Abstract
Baculovirus mediated gene transduction of mammalian cells (BacMam) is an emerging technique for rapid recombinant protein expression in mammalian cells. Towards this, we constructed two baculovirus transfer vectors that incorporate several mammalian transcriptional regulatory elements necessary for high level protein expression in mammalian cells. Using these vectors, we show that the BacMam system in combination with the 293 GnTI− cell line can be used for production of milligram quantities of soluble glycoproteins. Moreover, for crystallization trials, the purified glycoproteins are sensitive to EndoH treatment resulting in a loss of the bulk of the attached N-linked glycosylation. In addition, we also show that a combination of the BacMam system and 293GnTI− cell line can be used for producing milligram quantities of a GPCR-protein ligand complex suitable for crystallization trials.
Keywords: BacMam, 293 cells, GPCR, Parathyroid hormone receptor, Frizzled, US28, chemokine, Fractalkine, purification, crystallization
Introduction
The baculovirus system for insect cells is one of the most commonly used methods for expression of recombinant proteins for structural studies. Although this system has been used very successfully, there are still many cases of mammalian proteins that are refractory to expression in insect cells, and require a mammalian expression system. This could be due to the fact that insect cells are not natural hosts for mammalian proteins and therefore are unable to process these proteins into their correct functional conformations leading to low protein yields and/or non-functional proteins. Over a decade ago, it was demonstrated very elegantly by several groups that a number of mammalian cell lines are susceptible to transduction by baculovirus. Moreover, by the inclusion of mammalian transcriptional units within the baculovirus DNA, several mammalian cells grown as adherent cultures were shown to have the ability to recognize these transcriptional units when transduced by baculovirus [1–4]. This in turn leads to the very attractive option of using baculovirus mediated gene transduction for recombinant protein production in mammalian cells (now termed as the BacMam system). Given that many structural biology labs are adept at the use of baculovirus, the BacMam system offers a relatively easy seque into mammalian cells using pre-existing methodologies within the laboratory.
Although very attractive in theory, there were several limitations that precluded this approach from a structural biology perspective. One of the main reasons was that in order to produce the amounts of recombinant proteins needed for structural studies on a routine basis with minimal manual labor and maximal cost-effectiveness, one has to have the ability to grow and maintain mammalian cells such as 293S and CHO cells as high density suspension cultures. This was not feasible until recently due to lack of cell culture media that could promote long-term high-density suspension cultures of mammalian cells. However, with the recent introduction of several commercially available chemically defined media, it is now possible to do so.
The BacMam system has evolved rapidly over the last couple of years. It is currently being used by the pharmaceutical industry in a high-throughput setting for rapid protein production in mammalian cells for the purpose of drug screening [5]. However, there has been only one report where the BacMam system was used for producing recombinant proteins in quantities that are sufficient to support protein crystallization trials [6]. The authors were successful in being able to produce and purify functional mammalian proteases in the range of several milligrams per liter of culture.
In this report we demonstrate that the BacMam system can be used for producing soluble ligand binding domains of receptor glycoproteins in the range of several milligrams per liter of culture. Moreover by using the 293 GnTI− cell line [7] for protein production, we further show that for the purpose of crystallization trials, the proteins can be deglycosylated very efficiently under non-denaturing conditions. To demonstrate the ability of this system for purification of recombinant membrane proteins, we describe a simple strategy for the high-level expression and purification of a G protein-coupled receptor (GPCR)1 in complex with its protein ligand using the BacMam system. The purified GPCR-ligand complex is suitable for crystallization studies.
It is hoped that the results from this and the previous report [6] would help in persuading members of the structural biology community to explore this technically simple and robust system as part of their expression systems for routine high level protein production. While we find that neither baculovirus nor mammalian cells are uniformly successful at solving any expression problem, we can generally express the vast majority of proteins in one system or the other. This requires that both systems are simultaneously accessible within one laboratory. The advantage of BacMam is that it enables one laboratory to easily use both insect-based and mammalian cell lines for expression, through the core technology of vector delivery through the baculovirus particle. In this fashion, both expression systems can be used side-by-side in a synergistic fashion.
Materials and metods
The 293 GnTI− cell line was kindly provided by Prof H.G. Khorana (MIT, Cambridge, MA, USA) and Dr Philip J Reeves (MIT, Cambridge, MA, USA). pVL1393 vector was from BD Biosciences (San Jose, CA). pCI vector was from Promega (Madison, WI). Sapphire linearized baculovirus DNA was from Orbigen (San Diego, CA). Pro293s-CDM was from Cambrex (East Rutherford, NJ). DMEM, SF900 media, Cellfectin, Lipofectamine Penicillin/Streptomycin, Gentamycin, L-glutamine and Glutamax were from Invitrogen (Carlsbad, CA). All molecular biology reagents, EndoHf and PNGaseF were obtained from NEB (Ipswich, MA). EndoHf is a fusion protein between maltose binding protein and EndoH. Primers for PCR were synthesized by Operon (Huntsville, AL). Custom ordered double-stranded DNA was synthesized by Genscript (Piscataway, NJ). Sodium butyrate and Protein A-Sepharose were from Sigma (St. Louis, MO). The anti penta-his antibody was from Qiagen (Valencia, CA) and HRP-conjugated polyclonal rabbit anti-mouse antibody was from DAKO (Carpinteria, CA). PreScission 3C protease was from GE Healthcare (Piscataway, NJ) and pre-cast 12% SDS-PAGE gels were from Bio-Rad (Hercules, CA).
Construction of expression vectors pVLAD7 and pVLAD6
pCI vector was digested with BglII and BamHI and the 1kb fragment was gel purified. This fragment was ligated into pVL1393 that was digested with BglII and BamHI and transformed into DH5α competent cells. A positive clone with the insert in the right orientation was identified by restriction digest analysis of mini-prep DNA and was labeled as pVLAD1. pVLAD1 was digested with NotI and BamHI and ligated with Fragment A (Figure 1A). Fragment A was a custom synthesized double stranded DNA fragment coding for 5’-NotI–[3C protease site]-[Glycine-Serine]-[hIgG1 Fc tag]-[6-His tag]-[Stop Codon]-[WPRE]-[SV40 Late poly A]-BamHI-3’. The ligation mixture was transformed into DH5α competent cells and a clone containing the insert was identified by restriction digest analysis of mini-prep DNA. This final vector was labeled as pVLAD7. In another version, the Fc tag was replaced by a Protein C tag and the 6-His tag was replaced by a 8-His tag. This version of the vector was labeled as pVLAD6. The nucleotide sequence coding for the extra-cellular domain of the human parathyroid hormone receptor 1 (corresponding to amino acids 1–187, including the signal sequence) was PCR amplified and cloned into pVLAD7 (PTHR1ECD-pVLAD7) as an EcoRI/NotI fragment. A Kozak’s consensus sequence (CCACC) was included before the initiating Methionine codon in the forward PCR primer. A chosen clone was sequenced to ensure the absence of any unwanted mutations.
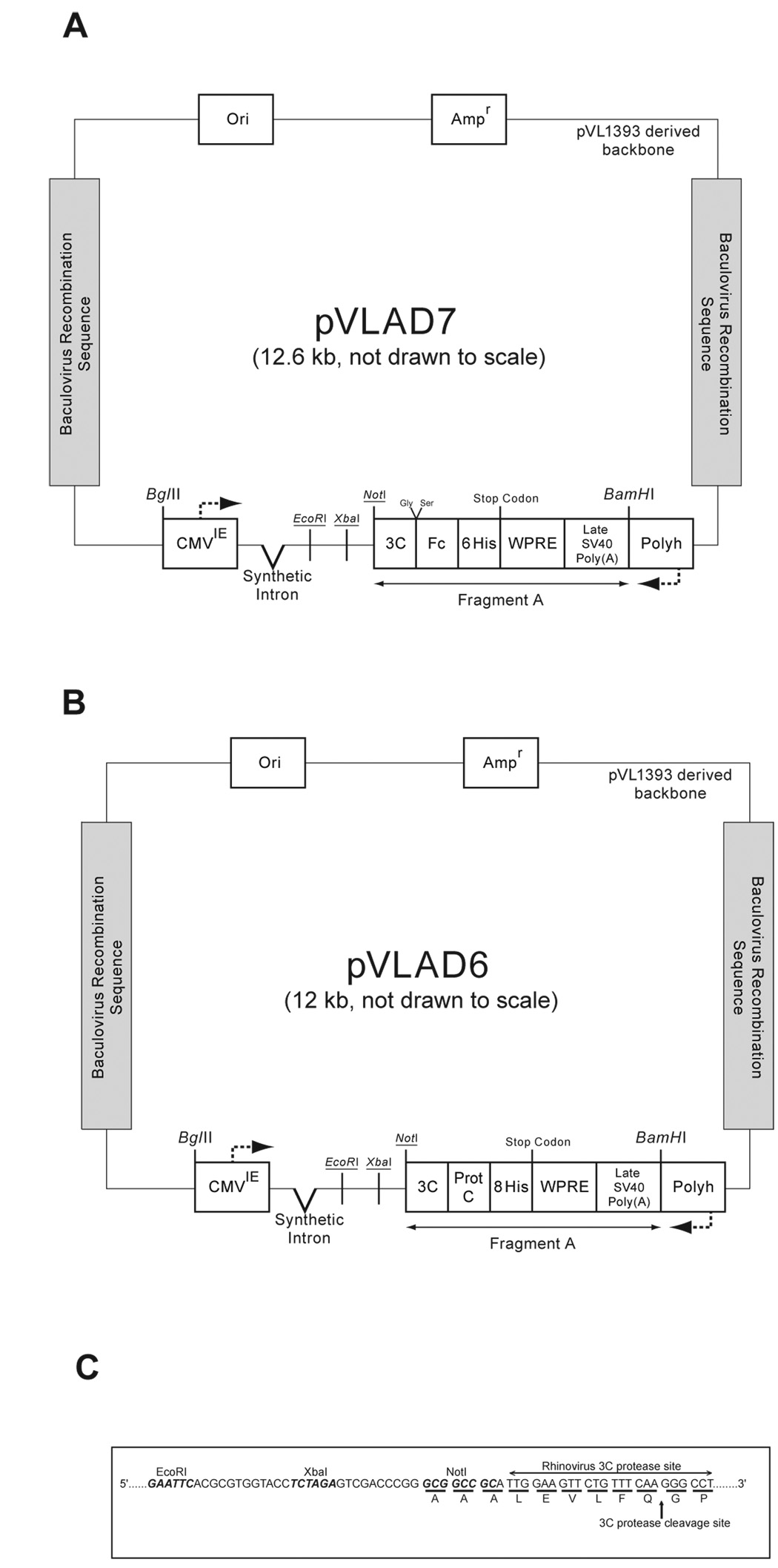
Figure 1. Schematic of pVLAD6 and pVLAD7 baculovirus transfer vectors.
