Summary
Sequenced genomes of dissimilatory sulfur-oxidizing and sulfate-reducing bacteria containing genes coding for DsrAB, the enzyme dissimilatory sulfite reductase, inevitably also contain the gene coding for the 12-kDa DsrC protein. DsrC is thought to have a yet unidentified role associated with the activity of DsrAB. Here we report the solution structure of DsrC from the sulfur-oxidizing purple sulfur bacterium Allochromatium vinosum determined with NMR spectroscopy in reducing conditions, and describe the redox behavior of two conserved cysteine residues upon transfer to an oxidizing environment. In reducing conditions, the DsrC structure is disordered in the highly conserved carboxy-terminus. We present multiple lines of evidence that in oxidizing conditions, a strictly conserved cysteine (Cys111) at the penultimate position in the sequence forms an intramolecular disulfide bond with Cys100, which is conserved in DsrC in all organisms with DsrAB. While an intermolecular Cys111-Cys111 disulfide-bonded dimer is rapidly formed under oxidizing conditions, the intramolecularly disulfide-bonded species (Cys100-Cys111) is the thermodynamically stable form of the protein under these conditions. Treatment of the disulfidic forms with reducing agent regenerates the monomeric species that was structurally characterized. Using a band-shift technique under non-denaturing conditions evidence was obtained for interaction of DsrC with heterohexameric DsrEFH, a protein encoded in the same operon. Mutation of Cys100 to serine prevented formation of the DsrC species assigned as an intramolecular disulfide in oxidizing conditions, while still allowing formation of the intermolecular Cys111-Cys111 dimer. In the reduced form this mutant protein still interacted with DsrEFH. This was not the case for the Cys111Ser and the Cys100Ser/Cys111Ser mutants, both of which also did not form protein dimers. Our observations highlight the central importance of the carboxy-terminal DsrC cysteine residues and are consistent with a role as a sulfur-substrate binding/transferring protein as well as with an electron-transfer function via thiol-disulfide interchanges.
Keywords: dissimilatory sulfite reductase, DsrC, sulfur oxidation, NMR solution structure, anoxygenic phototrophic sulfur bacteria
Introduction
In phototrophic sulfur-oxidizing bacteria, dissimilatory sulfite reductase (DsrAB, a tetramer composed of two alpha and two beta subunits) acting in reverse is believed to oxidize sulfide to sulfite to generate electrons for photosynthetic CO2 reduction.1–3 Very importantly, the enzyme is not vital for the oxidation of externally added sulfide. Instead, it has been shown for the purple sulfur bacterium Allochromatium vinosum that “reverse” sulfite reductase is essential for the oxidation of periplasmic sulfur globules formed as intermediates during growth on sulfide and thiosulfate.3 Sulfide as a substrate for the cytoplasmatically localized sulfite reductase is currently proposed to be generated by reduction of a perthiolic compound that serves as a carrier of sulfur from the periplasm into the cytoplasm.4–6 In sulfate-reducing bacteria and archaea, where sulfate is the terminal electron acceptor in anaerobic respiration, DsrAB catalyzes the reduction of sulfite to sulfide.7 All organisms with dsrAB also have the gene for DsrC, in some instances within the same operon as dsrAB and other dsr genes, and in other cases distant from dsrAB.4,8–11 Preparations of DsrAB isolated from sulfate-reducing bacteria have been reported to contain bound DsrC, and for this reason it has been called the gamma subunit of dissimilatory sulfite reductase.12,13 In other preparations of sulfite reductase, DsrC did not appear to be present.14 Along this line, direct interaction between DsrAB and DsrC from the phototrophic purple sulfur bacterium Ach. vinosum has not been reported, though all three genes occur in a large cluster, dsrABEFHCMKLJOPNSR.2,3
