; c. Lower RNA, steady protein; d. Higher RNA, steady protein (table 2).
The spatial locality principle used in loading is designed to reduce number of retrieval cycles and CPU idle time. It would result in simultaneous retrieval of batches of information, but whose retrievals to the next step vary in a wide temporal range. This would, in the cells, result in parallel production of RNA molecules whose translation into proteins lag to various extents (Table 2). For example, translation of many P. falciparum genes is delayed into next stage of its life cycle when compared with their mRNA abundance (Le Roch et al., 2004). At the translation level, protein production from RNA molecules belonging to one translational PTRO unit would always proceed simultaneously. And translation from functionally related translational PTRO units often proceeds in parallel. The set of cognate genes might be differentially partitioned into operons (or regulons) at the transcription level. And the cognate operons (or regulons) corresponding to a group of functional related translational PTRO units might be differentially regulated at transcription level. Elevated production of mRNA from the genes might not have happened, resulting in steady mRNA abundance while protein abundance increases (Table 2).
Nonetheless, the temporal principle used in purging would cause simultaneous discarding of batches of information units whose prospect for near future usage might vary – and potentially dramatically depends on purging policy implemented and the size of information body being purged. Optimal cache policy remains an active research topic. Depending on the cache policy used in a computer, information that is in the caches may well be removed from the memory, equivalent to reduced RNA abundance while protein abundance remains steady in the cells (Table 2). Similarly, batch degradation at protein level would lead to abundance reduction for proteins with varying prospects for near future usage. When they do not belong to one same degradation PTRO unit, decay of their cognate mRNA molecules might be differentially regulated, potentially leading to reduced protein abundance while cognate RNA abundance remains steady (Table 2).
It was further investigated whether the four types of discrepancy are all represented in experimental observation. Three published proteomic studies, where mRNA and protein abundance are measured in parallel in the same experiments, were examined. These studies were performed in P. falciparum, S. cerevisiae, and H. sapien, respectively (Griffin et al., 2002; Le Roch et al., 2004; Tian et al., 2004). All four scenarios happen in each of the three studies. Exemplary genes, along with references, are listed inTable 3. Most of observed discrepancies can be classified into one of the four trends (types of discrepancy). Additionally, two cases of reverse correlation between mRNA and protein abundance were observed by Tian and colleagues (Tian et al., 2004). Such cases can be explained by a combination of scenarios a (steady RNA, lower protein) and d (higher RNA, steady protein), or by a combination of scenarios b (steady RNA, higher protein) and c (lower RNA, steady protein).
Table 3.
Exemplary discrepancies observed in parallel examinations of RNA and protein abundance in P. falciparum, S. cerevisiae, and H. sapien. Four types of discrepancy are listed.
| Types of discrepancy | Steady RNA, lower protein | Steady RNA, higher protein | Lower RNA, steady protein | Higher RNA, steady protein | |
|---|---|---|---|---|---|
| P. falciparum | Genes | PF10_0159 | PF14_0598 | PFD0080c | PF14_0527 |
| Reference | Le Roch et al, 2004 (supplemental table 3). | ||||
| S. cerevisiae | Genes | YJL171C | YHR110W | YNR050C | YPR160W |
| Reference | Griffin et al, 2002 (supplemental table 1). | ||||
| H. sapien | Genes | Prdx3 | Myh9/C80049 | Nsf | Tubb3 |
| Reference | Tian et al, 2004 (supplemental table). | ||||
2.6: Final perspective
Discrepancy between RNA and protein abundance has often been described as surprises and/or technical difficulties in the literature. This discrepancy, however, occurs too frequently to be interpreted this way. Additionally, poor correlation between transcription rate (promoter activity) and mRNA abundance has also been reported in mammalian (Fan et al., 2002) and in yeast cells (Garcia-Martinez et al., 2004). This comparative study with computer has sketched, in the perspective of system operation, a bird’s-eye view of gene expression process and associated regulatory mechanisms. This systematic perspective suggests that these discrepancies are, in stead, experimental observations one should expect. Furthermore, these discrepancies might be, as have been suggested (Greenbaum et al., 2003), a rich body of information that can be used to improve our data mining efforts to decipher logics embedded in cellular gene expression regulation. For instance, it reveals deficiency of current practice in constructing biochemical network models - we generally perform data-mining of mRNA expression datasets and assume that co-expression of mRNA leads to co-expression of protein, which, as this study suggests, is questionable and inadequate.