(A) Shows a schematic of the pVLAD7 baculovirus transfer vector. The three enzyme restriction sites that are available for cloning are indicated in italicisized and underlined text. CMVIE= Cytomegalovirus Immediate/Early promoter and enhancer element, 3C= Rhinovirus 3C protease coding sequence, Fc= Human IGg1 Fc coding sequence, 6 His= 6 consecutive Histidine coding sequence, WPRE= Woodchuck post-transcriptional regulatory element, Polyh= Polyhedrin promoter. The direction of transcription initiated by the CMVIE and Polyhedrin promoters is indicated by the arrows. (B) Schematic of the pVLAD6 baculovirus transfer vector. Same as in (A) except that in pVLAD6, the Fc coding sequence is replaced by the Protein C (Prot C) epitope coding sequence and the 6 His is replaced by an 8 His coding sequence. (C) Nucleotide sequence corresponding to the region between the EcoRI site and the 3C protease site in pVLAD6 and pVLAD7 is shown.
Sf9 transfection and virus amplification
Sf9 cells were maintained as suspension cultures at 27°C in SF900 media supplemented with 10% heat-inactivated FCS, 2mM L-glutamine and 10 µg/ml Gentamycin. For Sf9 transfection, 0.5×106 cells were plated out per well in a 6-well plate. Twelve hours later, the cells were washed twice with serum and antibiotic free SF900 media. The PTHR1ECD-pVLAD7 construct was co-transfected with linearized baculovirus DNA using Cellfectin according to the manufacturers recommendations. The primary virus was harvested after 4 days and used for amplification. For viral amplification, 500 ml of Sf9 cells in suspension (2×106 cells/ml) were infected with 500µl of primary virus. Cells were diluted down to 2×106 cells/ml the next day with serum free SF900 media supplemented with 2mM L-glutamine. The amplified virus was harvested 4 days later by spinning down the cells at 1000 g and collecting the supernatant. Viral stocks were stored at 4°C in the dark.
Growth of 293 GnTI− cells in suspension and viral transduction
Adherent 293 GnTI− cells were grown in a humidified incubator (37°C, 5% CO2) in DMEM media supplemented with 2mM L-glutamine and 10% heat-inactivated FCS. When cells from two T-75 flasks reached 80% confluency, they were sloughed from the flasks using Pro 293s-CDM media supplemented with 0.1% FCS and 4mM Glutamax. The cells were pooled together, made up to 25 ml, transferred to a 125 ml square flask and placed on an orbital shaker (1 inch diameter of rotation) that was placed within the incubator. The cap was opened by a half turn, taped down and the speed of the shaker was set to 130 rpm [8]. After 24 hours, the cells were diluted down to 0.5×106 cells/ml. Typically, after each time the cells are diluted down, they exhibit a lag time of ~24 hours before they resume growth and start doubling every 24 hours. The suspension adapted cells were routinely maintained between 0.5–2×106 cells/ml during passaging. The maximum cell density we achieve under these growth conditions is ~ 4×106 cells/ml with greater than 90% cell viability. The culture volume was maintained at 25 ml in a 125 ml bottle. For scaling up, cells from four 125 ml bottles (2×106 cells/ml) were transferred to a 1L square flask, volume made upto 400ml and the cells were further grown in a 37°C warm room under identical agitation conditions. The growth profile of cells in a warm room is very similar to that in a 5% CO2, 37°C humidified incubator (see above). When the cell density reached 2×106 cells/ml, the cells were further split into 2L square bottles (1L culture per bottle) until the desired culture volume was reached. Cell density was measured using a haemocytometer and cell viability was measured by the Trypan blue dye exclusion method.
For viral transduction and protein expression, the appropriate volume of virus was added when the cells reached a density of 2×106 cells/ml. Sodium butyrate was added to a final concentration of 10mM and the cells were left shaking for 72 hours in the warm room. For transient transfections of adherent 293 GnTI− cells on a small scale, 1×106 cells/well in a 6 well plate were transfected with 2µg of plasmid DNA using Lipofectamine according to the manufacturer’s recommendations. The media was collected 60 hours post-transfection and subjected to Western blot analysis using the anti-his antibody (see below).
Protein purification using the Fc tag
For large-scale protein purification of secreted proteins containing the Fc tag, the transduced 293 GnTI− cells were spun down at 1000g and the supernatant was filtered through a 0.45 µm membrane. Two ml of protein A-Sepharose beads were packed into a column, the column was connected to a peristaltic pump and the beads were equilibrated with twenty bed volumes of HBS (20 mM HEPES, 150 mM NaCl, pH 7.1) at a flow rate of 1 ml/min. The filtered media was pumped over the column at a flow rate of 1 ml/min. The beads were washed with ten bed volumes of HBS and the protein was eluted with two bed volumes of 2M Arginine (pH 4.3) [9] at a flow rate of 0.5 ml/min. The beads were washed with HBS and stored in 20% ethanol for reuse. The protein containing fractions were immediately neutralized with one-tenth volume of 1M Tris.HCl, (pH 8.0), pooled and buffer exchanged into HBS using a PD-10 desalting column. Protein concentration was quantitated by measuring A280 of the purified sample in HBS and this was used to calculate the expression level per liter of culture (assuming A280 of 1 mg/ml=1.6).
Protein purification using the 8-His tag
For large-scale protein purification of secreted proteins containing the 8-His tag tag, the transduced 293 GnTI− cells were spun down at 1000g and the supernatant was filtered through a 0.45 µm membrane. The filtered media was concentrated down using a tangential flow concentrator and buffer exchanged into HBS. The media was incubated overnight with Ni-NTA agarose beads in the presence of 20mM immidazole. The beads were washed in a sintered glass filter with wash buffer (HBS+20mM immidazole), packed into a column and the bound protein was eluted with elution buffer (HBS+200mM immidazole).
Protein deglycosylation and 3C protease digestion
For preparative scale EndoHf digestion under non-denaturing conditions, the purified protein was incubated with 50K units of EndoHf at room temperature for 12 hours. SDS-PAGE gel analysis was used to verify deglycosylation. For 3C protease digestion, the sample was incubated with 80 units of Precission 3C protease for 12 hours at 4°C. For analytical deglycosylation experiments with PNGaseF under denaturing conditions, approximately 5µg of protein sample was incubated with the manufacturer supplied denaturing buffer at 95°C for 10 mins. The sample was cooled before adding the manufacturer supplied reaction buffer and NP-40 (final concentration 1%). 500 units of PNGaseF was then added and the sample was incubated at 37°C for 1 hour. Samples were analyzed by SDS-PAGE.
Gel filtration
In the case of Fc tagged proteins, the deglycosylated and 3C protease digested sample was incubated with the required volume of Protein A- resin and Glutathione Sepharose (GE Healthcare). The beads were spun down at 1000 rpm, the supernatant was collected and the beads were washed once with HBS. The supernatant and the wash solution were pooled together and injected in 500µl aliquots over a Superdex 200(10/30) gel filtration column connected to an AKTA FPLC purifier (GE Healthcare) and pre-equilibrated in HBS. The flow rate was maintained at 0.5 ml/min and 0.5 ml fractions were collected. Protein elution was monitored by measuring A280 and protein containing fractions were analyzed by SDS-PAGE.
Purification of US28-Fractalkine complex
The N-terminal chemokine domain of Fractalkine (residues 1–99, including the signal sequence) was PCR amplified, cloned into pVLAD7 and sequenced. Recombinant baculovirus were generated as described above. One liter of 293GnTI− cells was transduced with 100 ml of Fractalkine-pVLAD7 baculovirus and supplemented with 10mM sodium butyrate. The conditioned media was harvested 72 hours later, filtered and incubated overnight with 5ml of protein A-Sepharose in a batch mode. The beads were collected by vaccum filtration and washed with HBS and stored at 4°C in HBS with protease inhibitors till further use (usually within 24–36 hours). Based on small-scale elutions, we estimtate a density of ~2 mg of Fractalkine-Fc per ml of Protein A-Sepharose.