Some genes in this cluster, most notably dsrEFH, are specific to sulfur-oxidizing bacteria containing DsrAB and do not occur in sulfate-reducing prokaryotes.5,9,10 DsrEFH and also DsrC homologues are found in bacteria lacking DsrAB, including E. coli and many other Gammaproteobacteria. In E. coli, the DsrEFH homologues TusBCD interact with the DsrC homologue TusE in a sulfur relay system during 2-thiouridine biosynthesis.15 The crystal structure of Ach. vinosum DsrEFH has been determined, and shows that it is a hexamer containing two of each subunit.16 Recently, specific transfer of sulfur to Cys78 of Ach. vinosum DsrE by E. coli cysteine desulfurase IscS was experimentally verified (article submitted). Together, these findings indicate that sulfur transfer reactions involving DsrEFH and possibly also DsrC may be important during sulfur oxidation. On the other hand, DsrEFH and DsrC from Ach. vinosum were both co-purified with the membrane protein complex DsrMKJOP.2 This observation together with sequence similarities between DsrK and the catalytic subunit of archaeal heterodisulfide reductase have led to the suggestion that DsrK transfers electrons to or from DsrC via thiol-disulfide interchanges.2,7,17
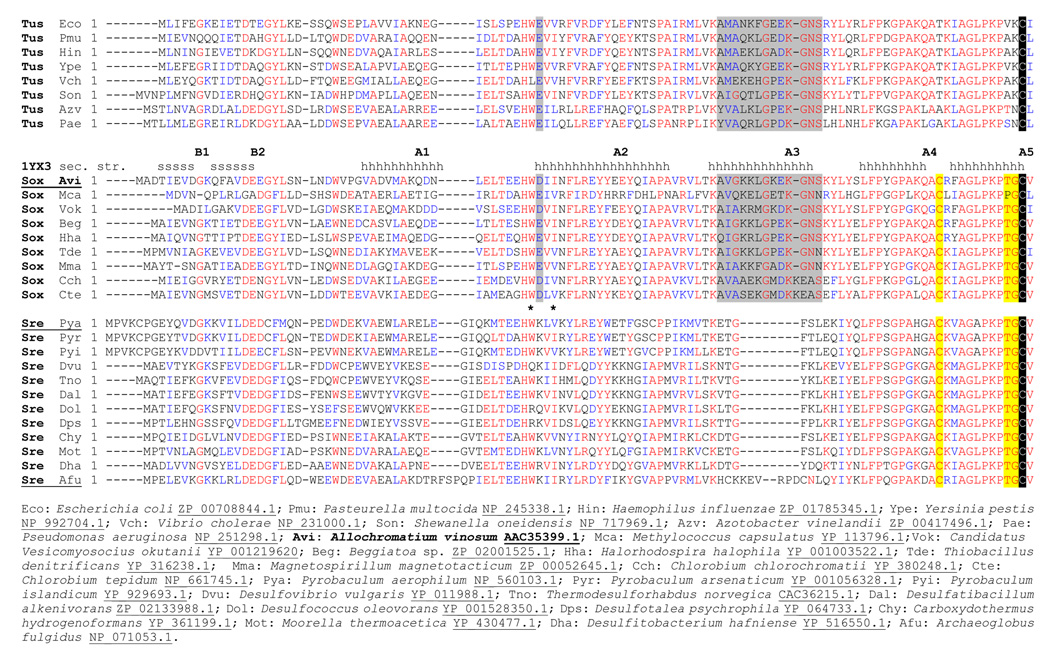
All DsrC sequences contain a strictly conserved Cys residue at the penultimate position in the carboxy-terminus (Fig. 1). Other residues at the carboxy-terminus are highly conserved, and the NMR structure of DsrC from the archaeon Pyrobaculum aerophilum showed these residues to be disordered,18 suggesting that this region interacts with other proteins as otherwise there would be no selective pressure against random mutations at these positions. DsrC sequences from organisms containing DsrAB, but not those bacteria lacking DsrAB (E. coli is an example for the latter) inevitably have an additional conserved Cys in the sequence (at a distance of 10 residues). The possible interaction of the two cysteines is suggested in the crystal structure of DsrC from the sulfate-reducing archaeon Archaeoglobus fulgidus, in which some molecules in the lattice have only 3.63 Å separation between the sulfur atoms of the two Cys residues.19
Fig. 1. Multiple sequence alignment of DsrC/TusE sequences.
Sequences are grouped according to whether the host organism’s genome has tusBCD but lacks dsrAB (Tus), has dsrEFH and dsrAB (Sox, sulfur oxidizers), or lacks dsrEFH but has dsrAB (Sre, sulfate or sulfite reducers). These groupings to correspond well to those seen in the phylogenetic tree cladogram, with the exception that DsrC sequences from Pyrobaculuum species were grouped more closely with sequences from dsrEFH-containing-sulfur oxidizers, rather than with sulfate-reducing bacteria. Below the sequence alignment are shown the full organism names and corresponding database accession numbers for each sequence. Positions of secondary structure elements in A. vinosum DsrC are indicated above its sequence. Residue insertions specific to sulfate reducers or TusBCD/DsrEFH-containing organisms are marked with asterisks (*) and grey shading, respectively. C-terminal residues specific to DsrAB-containing sulfur oxidizers or sulfate reducers are identified with yellow shading. The invariant penultimate cysteine is shaded black.
In our effort to attribute functions to the proteins encoded at the Ach. vinosum dsr locus and to clarify the dsr-encoded sulfur oxidation pathway, we collected not only structural information on DsrC via NMR but also studied disulfide bond formation within and between protein monomers for wild type DsrC as well as for mutant DsrC proteins lacking conserved cysteine residues. Furthermore, we obtained definite proof for interaction between DsrC and DsrEFH and show that this interaction is strictly dependent on the penultimate cysteine of DsrC.
Results
Chemical shift assignments and structure
Backbone amide 15N and 1H, alpha 13C and 1H, carbonyl 13C, and side chain 13C, 15N, and 1H chemical shift assignments were 97% complete over residues 2–112. Apart from residue M1, the only commonly assigned side chain atoms lacking assignments were Pro28 13Cγ, Lys71 13Cε, Lys79 1Hε and 13Cε, and Ser84 1Hβ and 13Cβ. Amide 15N and 1H assignments were missing for Glu44, Lys79, Ser84, and Cys111. All 13Cα shifts were assigned, and Ser85 1Hα was the only unassigned alpha proton. Chemical shifts were deposited to BioMagResBank with accession number BMRB-6518. A total of 1570 NOE distance restraints were extracted from the NOESY spectra and used together with 74 dihedral restraints and 70 H-bond restraints (2 per H-bond) to generate an ensemble of 20 structures. The backbone (Cα, N, C) r.m.s.d. to the average structure over residues 3–108 was 0.68 Å. Additional structure statistics are shown in Table 1. For the ensemble of structures, 96.9% of residues 3–108 occur in the most favored or additionally allowed region of the Ramachandran plot, according to PROCHECK. The structure was deposited to the Protein Data Bank with the accession number 1YX3.