Moreover, theoretical insights from simpler model systems have been vital in addressing issues in more complex system. In view of the daunting challenges in deciphering logics used in cellular gene expression regulation, such simpler model systems are urgently needed. The necessary parts for a theoretical understanding of cellular information retrieval process are coming into place. The production processes - transcription and translation - have long been extensively studied. The degradation processes at mRNA and protein levels, though lagging behind, has lately provoked intensive investigative interest, as exemplified by RNA interference and protein ubiquitination research respectively. More high-throughput research methods are developed to complement RNA and protein abundance measurement data. ChIp-on-chip data, for example, describe promoter binding events. The ribonomics technology (Tenenbaum et al., 2002) targets regulatory events at RNA level. Furthermore, the Gene Ontology system strives to provide an integrative functional context. But they remain largely disparate components. The proven computer optimization techniques, which we design and understand well, may conveniently catalyze our efforts to integrate them into cohesive, explanatory theoretical models.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The term Bandwidth is used here to describe the capacity (or maximum flow rate) of a path as opposed to its meaning of the range of usable parameter value, such as wave frequency, in telecommunication.
References
- Adleman LM. Molecular Computation of Solutions to Combinatorial Problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Bray D. Intracellular Signaling as a Parallel Distributed Process. Journal of Theoretical Biology. 1990;143:215–231. doi: 10.1016/s0022-5193(05)80268-1. [DOI] [PubMed] [Google Scholar]
- Bray D. Protein Molecules as Computational Elements in Living Cells. Nature. 1995;376:307–312. doi: 10.1038/376307a0. [DOI] [PubMed] [Google Scholar]
- Brent R, Bruck J. Can computers help to explain biology? Nature. 2006;440:416–417. doi: 10.1038/440416a. [DOI] [PubMed] [Google Scholar]
- Csete ME, Doyle JC. Reverse Engineering of Biological Complexity. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- Fan J, Cheadle C, Becker KG, Gorospe M. High-throughput analysis of the relative changes in gene transcription and mRNA turnover during T cell activation. Molecular Biology of the Cell. 2002;13:521A–521A. [Google Scholar]
- Fan XHC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Lee H, Bonneau R, Mallick P, Serikawa K, Morris DR, Aebersold R. Quantitative proteomic analysis of the budding yeast cell cycle using acid-cleavable isotope-coded affinity tag reagents. Proteomics. 2006;6:6146–6157. doi: 10.1002/pmic.200600159. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Molecular Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Molecular & Cellular Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Molecular and Cellular Biology. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Ji S. The cell as the smallest DNA-based molecular computer. Biosystems. 1999;52:123–133. doi: 10.1016/s0303-2647(99)00039-8. [DOI] [PubMed] [Google Scholar]
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Lager PJ. Post-transcriptional operons and regulons coordinating gene expression. Chromosome Research. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Florens L, Zhou YY, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Research. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Lu LJ, Xia Y, Paccanaro A, Yu HY, Gerstein M. Assessing the limits of genomic data integration for predicting protein networks. Genome Research. 2005;15:945–953. doi: 10.1101/gr.3610305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay VL, Li XH, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, et al. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics - Response of yeast to mating pheromone. Molecular & Cellular Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- Morris DR. Cis-acting mRNA structures in gene-specific translation control. In: Harford JB, Morris DR, editors. RNA Metabolism and Post-transcriptional Gene Regulation. New York: Wiley-Liss, Inc.; 1997. [Google Scholar]
- Searls DB. Reading the book of life. Bioinformatics. 2001;17:579–580. doi: 10.1093/bioinformatics/17.7.579. [DOI] [PubMed] [Google Scholar]
- Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai HY, Thorsson V, Eng J, et al. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Molecular & Cellular Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- Unger R, Moult J. Towards computing with proteins. Proteins-Structure Function and Bioinformatics. 2006;63:53–64. doi: 10.1002/prot.20886. [DOI] [PubMed] [Google Scholar]
- Wang DG. "Molecular gene": Interpretation in the right context. Biology & Philosophy. 2005;20:453–464. [Google Scholar]
- Wang DG, Gribskov M. Examining the architecture of cellular computing through a comparative study with a computer. Journal of the Royal Society Interface. 2005;2:187–195. doi: 10.1098/rsif.2005.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng WJJ. Engineering approaches toward biological information integration at the systems level. Current Bioinformatics. 2006;1:85–93. [Google Scholar]