The US28 coding sequence (Genbank accession no AY174271) was PCR amplified to include a C-terminal 1D4 epitope tag followed by a stop codon and cloned into pVLAD6 (see above) as an EcoRI fragment. A clone containing the insert in the right orientation was identified by restriction digest analysis of mini-prep DNA and sequenced. This clone was labeled as US28-1D4-pVLAD6. Note that the presence of a stop codon immediately after the 1D4 epitope prevents translation beyond the 1D4 epitope into the 3C site. A Kozak’s consensus sequence (CCACC) before the first ATG codon was included in the forward PCR primer. Recombinant baculovirus for US28-1D4-pVLAD6 were generated as described above. Two liters of 293 GnTI− cells (~1.5×106 cells/ml) were transduced with 200 ml of US28-1D4-pVLAD6 baculovirus and sodium butyrate was added to a final concentration of 10 mM. The cells were harvested 30 hours later by spinning down at 1000g and washed with HBS (supplemented with 5mM EDTA, pH 8.0). All steps from here on were carried out at 4°C. The washed cells were re-suspended in 200 ml of lysis buffer, (20mM Tris.HCl, pH 8.0, 5mM EDTA), incubated on ice for 30 mins and dounce homogenized (20 strokes using a tight fitting pestle). The unlysed cells and cell debris were spun down at 1000g, supernatant collected and the pellet resuspended in 50 ml of lysis buffer and redounced. The suspension was centrifuged again at 1000g, the supernatants pooled together and spun at 48,000g for 1 hour. The crude membrane pellet was solubilized in 200 ml of solubilization buffer (20mM HEPES, pH 7.1, 200mM NaCl, 10% glycerol, 0.5% DDM, 0.1% CHS, complete EDTA free protease inhibitor tablets) for 2 hours by gentle end over end mixing followed by centrifugation at 48,000g for 1 hour. The clarified supernatant was collected and batch incubated with 5ml of protein A-Sepharose bound to Fractalkine-Fc (see above) by gentle end over end mixing for 36 hours. The beads were packed into a column, connected to a peristaltic pump and washed at a flow-rate of 1ml/min with 20 ml of wash buffer (20mM HEPES, pH 7.1, 200mM NaCl, 10% glycerol, 0.02% DDM/ 0.004% CHS). Protein was eluted with 3 bed volumes of elution buffer (100mM Glycine, pH 3.1, 200mM NaCl, 10% glycerol, 0.02% DDM/0.004% CHS) at the same flow rate. The collected fractions (8 ml each fraction) were neutralized immediately with one-tenth volume of neutralization buffer (1M Tris.HCl, pH 8.0, 200mM NaCl, 10% glycerol. 0.02% DDM/0.004% CHS). The resin was washed with 50 bed volumes of wash buffer and stored in wash buffer with 0.02% sodium azide. Protein containing fractions were identified by SDS-PAGE analysis, pooled together and incubated overnight with 50 units of 3C protease. The sample was depleted of free Fc and any undigested Fractalkine-Fc by passing through a 1 ml protein A-Sepharose column. The flow-through was concentrated down to 0.5 ml using a 50 kDa MWCO centrifugal concentrator and further purified over a Superose 6(10/30) gel filtration column (GE Healthcare) connected to a BioLogic Duo flow FPLC system (Bio-Rad) and equilibrated in gel filtration buffer [20 mM HEPES, pH 7.1, 150 mM NaCl, 10% glycerol, 0.02% DDM (Anagrade)/0.004% CHS]. Flow rate was maintained at 0.3 ml/min and 0.5 ml fractions were collected. Protein elution was monitored by measuring A280 (0.5 cm path-length UV detector) and protein containing fractions were analyzed by SDS-PAGE.
SDS-PAGE and Western blotting
For SDS-PAGE analysis, samples were diluted 1:1 with reducing or non-reducing Laemmli sample buffer. Samples containing reducing buffer were heated to 95°C for 3 mins before loading onto 12% SDS-PAGE gels. Samples containing US28-1D4 were diluted 1:1 with sample buffer incubated at room temp for 10 mins and loaded onto 12% SDS-PAGE gels. The gels were stained with Coomassie blue. For Western blotting, protein was transferred onto PVDF membranes for 1 h at 350 mA in ice-cold transfer buffer (25 mM Tris, 192 mM Glycine and 10 % methanol). The blot was incubated for 30 min in TBST (20 mM Tris.HCl, pH 7.4, 150 mM NaCl, 0.2% Tween) containing 5% non-fat dry milk powder (Carnation) and 1µg/ml of anti-penta His antibody (or 100 ng/ml of 1D4 antibody). The blot was washed for 10 min with TBST and incubated with 200 ng/ml of an HRP-conjugated polyclonal rabbit anti-mouse secondary antibody in TBST for 30 min. The blot was washed thrice for 10 min each with TBST and developed using an enhanced chemiluminiscent detection kit (GE Healthcare).
Results
293GnTI− cell line
The 293 GnTI− cell line developed by Khorana and collegues is a HEK293S derived cell line that lacks the capability to process N-linked glycans on glycoproteins from the high mannose to the complex mature form due to inactivation of both alleles of the GnTI gene [7]. The resulting N-linked high mannose glycans are homogenous. This feature of the cell line makes it extremely attractive for recombinant protein production from a structural biology perspective since one of the major obstacle to obtaining high resolution diffraction quality crystals of glycoproteins expressed and purified from eukaryotic hosts is the heterogenous nature of the attached N-linked glycans. In addition to the homogeneity, the high mannose glycans are susceptible to EndoH enzyme treatment. EndoH is a glycosidase that cleaves within the chitobiose core of high mannose glycans in N-linked glycoproteins. The enzyme cleaves the linkage between the 1st and 2nd GlcNAc of the high mannose glycan moiety. Thus, the EndoH treated glycoprotein from 293GnTI− cells in principal should be devoid of the bulk of N-linked glycosylation while at the same time the single GlcNAc’s still attached to the Asparagine/s would be presumably sufficient for maintaining protein solubility and activity.
We are able to maintain long-term suspension growth of this cell line in Pro293s-CDM media in the presence of 0.1% serum. Although the stock cells are passaged in suspension in a CO2 incubator, the dual buffering capability of Pro293s-CDM allows us to grow large scale cultures (>400ml) in a regular 37°C warm room environment. This relieves a lot of the space constraints for large-scale cultures. In our experience, there is no significant difference in growth rates between suspension cultures passaged in a CO2 incubator and those that have been passaged in the warm room as long as the cells are passaged every 3 days.
pVLAD7 and pVLAD6 baculovirus transfer vector
For the purpose of baculovirus mediated gene transduction of mammalian cells, we constructed two pVL1393 derived vectors, pVLAD7 and pVLAD6 (Figure 1A and B). Both vectors incorporate several elements necessary for mammalian transcription initiation (CMV promoter), transcription termination (SV40 poly A late signal), RNA splicing (synthetic intron), mRNA processing, stability and export to the cytoplasm for translation (WPRE and synthetic intron) [Figure 1A and B]. The CMV promoter is one of the strongest promoter that is constitutively active in a number of mammalian cell lines while the SV40 poly A late signal is a strong transcription termination signal. A combination of these two elements should ensure that the recombinant cDNA is constitutively transcribed while at the same time transcription termination is tightly regulated. The effect of splicing of the nascent RNA transcript on protein expression from transfected cDNA’s in mammalian cells has been extensively studied over the last two decades. The general outcome from these studies points to the phenomenon of increased recombinant protein expression when a splicing element is included 5’ to the coding sequence of the cDNA [10, 11]. For robust protein expression, the mRNA needs to be further processed and exported from the nucleus to the cytoplasm in a stable form that is recognized by the ribosomal translation machinery. These processes are enhanced by the Woodchuck hepatitis virus post-transcriptional regulatory element [WPRE] (and also to a certain extent by the process of splicing of the nascent RNA) [12]. To summarize, we have included in pVLAD7 and pVLAD6 several features necessary for robust recombinant gene transcription and mRNA processing in mammalian cells. For the purpose of protein purification, the pVLAD7 vector encodes downstream of the multiple cloning site for a 3C protease site followed by the human IgG1 Fc tag (amino acids Asp104 to Lys330 of human IgG1 heavy chain) and a 6-His tag (Figure 1A). In the case of pVLAD6, the Fc fragment was replaced by the Protein C epitope and the 6-His tag was replaced with an 8-His tag (Figure 1B). For cloning purposes, the cDNA is cloned in frame with the 3C protease sequence in both vectors (Figure 1C). Cleavage of the recombinant fusion protein with 3C protease permits separation of the purification tag from the native protein.
As a proof of principle that a combination of the BacMam system and 293GnTI− cells could provide sufficient amounts of deglycosylated recombinant soluble glycoproteins for structural studies, we describe here the purification of the extra-cellular ligand binding domain of a class B GPCR, the Parathyroid hormone receptor 1 (PTHR1). This receptor mediates most of the physiological role of PTH in mineral ion homeostasis, particularly in bones and kidneys [13]. Previous reports have shown that the ~170 amino acid extra-cellular domain (excluding the signal sequence) confers ligand binding activity and moreover glycosylation of the receptor at the 4 potential sites in the extra-cellular domain is not necessary for ligand binding [14–16].