Table 1.
Structural statistics for the 20-member ensemble of Ach. vinosum DsrC structures
| Distance restraints | |
| Total | 1570 |
| Intraresidue | 447 |
| Sequential | 335 |
| Medium range (1 < |i-j| < 5) | 333 |
| Long range | 455 |
| Hydrogen bonds | 2*35 |
| Dihedral angle restraints | |
| Total | 74 |
| phi | 37 |
| psi | 37 |
| chi-1 | 0 |
| Total restraints | 1714 |
| Restraints per restrained residue (106 residues) | 16.2 |
| Restraints, long range, per residue (106 residues) | 4.3 |
| Average restraint violations per structure | |
| Distance restraints (all > 0.0 Å) | 33.8±3.5 |
| maximum violation (Å) | 0.04 |
| Dihedral restraints (all > 0.00) | 0.65±0.67 |
| maximum violation (°) | 0.58° |
| R.m.s.d to average structures (Å) | |
| Residues 3–108 (106 residues) | |
| Backbone atoms (N,Cα,C’) | 0.68 |
| All heavy atoms | 1.17 |
| Residues 5–44,52–93 (101 residues—78–82 loop excluded) | |
| Backbone atoms (N,Cα,C’) | 0.61 |
| All heavy atoms | 1.09 |
| Ramachandran (PROCHECK): residues 3–108 inclusive (106 residues), 20 structures | |
| most favored region (%) | 83.3 |
| additional allowed region (%) | 13.6 |
| generously allowed region (%) | 1.7 |
| unallowed region (%) | 1.4 |
Description of the structure
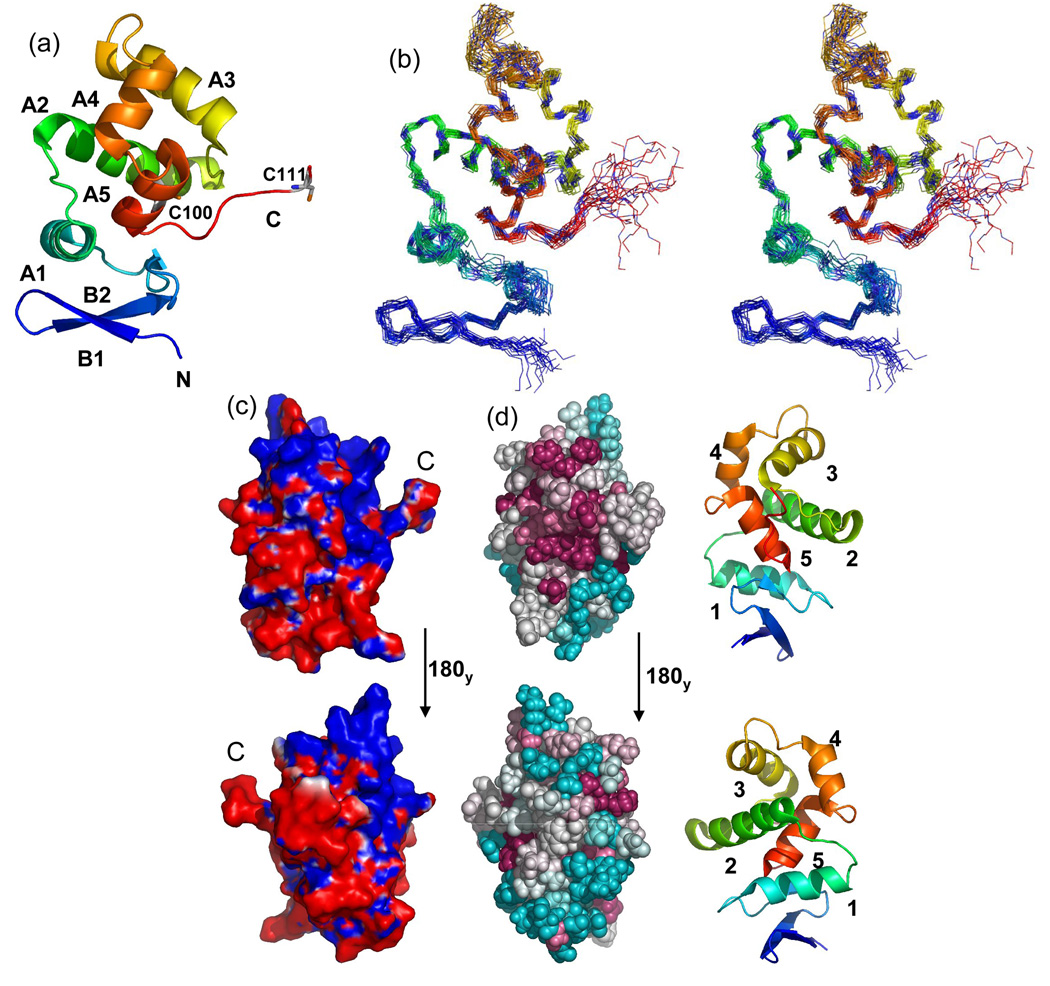
The Ach. vinosum DsrC tertiary structure consists of a two strand β-hairpin occurring at the amino-terminus, followed by five helices that form an adjoining orthogonal bundle encircling the fifth helix (Fig. 2a,2b). The conserved carboxy-terminal residues exit the bundle from the end of the fifth helix, forming a flexible arm. Mapping of the calculated electrostatic potential on the molecular surface showed a positively charged region of the structure centered around helices 3 and 4 that includes the point of exit of the carboxy-terminal arm from the globular protein structure (Fig. 2c). Furthermore, conserved residue Cys100 is partially buried within this region.