The extra-cellular domain of PTHR1 (PTHR1ECD) including the native signal sequence was PCR amplified and cloned into pVLAD7. Before undertaking large-scale baculovirus production, in order to determine whether this protein if at all could be expressed in 293 GnTI− cells, the PTHR1ECD-pVLAD7 vector was transiently transfected on a small-scale into adherent 293GnTI− cells. A Western blot analysis of the conditioned media after 60 hours shows that PTHR1ECD-Fc is indeed synthesized and secreted into the culture medium (Figure 2A) with an apparent molecular weight of 60 kDa on an SDS-PAGE gel. After confirming protein expression by transient transfection of 293 cells on a small-scale, recombinant baculovirus for PTHR1ECD-pVLAD7 were produced and amplified as described in materials and methods.
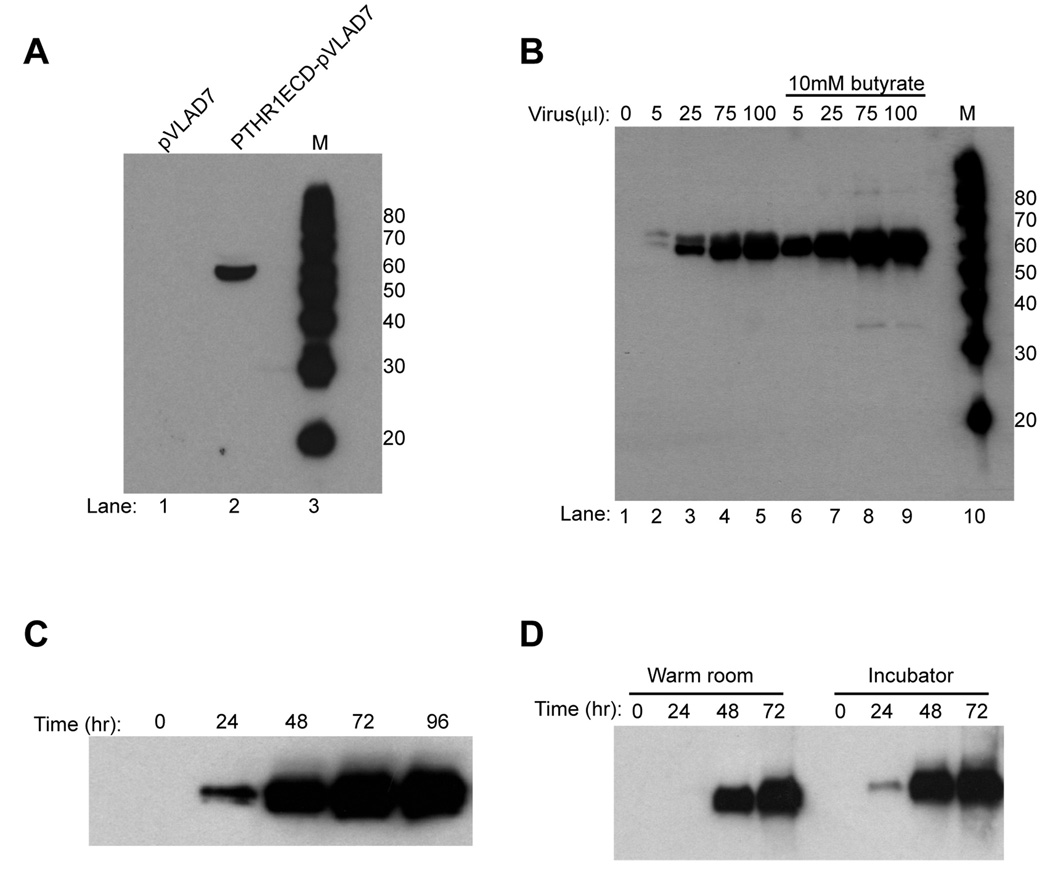
Figure 2.
(A) Transient transfection of pTHR1ECD-pVLAD7. Lane 1= supernatant from cells transfected with wild-type pVLAD7 vector, lane 2= supernatant from cells transfected with PTHR1ECD-pVLAD7 vector, lane 3= magic marker. (B) Immunoblot analysis of protein expression in 293GnTI− cells transduced with PTHR1ECD-pVLAD7 baculovirus. One milliliter of suspension 293GnTI− cells (1×106 cells/ml) were plated out in a 12 well-plate and transduced with the indicated volumes of virus in the absence (lanes 1–5) and presence of 10mM sodium butyrate (lanes 6–9). Media was harvested 72 hours after virus addition and protein expression was analyzed by immunoblot analysis with an anti-penta his antibody. M= Magic marker. (C) Time course of protein expression. Aliquots of media from PTHR1ECD-pVLAD7 transduced cultures was harvested at the indicated time points after virus addition and analyzed by immunoblotting with an anti-penta his antibody. (D) Analysis of protein expression in cultures maintained in a warm room. Suspension cultures of cells maintained in a 37°C warm room and CO2 regulated cell-culture incubator were transduced with PTHR1ECD-pVLAD7 and supplemented with 10mM butyrate. Media was harvested at the indicated time points after virus addition and analyzed by by immunoblotting with an anti-penta H his antibody.
Optimization of transduction conditions
Initial small-scale analytical experiments with 1 ml cultures in 12 well-plates were performed to optimize transduction conditions of suspension adapted 293GnTI− cells with the pVLAD7 recombinant baculovirus. The parameters that were examined were volume of virus for transduction, time course of protein expression and effect of histone deacetylase inhibitors on protein expression. We show here the results for the PTHR1ECD-pVLAD7 baculovirus. The results obtained with pVLAD7 and pVLAD6 baculoviruses for most other proteins are qualitatively similar to those of PTHR1ECD-pVLAD7.
The first parameter that was optimized was the volume of virus to be added to the cells to detect protein expression. As shown in figure 2A, lanes 1–5, protein expression is detected when 5 µl of virus is added to 1 ml of cells and the expression level increases with increasing volumes of virus added (Figure 2B, lanes 2–5). We typically do not use a ratio greater than 1:10 of virus added to culture volume as we noticed that cells tend to aggregate at higher ratios. To further boost protein expression, we examined the effect of sodium butyrate, a histone deacetylase inhibitor, on protein expression. As can be clearly seen in Figure 2B, lanes 6–10, addition of 10mM butyrate to the cultures results in significantly higher protein expression at all virus to culture ratios tested (Figure 2B, compare lanes 2–5 vs 6–9). A time course of protein expression analysis by Western blotting of the butyrate supplemented media is shown in Figure 2C. A 25 ml suspension culture of 293 GnTI− cells (1×106 cells/ml) was transduced with 2.5 ml of PTHR1ECD-pVLAD7 baculovirus and supplemented with 10mM sodium butyrate. Small aliquots of the media was harvested at the indicated time points (Figure 2C) after initial virus addition and analyzed by immunoblot analysis with an anti-penta his antibody. The result indicates that protein accumulation in the media peaks at 72 hours after virus addition. Finally, we compared time course of protein expression of cultures maintained in a 37°C warm room versus in a 37°C, 5% CO2 humidified incubator (Figure 2D). A 25 ml suspension culture of cells (1×106 cells/ml) that had been maintained for 3 passages in a 37°C warm room environment without any CO2 regulation was transduced with 2.5 ml of PTHR1ECD-pVLAD7 baculovirus and supplemented with 10mM butyrate. For comparison sake, a parallel culture that had been maintained in a regular humidified CO2 regulated cell-culture incubator was also transduced with the PTHR1ECD-pVLAD7 baculovirus and supplemented with butyrate. Both transduced cultures were shaken under identical agitation conditions in their respective culturing environment. Media was harvested at the indicated time points after virus addition and analyzed for protein expresssion by immunoblot analysis with an anti-penta his antibody. The results indicate that there is a slight lag in the onset of protein expression when cultures are maintained in a warm room environment. However, at the end of the 72 hour incubation period, protein expression levels are similar in both cases as judged by Western blot analysis.
We have also determined that the optimal cell density for viral transduction and protein expression is 1–2×106 cells/ml (data not shown). When transduction is performed at higher cell densities, there is significantly reduced protein expression and this could have to do with a shift in cell metabolism or a change in the culture media composition (such as lactate accumulation or change in glucose concentration). In addition, amongst the various synthetic chemically defined media tested, we have found that the 293GnTI− cells grow most robustly in suspension with significantly higher protein expression levels in Pro293s-CDM (data not shown).
Thus a large-scale viral transduction and protein expression involves, adding the amplified recombinant baculovirus to the culture at a particular virus to culture ratio determined from small-scale titrations. It is critical that for maximal protein expression, the cell density at the time of virus addition be 1–2×106 cells/ml. The culture is further supplemented with 10mM butyrate and incubated in a 37°C warm room for 72 hours before harvesting the media.
Large-scale purification of PTHR1ECD
One liter of 293GnTI− cells were transduced with 100 ml of baculovirus for PTHR1ECD-pVLAD7 and sodium butyrate was added to a final concentration of 10mM. The cells were incubated shaking in a warm room at 37°C. The supernatant was harvested 72 hours later and the Fc fusion protein was purified using protein A-Sepharose (see materials and methods). A sample of the purified protein analyzed by SDS-PAGE under reducing and non-reducing conditions is shown in Figure 3A. Majority of the PTHR1ECD-Fc fusion protein migrates with an apparent molecular weight of 60 kDa (Figure 3A, lane1) and as a disulfide linked dimer of ~120 kDa (Figure 3A, lane 2) under reducing and non-reducing conditions respectively. This is consistent with our expectation that in solution, the PTHR1ECD domain contains no inter-molecular disulfides while the Fc domain exists as an inter-molecular disulfide linked dimer. The theoretical molecular weight of the mature fusion construct without taking into account any post-translational modifications is 48 kDa. The total protein yield is 14 mg per liter with >90% purity of the PTHR1ECD-Fc recombinant protein.