Fig. 2. DsrC ribbon structures, ensemble stereoview, electrostatic surface, and structural map of conserved residues.
(a) Ribbon cartoon of A. vinosum DsrC. The amino-terminus is blue, the carboxy-terminus is red. The amino-terminal hexa-His tag is not shown. Alpha helices and beta strands are labeled. Positions of cysteines are shown. Only a single conformation of the dynamic carboxy-terminal segment (residues 107–112) is shown. (b) Stereoview of A. vinosum DsrC ensemble. The amino-terminus is blue; the carboxy-terminus is red. The amino-terminal hexa-His tag is not shown. (c) Molecular surface colored by electrostatic potential. Red and blue colors correspond to units of -1 and +1 kbT/ec. The perspectives are related by a 180 deg. rotation about the y-axis. The left view is the same as in Figs. 3a and 3b. (d) ConSurf35 depiction of sequence conservation in the DsrC/TusE multiple sequence alignment mapped onto the Ach. vinosum DsrC structure. For reference, numbered ribbon cartoon structures appear to the right of each ConSurf map. The upper structures are rotated 90 degrees about the y-axis from that the orientation in Fig. 3a, so as to look directly upon the carboxy-terminus. Lower structures are rotated 180 degrees again about the y-axis. Darker, warmer colors indicate greater residue conservation.
Carboxy-terminal tail dynamics
Interpretation of the observation of disorder at the carboxy-terminus in the P. aerophilium DsrC NMR structure was confounded by the presence of a carboxy-terminal 8-residue hexa-His purification tag, which could have disrupted native interactions at the carboxy-terminus in a way that caused the disorder in this region.18 To alleviate this concern, the Ach. vinosum DsrC construct had its His tag placed at the amino-terminus of the protein, away from the carboxy-terminus. One bond amide heteronuclear NOE data (not shown) were qualitatively smaller than the average value for the rest of the protein, excluding two residues at the amino-terminus. These data indicate greater mobility in the carboxy-terminus beginning at residue 106. Random coil chemical shifts and absence of long range correlations in NOESY spectra for the carboxy-terminal residues also imply greater mobility in this part of the protein. Somewhat greater backbone mobility may also occur in the segment between helices 3 and 4, indicated by fewer NOESY peaks, more unobserved resonances, and sharper resonance lines. The effects of these characteristics can be seen as increased disorder in the structure superposition of the NMR ensemble (Fig. 2b).
Comparative analysis to similar structures
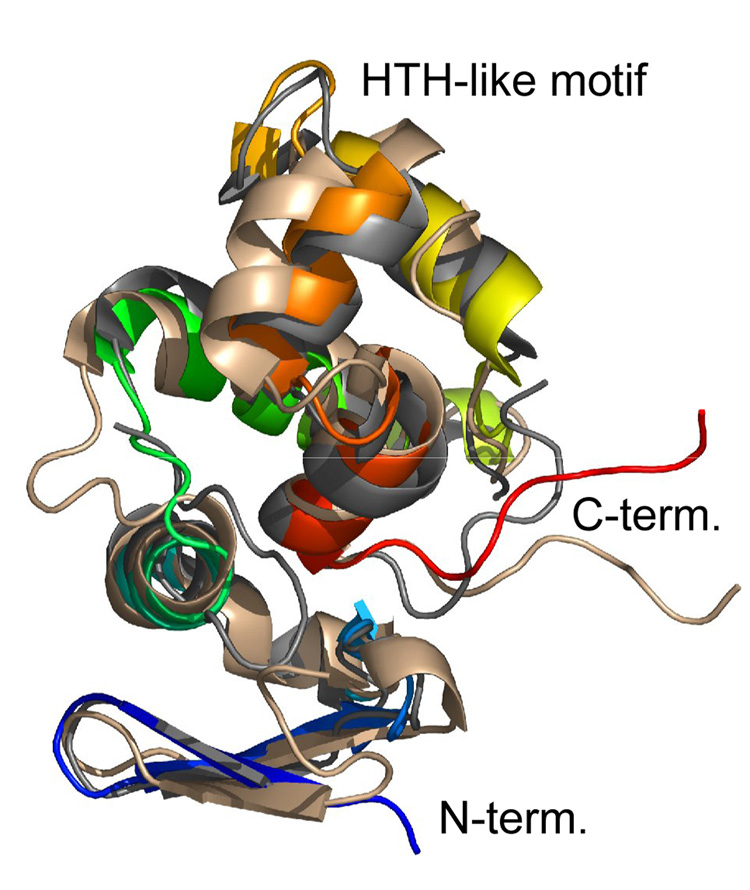
As expected, the structure of Ach. vinosum DsrC is quite similar to that of two archaeal DsrC structures determined previously: the NMR structure of Pyrobaculum aerophilum DsrC18 and the X-ray structure of Archeoglobus fulgidus DsrC.19 Ach. vinosum DsrC has 40% and 46% sequence identity to these other two DsrCs, respectively. Structure similarity searches with Dali20 identify only other DsrC proteins as matches to the entire structure. Superpositions of these structures’ CA atoms with those of Ach. vinosum DsrC using pairwise Dali indicated reasonable similarity: Cα atoms of the P. aerophilum structure superimpose with r.m.s.d 2.9 Å over 104 residues with a Z-score of 12.0, while the Agl. fulgidus structure superimposes with r.m.s.d 1.9 Å over 106 residues with Z-score 15.4 (Fig. 3).