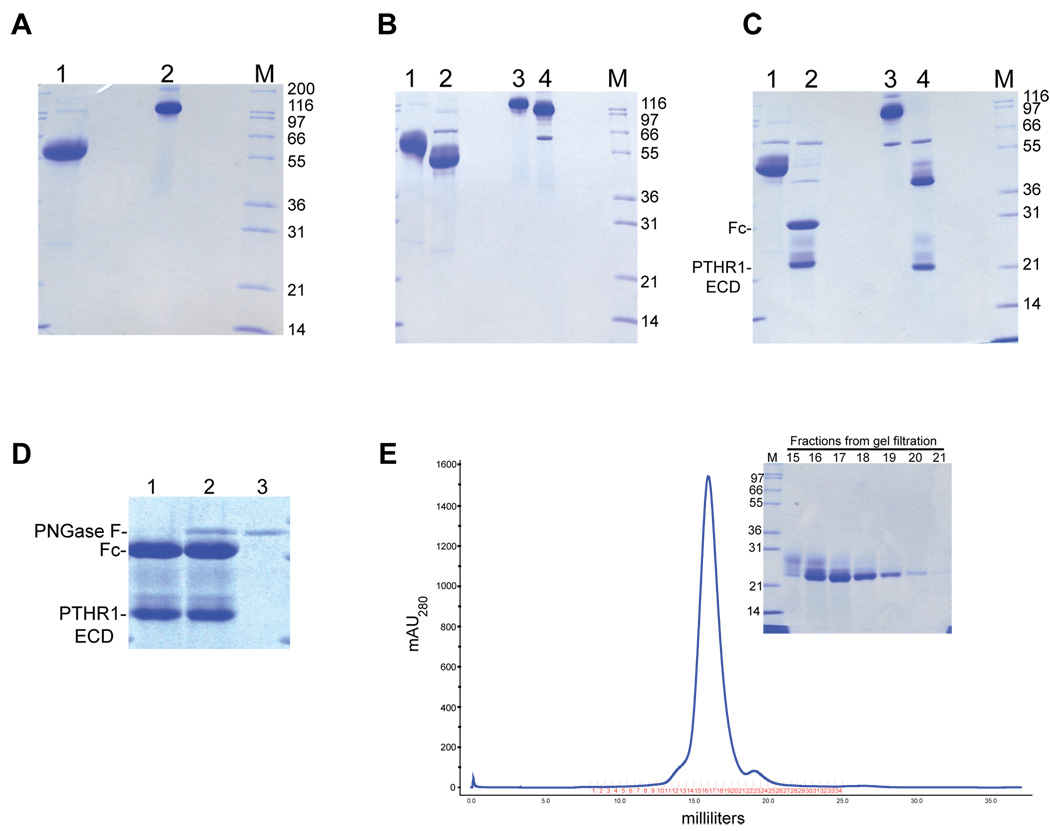
Figure 3. PTHR1ECD purification.
(A) Purification of PTHR1ECD-Fc. Lane 1 shows a sample of the elution from the protein A column. The sample was prepared under reducing conditions for SDS-PAGE. Lane 2, same as in lane 1 except that the sample was prepared under non-reducing conditions for SDS-PAGE. (B) EndoHf treatment of PTHR1ECD-Fc. The purified PTHR1ECD-Fc fusion protein was treated with EndoHf under non-denaturing conditions and analyzed by SDS-PAGE. Lanes 1 and 2 show the untreated and EndoHf treated samples respectively prepared under reducing conditions for SDS-PAGE. The band migrating at 66 kDa in lane 2 corresponds to EndoHf. Lanes 3 and 4 are identical to samples in lanes 1 and 2 respectively except that they were prepared under non-reducing conditions for SDS-PAGE. (C) Lane 1 shows a sample of the EndoHf treated fusion protein and lane 2 shows a sample that was further treated with 3C protease. The positions of free Fc and PTHR1ECD in lane 2 are indicated. The band migrating at 48 kDa in lane 2 corresponds to 3C protease. The samples in lane 1 and 2 were prepared under reducing conditions for SDS-PAGE. Samples in lanes 3 and 4 are identical to samples in lanes 1 and 2 except that they were prepared under non-reducing conditions for SDS-PAGE. (D) Lane 1 shows a sample of the EndoHf and 3C treated sample. Lane 2 shows a sample that was further treated with PNGase under denaturing conditions. Lane 3 shows a sample of PNGase F only. (E) Size exclusion chromatographic purification of PTHR1ECD. The EndoHf and 3C treated sample was depleted of EndoHf and 3C and purified over a Superdex 200(10/30) size exclusion column. The main panel shows the elution profile of PTHR1ECD from the column. The inset shows an SDS-PAGE analysis of the elution fractions across the main peak.
The purified protein was treated with EndoHf under non-denaturing conditions (see materials and methods) resulting in a drop in the molecular weight of the protein to 51 kDa and 100 kDa under reducing and non-reducing conditions respectively (Figure 3B). The PTHR1ECD-Fc fusion possesses 5 potential N-linked glycosylation sites (4 in PTHR1ECD and 1 in the Fc domain). It has been shown in previous work that some if not all 4 potential glycosylation sites in PTHR1ECD are utilized in 293 cells and insect cells. An increase in incubation time, amount of enzyme used or increasing the incubation temperature to 37°C does not result in any further shift (data not shown). It should be noted that the EndoHf treated sample migrates as a sharp band with a trailing smear suggesting that the sample either possesses N-linked glycosylation sites that are resistant to EndoHf treatment under non-denaturing conditions or the sample has post-translational modifications other than N-linked glycosylation. This is further clarified upon cleavage of the sample with 3C protease thus separating the PTHR1ECD from the Fc domain. Treatment with 3C protease converts the 51 kDa fusion protein into two main bands, a 29 kDa band corresponding to the Fc moiety and a 22 kDa band corresponding to PTHR1ECD (Figure 3C). The Fc band migrates as a sharp homogenous band while the PTHR1ECD migrates as a sharp band with a trailing smear and also an additional fuzzy band migrating just below the Fc under reducing conditions. To verify whether the heterogeneity in PTHR1ECD was due to incomplete deglycosylation by EndoHf, the sample was treated with PNGaseF under denaturing conditions. If the heterogeneity was due to N-linked glycosylation, we would expect that treatment with PNGaseF under denaturing conditions would convert the heterogenous forms of PTHR1ECD into one form. However, as can be clearly seen in Figure 3D, treatment with PNGaseF does not result in any change in the migration pattern of the heterogenous forms of PTHR1ECD. This result leads us to conclude that additional post-translational modifications such as O-linked glycosylation contribute to the heterogeneity of PTHR1ECD and the sample is deglycosylated upon treatment with EndoHf under non-denaturing conditions.
The deglycosylated sample was depleted of free Fc, undigested PTHR1ECD-Fc and 3C protease as described in materials and methods and further purified over a gel filtration column (Figure 3E). The PTHR1ECD elutes from the gel filtration column as a well-defined peak at 16 ml with the trailing smear eluting slightly earlier compared to the main band of PTHR1ECD as judged by SDS-PAGE analysis (Figure 3E inset). Based on molecular weight calibration of the gel filtration column using protein standards of known molecular weight, the elution profile of PTHR1ECD suggests that it is monomeric in solution in the absence of ligand.
Previous attempts at expressing PTHR1ECD in insect cells resulted in expression levels of 0.6 mg/ml [16]. Use of the BACMAM system results in significantly higher yields of 14 mg/liter of the PTHR1ECD-Fc fusion (corresponding to ~6 mg/liter of PTHR1ECD) and moreover by using the 293GnTI− cells, we were able to quantitatively deglycosylate bulk of the N-linked glycosylation present on PTHR1ECD which is an added advantage in crystallization attempts of this molecule.
Purification and crystallization of the mouse Frizzled-4 receptor ligand binding domain
The Frizzled family of proteins are receptors for secreted Wnt proteins and other ligands. These receptors belong to the GPCR superfamily and play an important role in regulation of cell polarity, formation of neural synapses and the regulation of proliferation and several other processes in the developing and adult organisms [17]. All members of this family share several conserved features, including an extracellular ligand binding Cysteine rich domain and seven hydrophobic transmembrane segments. One member of this family, human Frizzled-4, has been implicated in familial exudative vitreoretinopathv [18]. In order to begin understanding the molecular basis for ligand recognition by mammalian Frizzled-4, we expressed, purified and crystallized the ligand binding domain of mouse Frizzled-4 using the BacMam system.
The mouse Frizzled-4 receptor extracellular Cysteine rich ligand binding domain (mFz4 ECD) corresponding to residues 1–162 (including the signal sequence) was cloned into pVLAD6. Recombinant baculovirus were generated and the secreted protein was purified from 500 ml of media from a transduced 293GnTI− culture using Ni-NTA beads as described in materials and methods. The purified protein runs as a doublet between 21 and 24 kD (Figure 4A) and the approximate yield of mFz4 ECD is 1 mg/L. Further treatment under non-denaturing conditions with EndoHf collapses this doublet into a single band that migrates at ~21 kDa (Figure 4B, lane 3). The deglycosylayed sample was further purified over a Superdex 75(10/30) gel filtration column. The mFz4 ECD elutes from the column in a peak centered around 14.02 ml (Figures 4D) and an SDS-PAGE analysis shows that the purified protein is >90% pure (Figure 4D inset, lanes 4–7). The purified protein from the gel filtration column was concentrated down to 3 mg/ml for crystallization trials using the hanging drop method. Crystals were obtained after 2 days in 1M Sodium Malonate, 0.1% Jeffamine and 100mM HEPES, pH 6.8 (Figure 4E). Crystals were harvested after 7 days and flash frozen using 20% glycerol as a cryoprotectant. These crystals diffracted to a resolution of 2.1 Å (Figure 4E, inset) at a synchrotron light source enabling us to solve the structure of mFz4 ECD, the details of which will be published elsewhere (manuscript in preparation).
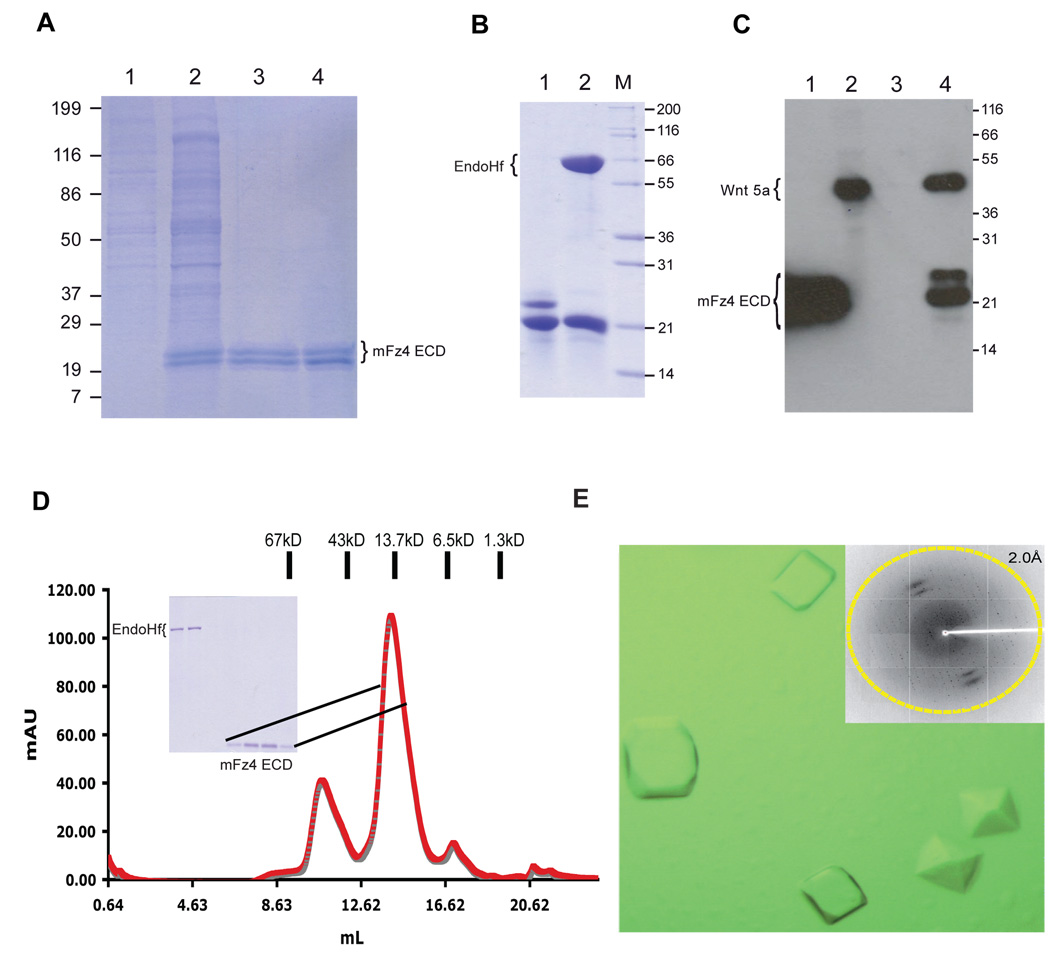
Figure 4.
(A) Lanes 1–4 show the elution of mFz4 ECD from the Ni-NTA agarose column. The mFz4 ECD doublet is indicated. (B) Lane 1 shows a sample of the Ni-NTA purified mFz4 ECD. Lane 2 shows a sample that is further treated with Endo Hf. The band that migrates at 66kDa corresponds to Endo Hf. (C) mFZ4 ECD is capable of binding Wnt 5a. Lane 1, cell culture supernatant from pVLAD6 mFz4 ECD transduced cells. Lane 2, soluble detergent extract of the cell pellet from pVLAD6 Wnt 5a transduced cells. Lane 3, Elution from Ni-NTA beads after incubating the resin with the detergent extract from lane 2. Lane 4, elution from the Ni-NTA beads that had been incubated with a detergent extract of cells that had been co-transduced with pVLAD6 mFz4 ECD and pVLAD6 Wnt 5a. (D) The EndoHf and 3C protease treated mFz4 ECD was further purified over a Superdex 75(10/30) column. The inset shows an SDS-PAGE analysis of the proteins eluting in the peaks at 10.75 and 14.02 mls. (D) Crystals of mFz4 ECD are shown. The inset shows the diffraction pattern of a crystal at a synchrotron light source.
In order to determine whether the expressed mFz4 ECD is capable of binding it cognate ligand Wnt 5a, analytical pull down experiments were performed. For these experiments, a version of Wnt 5a that has a C-terminal Protein C epitope and lacking a 8-His tag was used. mFz4 ECD when expressed by itself is predominantly found in the cell culture supernatant whereas Wnt 5a expressed by itself is found in the soluble detergent extract of the cell pellet (Figure 4C, lanes 1 and 2) as determined by Western blot using an antibody directed against the Protein C epitope. Moreover, Wnt 5a cannot be purified from this detergent extract using Ni-NTA beads (Figure 4C, lane 3), which is consistent with this construct lacking a 8-His tag. However when pVLAD6 mFz4 ECD and Wnt 5a are co-expressed and the soluble detergent extract of the cell pellet was subjected to pull-down assays using Ni-NTA beads, it can be clearly seen that mFz4 ECD can be purified from this extract and in addition, Wnt 5a also co-purifies (Figure 4C, lane 4). This result indicates that mFz4 ECD is bioactive.
Purification of a GPCR- protein ligand complex
US28 is a constitutively active HCMV encoded GPCR that displays promiscuous binding with members of the CC-class of chemokines [19, 20]. US28 also binds the membrane bound CX3CL1 chemokine (also known as Fractalkine) with a sub nanomolar affinity [21]. Fractalkine consists of an N-terminal chemokine domain that is separated from the transmembrane region by a mucin-like stalk [22]. Numerous biochemical studies have demonstrated that just the N-terminal chemokine domain of Fractalkine is sufficient to bind US28 as well as its endogenous receptor CX3CR1 in heterologous expression systems [21, 23]. US28 has been implicated in helping the virus hijack the infected host cellular transcriptional machinery, promote cell to cell fusion, cell migration and also help infected cells evade detection by the immune system by virtue of its ability to “scavenge” a broad range of CC-chemokines such as CCL5 (RANTES) and CCL3 (MIP-1α). Moreover, US28 has also been show to be a co-receptor for HIV entry [24]. Thus an understanding of the molecular basis for the interaction of US28 with its ligands is important from a physiological as well as drug development point of view. From here on, the term Fractalkine refers to the N-terminal chemokine domain of full-length Fractalkine.
Our success in purifying milligram quantities of soluble secreted proteins using the BacMam system prompted us to examine whether this system could be used for purification of functional membrane proteins such as GPCRs for structural studies. We decided to focus on GPCRs that have protein ligands. There were two main reasons for doing so. The first being that in the case of GPCRs which have protein ligands, we can express and purify the ligands as Fc fusion molecules using the BacMam system. The second reason being we can use the protein ligand-Fc fusion bound to protein A-Sepharose as a ligand affinity column for a single step purification of functional GPCRs from the solubilized membranes. Here we provide evidence for the feasibility of such a strategy by describing the purification of the viral chemokine GPCR US28 in complex with Fractalkine.
Western blot analysis of conditioned media from large-scale cultures transduced with Fractalkine-pVLAD7 baculovirus shows the presence of two major immunoreactive bands when the sample is reduced and heated to 95°C in sample buffer before loading onto the SDS-PAGE gel (Figure 5A, lane 1). The band migrating at ~45 kDa corresponds to full-length Fractalkine-Fc while the band migrating at ~35 kDa corresponds to the Fc domain due to proteolysis between the Fractalkine and Fc domains during cell culture. Surprisingly, when the sample preparation conditions are changed to just reducing conditions in sample buffer without heating to 95°C, three immunoreactive bands are observed (Figure 5A, lane 2). The band migrating at 50 kDa corresponds to the Fc domain while the full-length Fractalkine-Fc now runs as two bands at ~ 60 and 70 kDa bands. This anomalous migration of full-length Fractalkine-Fc is most probably due to incomplete denaturation of the chemokine domain when the sample is not heated before SDS-PAGE. Note that from here on, all samples for SDS-PAGE gels are prepared under reducing conditions in sample buffer without heating to 95°C. This is necessary for the reason that in the experiments described below, boiling the sample prior to SDS-PAGE results in aggregation of US28-1D4 preventing it from entering the gel. This is a common phenomenon observed for other chemokine GPCRs too [25, 26].