Fig. 3.
Superposition of DsrC structures Ach. vinosum (1YX3, rainbow), P. aerophilum (1JI8, wheat), and Agl. fulgidus (1SAU, grey).
Among the structures of the three DsrC proteins from different organisms, the most variable regions are a helix-turn-helix-like structural motif between helices 3 and 4, and the segment connecting helices 1 and 2 (Fig. 1). These regions are the sites of the most sequence variation due to residue insertions and deletions. Between helices 1 and 2, Agl. fulgidus DsrC has a 5-residue insertion that forms a loop at the end of helix 1, while P. aerophilum DsrC has a two-residue insertion that forms part of a bulge in the connecting segment; this insertion is shared only with DsrC from other Pyrobaculuum species, and is not present in DsrC sequences from sulfate reducers. In the region around helices 3 and 4, P. aerophilum DsrC and homologues from sulfite or sulfate-reducing organisms have 7 fewer residues than DsrC proteins form sulfur-oxidizing bacteria and bacteria lacking dsrAB genes. Notably all organisms that have the 7-residue insertion (which is 8 residues in Chlorobium sp.) also have dsrEFH genes or their homologues tusBCD (Fig. 1). These groups correspond to dsrAB-containing sulfur oxidizers (the dsrEFH group) or to bacteria lacking dsrAB that have the dsrC and dsrEFH homologues tusE and tusBCD. A single exception to this pattern is Agl. fulgidus DsrC, which has a six-residue insertion in this region, but the insertion sequence is not similar to the other sequences in the group.
This 7-residue insertion that is found in DsrC from all sulfur oxidizers and in TusE manifests itself structurally in Ach. vinosum DsrC as a longer helix 3 and extra turn residues prior to helix 4, when compared to the P. aerophilum DsrC structure. Because of differences in the turn, the relative orientations of helix 4 in the two proteins are somewhat different in the global superposition (Fig. 3). In Ach. vinosum DsrC, the insertion provides four additional lysine residues, making that part of the surface markedly positively charged (Fig. 2c). Other sequences have fewer lysine residues in the insertion, although one (K81) is strictly conserved in all DsrC sequences that have the insertion. In general however, sequence conservation within the insertion is lower than elsewhere in the proteins. The Agl. fulgidus DsrC, whose sequence seems to be in a class by itself, has a 5-residue insertion that differs from the other insertion sequences. However, the Agl. fulgidus DsrC structure is similar to the Ach. vinosum structure in this region, despite the limited sequence similarity, as it is elsewhere in the structure where sequence is more conserved.
Mapping conserved residues onto the structure of Ach. vinosum DsrC shows that the conserved residues on the surface cluster at the side of the protein on which the carboxy-terminal arm exits the globular structure. This pattern holds whether all DsrC and TusE sequences are included in the alignment used for the mapping, or whether subgroups of sequences clustered in the manner described above are used. Of course, subgroups have greater similarity to each other, so additional patches of conserved surface-exposed residues are seen, but they still appear for the most part on the same side of the protein. An exception to this is seen in the insertion sequence of DsrC from sulfur oxidizers and in TusE centered around helices 3 and 4 that is absent in DsrC from sulfate/sulfite reducers; conserved residues in this insertion occur at the top of the protein structure as depicted, and the conserved lysine residue (Lys81) is found on the opposite side of the protein.
The DsrC fold was first observed in P. aerophilum DsrC18, and at the time of writing has only been observed in other DsrC proteins. However, as noted previously for P. aerophilum DsrC18, the substructure centered around helices 3 and 4 is found to be strikingly similar to the DNA-binding domains of bacterial transcriptional regulatory proteins; in Ach. vinosum DsrC for example the backbone N, Cα, and C’ atoms of 48 residues spanning helices 2, 3, and 4 in Ach. vinosum DsrC superimpose on a structurally similar portion of the DNA-binding domain of the TetR/CamR family repressor protein EthR21 from M. tuberculosis with 2.2 Ǻ r.m.s.d.
Disulfide bond formation in oxidizing conditions
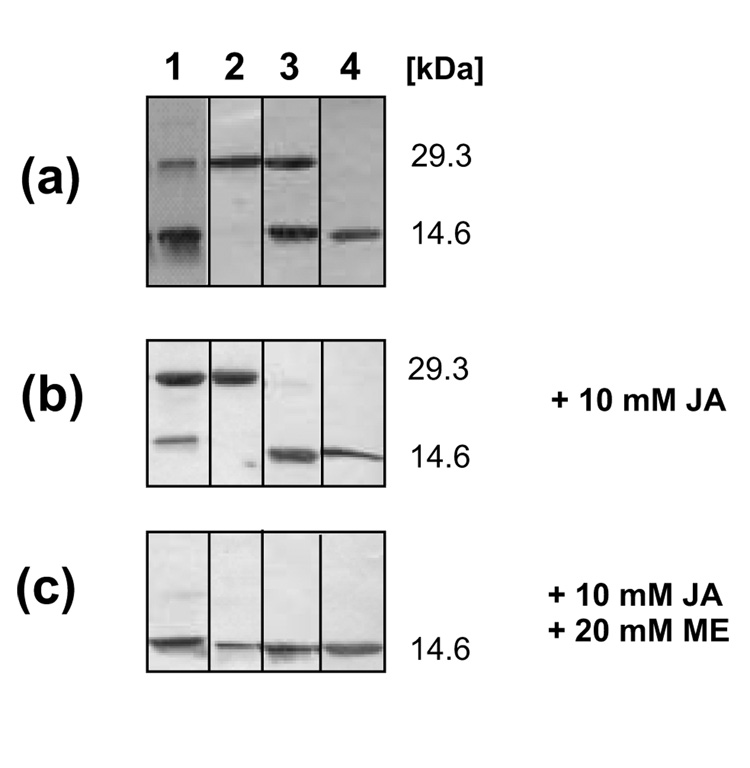
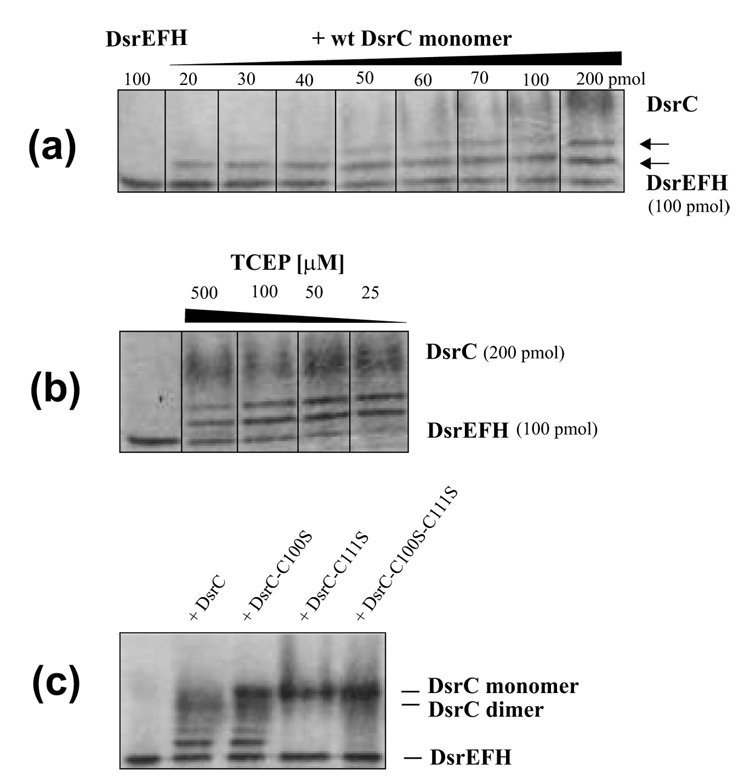
In the presence of a reducing agent (1 mM TCEP) recombinant DsrC eluted from gel filtration on Sephadex G75 at a volume corresponding to the monomeric recombinant protein (14,638 Da). In contrast, two peaks at 29 and 14.6 kDa, corresponding to dimeric and monomeric DsrC, were observed in the absence of a reducing agent. SDS-PAGE under reducing conditions yielded a single band of 14.6 kDa for both fractions (not shown), indicating that dimerization of DsrC in the absence of a reducing agent was due to formation of a disulfide bond between two DsrC monomers. When dimeric DsrC was prepared for SDS-PAGE by boiling in non-reducing buffer in the absence of the alkylating agent iodoacetamide (Fig. 4a, lane 1) intensity of the 29 kDa band decreased significantly in comparison with samples in which free cysteine had been alklyated before boiling (Fig. 4b, lane 1). This observation can be explained by the assumption that free cysteine residues not engaged in disulfide bonds resolved the pre-existing interprotein disulfide upon incubation at elevated temperatures. When these free cysteine residues are alkylated they are no longer able to resolve the disulfide (Fig. 4b, lane 1). Treatment with excess reducing agent after alkylation of dimeric DsrC resulted in the monomeric protein (Fig. 4c, lane 1) and provided final proof for the existence of an interprotein disulfide. To assess the role of each individual conserved cysteine residue in disulfide bond formation and resolution we generated mutated DsrC with either one or both, Cys100 and Cys111, replaced by serine. When both cysteines were replaced all protein behaved as a monomer in size exclusion chromatography and upon SDS-PAGE analysis (Fig. 4a, lane 4) proving that disulfide bond formation is the sole reason for dimerization of DsrC. In the absence of Cys111, the native protein also behaved exclusively as a monomer but part of it was turned into a Cys100-Cys100 dimer after boiling under non-reducing conditions (Fig. 4a, lane 3). However, alkylation with iodoacetamide prevented formation of this bond (Fig. 4b, lane 3). When Cys100 was turned into a serine almost all of the native protein was present as a dimer, obviously due to Cys111-Cys111 disulfide bond formation. This dimer was no longer transformed into a monomer upon non-reductive SDS-PAGE due to the lack of possible resolving Cys100 residues (lane 2 in Fig. 4a and 4b). Summarizing these experiments it appears that DsrC purified in the absence of reducing agents has a strong tendency to form dimers by disulfide bond formation involving the carboxy-terminal cysteine residue at position 111. Cysteine 100 is not involved in intermolecular disulfide bonds.