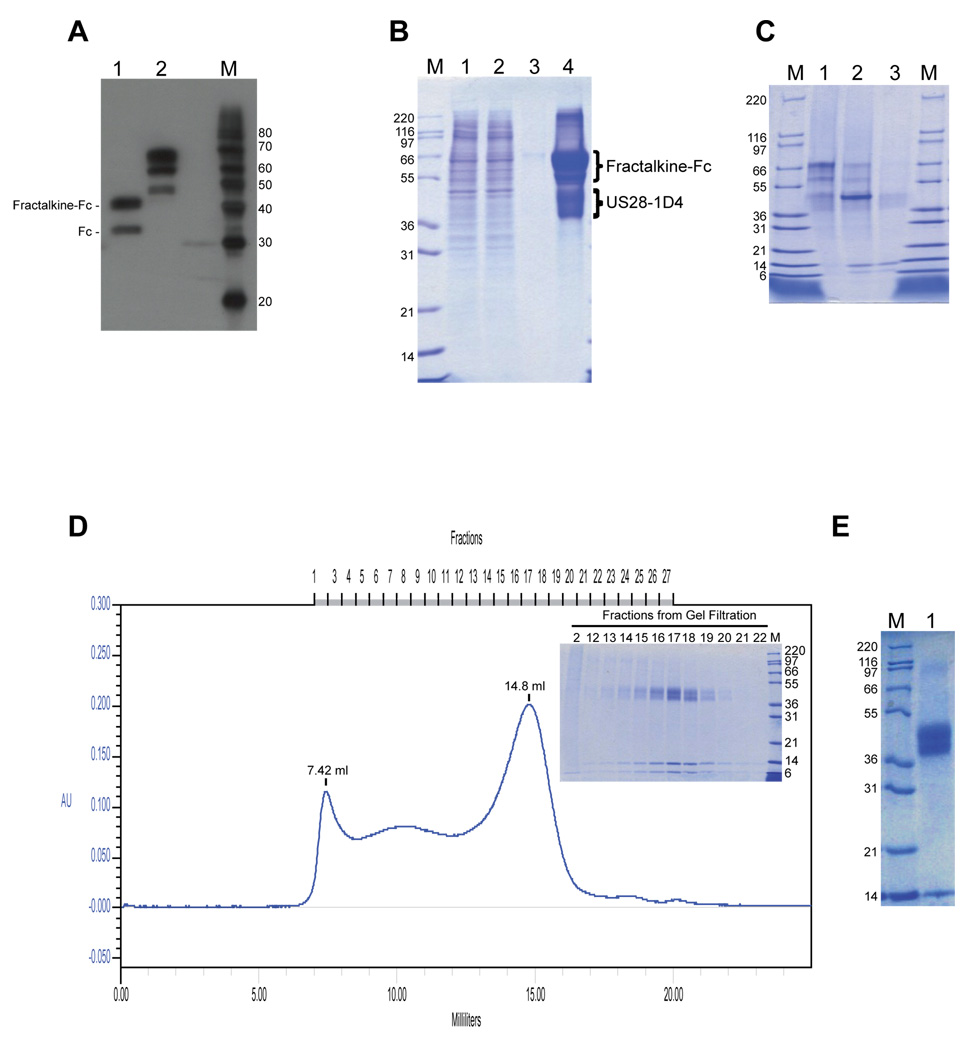
Figure 5. Purification of US28-Fractalkine complex.
(A) Immunoblot analysis of Fractalkine-Fc under different sample preparation conditions. Aliquots of media from a culture of 293GnTI− cells transduced with baculovirus for Fractalkine-pVLAD7 were prepared under reducing conditions and heated to 95°C (lane 1) or not heated (lane 2) before SDS-PAGE and immunodetection. The bands corresponding to free Fc and Fractalkine-Fc in lane 1 are indicated. M= magic marker. (B) Ligand affinity purification of US28-1D4. Lane 1 shows the solubilized membranes before incubation with the Fractinlkine-Fc/Protein A resin. Lanes 2 and 3 show a sample from the unbound fraction and the last wash fraction respectively. Lane 4 shows a sample from the main elution fraction. The bands corresponding to Fractalkine-Fc and US28-1D4 in lane 4 are indicated. (C) 3C digestion of Fractalkine-Fc. The eluted sample shown in lane 1 was treated with 3C protease. A sample of the 3C treated material is shown in lane 2. The 3C treated material was passed through a protein A-Sepharose column. Lane 3 shows the flow-through from the protein A column. (D) Size exclusion chromatographic purification of US28-1D4/Fractalkine complex. The sample from lane 3 in (C) was concentrated and injected over a Superose 6(10/30) size exclusion column. The main panel shows the elution profile from the Superose column. The inset shows an SDS-PAGE analysis of the peak fractions at 7.42 ml and 14.8 ml. (E) Concentrated sample used for crystallization trials is shown.
For the purpose of US28-1D4 purification, a ligand affinity column composed of Fractalkine-Fc bound to protein A-Sepharose was prepared as detailed in the materials and methods. We prefer to use a batch mode of binding Fractalkine-Fc to protein A-Sepharose rather than a column format (as in the case of PTHR1ECD-Fc) due to the fact that in a batch mode, one would expect a more uniformly distributed binding of the protein to the resin in contrast to when the protein is bound to the resin in a column format. Based on protein elution and quantification from small-scale elutions, we estimate a protein density of ~2 mg of Fractalkine-Fc/ml of resin.
For expression of US28-1D4, two liters of 293 GnT1− cells were transduced with 200 ml of US28-1D4-pVLAD6 baculovirus. The cells were harvested 30 hours later and the membranes prepared as described. The membranes were solubilized with a combination of DDM/CHS for 2 hours and the clarified soluble supernatant was batch incubated with the Fractalkine-Fc bound to protein A-Sepharose beads (see above). After washing the beads, protein was eluted from the protein A column using a low pH buffer (see materials and methods). An SDS-PAGE analysis of the eluted protein indicates the presence of two major protein bands in the elution fraction (Figure 5B, lane 4). The major overloaded band migrating as a doublet between 55–70 kDa corresponds to full-length Fractalkine-Fc, which is consistent with the previously described Western blot analysis of the conditioned media (Figure 5A, lane 2). In addition, the other major protein band that migrates as a doublet between 42–50 kDa corresponds to US28-1D4. The identity of this band was confirmed by a Western blot analysis with the 1D4 antibody (data not shown). Based on the intensity of Commassie staining, we estimate a total yield of ~2 mg of US28-1D4 from a two liter culture.
The eluted protein was treated with 3C protease to cleave the linkage between Fractalkine and the Fc domain. SDS-PAGE analysis of the 3C treated sample shows a marked decrease in the intensity of the Fractalkine-Fc bands at ~60 and 70 kDa and at the same time the appearance of three new bands (Figure 5C, compare lanes 1 and 2). The band migrating at ~50 kDA corresponds to Fc and the bands migrating at ~14 and 8 kDa corresponds to Fractalkine. As mentioned above, Fractalkine runs as two bands on SDS-PAGE gels under these sample preparation conditions most probably due to incomplete denaturation. The 3C treated sample was further passed over a protein A-agarose column and an SDS-PAGE analysis of the flow-through from the column clearly shows that the sample is depleted of the Fc domain and also any remaining undigested Fractalkine-Fc, leaving mainly US28-1D4 and Fractalkine in the flow-through solution (Figure 5C, compare lanes 2 and 3).
The flow-through sample was concentrated down to 0.5 ml using a 50 kDa MWCO concentrator and applied to a Superose 6(10/30) gel filtration column. Protein elution was monitored using A280 (0.5 cm path length UV detector). The applied sample elutes from the column in two major peaks (Figure 5D). The first peak centered around 7.42 ml (fraction 2) corresponds to the void volume of the column and a sample from the collected fraction 2 when run on a gel indicates the presence of background aggregated proteins (Figure 5D inset). The second major peak centered around 14.8 ml (fraction 17) corresponds to a molecular weight of 230 kDa based on comparison with the elution volumes of known molecular weight standards used for column calibration. The collected sample fractions 12–22 when analyzed by SDS-PAGE clearly shows the co-elution of US28-1D4 doublet and Fractalkine across this peak with the staining intensity of both US28-1D4 and Fractalkine being the highest in fraction 17 (Figure 5D inset). This result indicates that US28-1D4 and Fractalkine form a stable, monodispersed complex and based on the elution position of the complex on the Superose 6(10/30) column, we conclude that the US28-1D4/Fractalkine complex exists as a dimer under these purification conditions. The final yield of the US28-1D4/Fractalkine complex from the gel filtration column is ~1 mg. For crystallization trials, fractions 15–19 were pooled together and concentrated down to a final protein concentration of ~10 mg/ml using a 100 kDa MWCO centrifugal concentrator. An aliquot of the concentrated sample analyzed by SDS-PAGE shows that the final purity of the sample is >90% (Figure 5E). In addition to the main US28-1D4 band running at 42–50 kDa and Fractalkine migrating at 14 kDa, a small amount of undissociated receptor dimer can also be seen running at ~97 kDa. Current nanovolumetric protein crystallization technologies permit screening of approximately 600 crystallization conditions of the sample from a single two-liter prep. We note the heterogenous nature of the purified US28-1D4 band on an SDS-PAGE gel and this is due to post-translational modifications at the multiple potential O-linked glycosylation sites, single Tyrosine sulfation site in the N-terminus and multiple Serine phosphorylation sites in the C-terminus of the receptor (data not shown). The presence of O-linked glycosylation and Tyrosine sulfation on chemokine receptors has been shown to be important for high affinity ligand binding [27, 28]. In addition, US28 has been shown to be constitutively phosphorylated in other recombinant expression systems [29].