Fig. 4. SDS-PAGE analysis with non-reducing sample buffer of wild-type and mutated DsrC proteins purified under aerobic conditions in the absence of reducing agent.
Lane 1, wt DsrC dimeric, Lane 2, DsrC-Cys100Ser dimeric, Lane 3, DsrC-Cys111Ser monomeric, Lane 4 DsrC-Cys100Ser/Cys111Ser monomeric (a) proteins as isolated (b) proteins pretreated with iodoacetamide leading to alkylation of free cysteine residues (c) proteins reduced with 2-mercaptoethanol after treatment with iodoacetamide
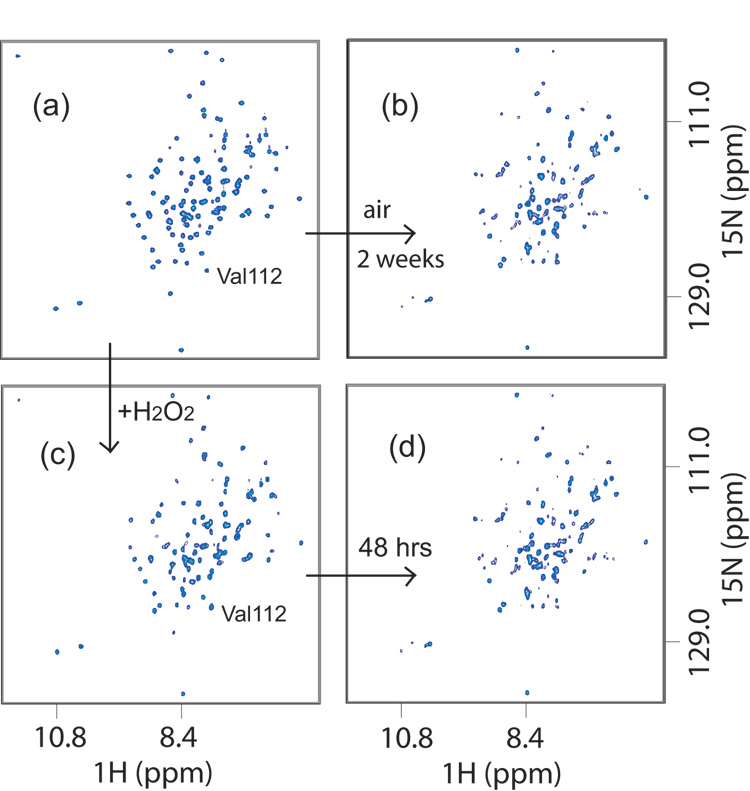
Disulfide bond formation of DsrC molecules was further analyzed by NMR. We initially observed that a small volume (< 1 mL) of ca. 1 mM DsrC in NMR buffer stored for several weeks at 4 °C in a 15 mL conical tube displayed dramatically different 1H-15N-HSQC spectra before and after this period of aging (Fig. 5). No precipitate was observed in the tube, nor was degradation of the protein apparent in SDS-PAGE gels. However, addition of 5 mM DTT apparently restored this sample to its original state, as the resulting 1H-15N-HSQC spectrum was identical to the original. We duplicated this behavior by incubating a reduced sample with hydrogen peroxide (ca. 10–30 mM, or 0.02–0.05% wt. %) and monitoring the changes in its 1H-15N-HSQC spectrum over time (Fig. 5). Initially, only the peaks for the residues Thr109 and Val112 were perturbed (Fig. 5c). Subsequently, other peaks moved or acquired a scratchy, distorted appearance, while others remained unchanged. The spectra of H2O2-treated DsrC after 48 hours (Fig. 5d) and 120 hours (not shown) were very similar to the one originally obtained from a sample stored for weeks under air (Fig. 5b).