Discussion
Recombinant protein expression in mammalian cells for the purpose of structural studies has been lagging behind other more commonly used systems such as E.Coli and the baculovirus system for insect cells. This can be mainly attributed to the rather time consuming and laborious process of generation, selection and maintanance of stable cell lines expressing high levels of the recombinant protein. Recently, PEI mediated transient transfection of mammalian cells has been used as a means for expressing recombinant proteins at the milligram level [30]. However, PEI mediated transfection suffers from the drawbacks that large quantities (in the order of milligrams) of high purity sterile plasmid DNA is required for large scale cultures and also a number of manual handling steps are involved during the transfection process of adherent as well as suspension cells. This could potentially give rise to large variations in transfection efficiencies and protein expression levels when the culture volumes are large.
An attractive alternative to this would be the use of baculovirus mediated protein expression in mammalian cells. We have constructed pVLAD7 and pVLAD6, novel baculovirus transfer vectors that incorporate several mammalian transcriptional elements necessary for this purpose. The whole process of transfection of Sf9 cells with only 2 µg of plasmid DNA, virus amplification and large-scale transduction and protein purification from 293 GnTI− cells takes approximately two weeks, which is comparable to the time frame of using the baculovirus system in insect cells. Plasmid DNA transfection into Sf9 cells is done on a very small scale thus enabling very consistent transfection efficiencies. There is very little human intervention involved during the course of virus amplification, virus transduction and protein expression. Most of the human effort is directed towards the relatively simple task of passaging cells in suspension every 3 days. We can grow large-scale cultures of these mammalian cells in a warm room to a relatively high density thus obviating the need for expensive multiple cell culture incubators. Once a large volume of the recombinant baculovirus has been generated, it can be stored and used multiple times over at least a 6-month period of time. All these factors account for a very high consistency of expression levels from prep to prep for large-scale cultures. Over the course of this study, the BacMam system in combination with the 293GnTI− cell line has been used successfully in our lab for expression and purification of a number of functional soluble glycoproteins that were previously refractory to expression or misfolded in other expression systems (unpublished data). In addition to the high expression levels, use of the 293GnTI− cells permits a very convenient way for deglycosylating bulk of the N-linked glycosylation on the purified glycoproteins. We have also shown that the BacMam system in conjunction with the 293 GnTI− cell line can be used for the purification of milligram quantities of a functional GPCR for structural studies.
In summary, we present results showing that any lab currently using baculovirus infection of insect cells, such as Sf9 and Hi5, can seamlessly incorporate mammalian cell expression methodologies since both baculovirus and BacMam both rely on the central methods of virus production, amplification, and titering, for which we have described highly streamlined methods here.
Acknowledgements
We are grateful to Dr. Frederick M. Boyce (Massachusetts General Hospital, Boston) for his advice during the initial stages of this work. This work was supported by grants from HHMI and W.M Keck foundation to KCG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: GPCR, G protein-coupled receptor; DDM, n-dodecyl-β-D-maltoside; CHS, cholesteryl hemisuccinate tris salt; Sf9, Spodoptera Frugiperda; FPLC, fast protein liquid chromatography; A280, absorbance at 280 nanometer wavelength; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element; GlcNAc, N-acetylglucosamine; PTH, parathyroid hormone; FCS, foetal calf serum; GnTI, N-acetylglucosaminyltransferase-I; MWCO, molecular weight cut-off; PTH, parathyroid hormone
References
- 1.Boyce FM, Bucher NL. Baculovirus-mediated gene transfer into mammalian cells. Proc Natl. Acad. Sci. U S A. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condreay JP, Witherspoon SM, Clay WC, Kost TA. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector, Proc. Natl. Acad. Sci. U S A. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. U S A. 1992;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I, Miyamura T, Matsuura Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 1997;78(Pt 10):2657–2664. doi: 10.1099/0022-1317-78-10-2657. [DOI] [PubMed] [Google Scholar]
- 5.Kost TA, Condreay JP, Ames RS, Rees S, Romanos MA. Implementation of BacMam virus gene delivery technology in a drug discovery setting. Drug Discov. Today. 2007;12:396–403. doi: 10.1016/j.drudis.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Scott MJ, Modha SS, Rhodes AD, Broadway NM, Hardwicke PI, Zhao HJ, Kennedy-Wilson KM, Sweitzer SM, Martin SL. Efficient expression of secreted proteases via recombinant BacMam virus. Protein Expr. Purif. 2007;52:104–116. doi: 10.1016/j.pep.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl. Acad. Sci. U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller N, Girard P, Hacker DL, Jordan M, Wurm FM. Orbital shaker technology for the cultivation of mammalian cells in suspension. Biotechnol. Bioeng. 2005;89:400–406. doi: 10.1002/bit.20358. [DOI] [PubMed] [Google Scholar]
- 9.Arakawa T, Philo JS, Tsumoto K, Yumioka R, Ejima D. Elution of antibodies from a Protein-A column by aqueous arginine solutions. Protein Expr. Purif. 2004;36:244–248. doi: 10.1016/j.pep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang MT, Gorman CM. Intervening sequences increase efficiency of RNA 3' processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potts JT. Parathyroid hormone: past and present. J. Endocrinol. 2005;187:311–325. doi: 10.1677/joe.1.06057. [DOI] [PubMed] [Google Scholar]
- 14.Bisello A, Greenberg Z, Behar V, Rosenblatt M, Suva LJ, Chorev M. Role of glycosylation in expression and function of the human parathyroid hormone/parathyroid hormone-related protein receptor. Biochemistry. 1996;35:15890–15895. doi: 10.1021/bi962111+. [DOI] [PubMed] [Google Scholar]
- 15.Grauschopf U, Lilie H, Honold K, Wozny M, Reusch D, Esswein A, Schafer W, Rucknagel KP, Rudolph R. The N-terminal fragment of human parathyroid hormone receptor 1 constitutes a hormone binding domain and reveals a distinct disulfide pattern. Biochemistry. 2000;39:8878–8887. doi: 10.1021/bi0001426. [DOI] [PubMed] [Google Scholar]
- 16.Monaghan P, Woznica I, Moza B, Sundberg EJ, Rosenblatt M. Recombinant expression and purification of the N-terminal extracellular domain of the parathyroid hormone receptor. Protein Expr. Purif. 2007;54:87–93. doi: 10.1016/j.pep.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang HC, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 2004;5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, Zhang LH, Singaraja RR, Guernsey DL, Zheng B, Siebert LF, Hoskin-Mott A, Trese MT, Pimstone SN, Shastry BS, Moon RT, Hayden MR, Goldberg YP, Samuels ME. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat. Genet. 2002;32:326–330. doi: 10.1038/ng957. [DOI] [PubMed] [Google Scholar]
- 19.Gao JL, Murphy PM. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J. Biol. Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 20.Kuhn DE, Beall CJ, Kolattukudy PE. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem. Biophys. Res. Commun. 1995;211:325–330. doi: 10.1006/bbrc.1995.1814. [DOI] [PubMed] [Google Scholar]
- 21.Kledal TN, Rosenkilde MM, Schwartz TW. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett. 1998;441:209–214. doi: 10.1016/s0014-5793(98)01551-8. [DOI] [PubMed] [Google Scholar]
- 22.Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 23.Mizoue LS, Sullivan SK, King DS, Kledal TN, Schwartz TW, Bacon KB, Handel TM. Molecular determinants of receptor binding and signaling by the CX3C chemokine fractalkine. J. Biol. Chem. 2001;276:33906–33914. doi: 10.1074/jbc.M101348200. [DOI] [PubMed] [Google Scholar]
- 24.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 25.Blackburn PE, Simpson CV, Nibbs RJ, O'Hara M, Booth R, Poulos J, Isaacs NW, Graham GJ. Purification and biochemical characterization of the D6 chemokine receptor. Biochem. J. 2004;379:263–272. doi: 10.1042/BJ20031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dukkipati A, Vaclavikova J, Waghray D, Garcia KC. In vitro reconstitution and preparative purification of complexes between the chemokine receptor CXCR4 and its ligands SDF-1alpha, gp120-CD4 and AMD3100. Protein Expr. Purif. 2006;50:203–214. doi: 10.1016/j.pep.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Bannert N, Craig S, Farzan M, Sogah D, Santo NV, Choe H, Sodroski J. Sialylated O-glycans and sulfated tyrosines in the NH2-terminal domain of CC chemokine receptor 5 contribute to high affinity binding of chemokines. J. Exp. Med. 2001;194:1661–1673. doi: 10.1084/jem.194.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farzan M, Babcock GJ, Vasilieva N, Wright PL, Kiprilov E, Mirzabekov T, Choe H. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1 alpha association and HIV-1 entry. J. Biol. Chem. 2002;277:29484–29489. doi: 10.1074/jbc.M203361200. [DOI] [PubMed] [Google Scholar]
- 29.Mokros T, Rehm A, Droese J, Oppermann M, Lipp M, Hopken UE. Surface expression and endocytosis of the human cytomegalovirus-encoded chemokine receptor US28 is regulated by agonist-independent phosphorylation. J. Biol. Chem. 2002;277:45122–45128. doi: 10.1074/jbc.M208214200. [DOI] [PubMed] [Google Scholar]
- 30.Geisse S, Jordan M, Wurm FM. Large-scale transient expression of therapeutic proteins in mammalian cells. Methods Mol. Biol. 2005;308:87–98. doi: 10.1385/1-59259-922-2:087. [DOI] [PubMed] [Google Scholar]