Fig. 5. 1H-15N-HSQC NMR spectra of DsrC in oxidizing conditions.
(a) Reduced DsrC. (b) Aged two weeks under air. (c) Oxidized by addition of ~20 mM H2O2 to reduced DsrC. (d) After 48 hrs in ~20 mM H2O2. In (b) and (d), non-uniform peak appearance suggests partially disordered structure, though some peaks remain unchanged. In (c) versus (a), slight upfield 15N shift of V112 peak indicates rapid Cys111-Cys111 disulfide bond formation.. Similarity of (b) and (d) show that the oxidized state is reached by air or H2O2 oxidation. Re-reduction of the oxidized state with DTT yields spectra indistinguishable from (a).
NMR spectroscopy of DsrC mutant proteins in oxidizing conditions
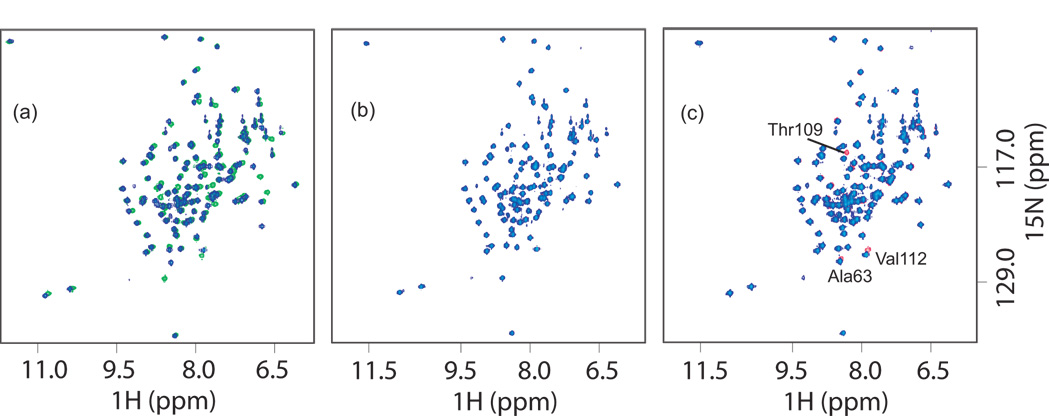
Chemical shift dispersion and line widths of peaks in the 1H-15N-HSQC spectra of DsrC mutants are similar to that seen for the native protein. Superposition of spectra of mutant DsrC proteins with native DsrC show that changes are localized to sequence neighbors and proximal residues in the tertiary structure (Fig. 6). These results indicate that the mutant proteins are properly folded. Following on the observation that addition of hydrogen peroxide to reduced native DsrC produced identical changes in the NMR spectra to those seen when solutions of the protein sat for many days under air, H2O2 (approximately 0.05% v/v) was used to accelerate protein disulfide formation in DsrC cysteine mutants. Resulting changes in amide 1H and 15N chemical shifts relative to the shifts observed in reducing conditions were used to infer local perturbations in the chemical environment around specific residues caused by the oxidizing environment. In the Cys100Ser mutant, changes in amide 1H-15N-HSQC peaks for residues Ala63, Thr109, and Val112 were observed. This is similar to what is seen initially in native DsrC. Residue 110 is proline, and lacking an amide proton therefore does not yield a peak in this experiment. The 1H-15N-HSQC peak for Cys111 is not observed for native DsrC, presumably due to intermediate conformational exchange at this residue, and a Ser111 peak is also not observed in this mutant. These observations suggest formation of an intermolecular disulfide between Cys111 residues of two molecules, forming a dimer of two independent domains connected by a flexible linker. In the disulfide dimer state, this linker must perturb residue Ala63 which is adjacent to the point at which the polypeptide chain exits the globular portion of the protein near the carboxy-terminus at residue Pro106. There is no evidence for interaction between the two globular domains other than at Ala63 and in the linker itself. Unlike native DsrC however, upon aging over time, no further changes in the spectra are observed, suggesting that the absence of Cys100 in this mutant precludes the substantial changes seen in the 1H-15N-HSQC spectra of native DsrC as the protein ages in oxidizing conditions that result from slow intramolecular disulfide bond formation (Fig. 5). With the Cys111 to Ser mutant, there is no evidence of intermolecular dimer formation via a Cys100-Cys100 disulfide between two protein molecules. Presumably, this is because Cys100 is not located on an exposed convex surface of the protein, and its thiol cannot approach the Cys100 thiol of another DsrC molecule. Likewise, in the Cys100Ser/Cys111Ser double mutant, there is no evidence in the 1H-15N-HSQC spectrum for any effect from hydrogen peroxide; the spectrum is identical to that of the mutant protein in reducing conditions.
Fig. 6. 1H-15N HSQC spectra of DsrC and mutants.
(a) Native DsrC (green) and DsrC Cys100Ser/Cys111Ser (blue) in reducing conditions showing effects of introducing Cys mutations. Shifts in peaks are due to neighbor effects near mutated residues. (b) DsrC Cys111Ser with DTT (blue) and without DTT (red, not visible because there is no change in the spectra and they are underneath the blue peaks). (c) DsrC Cys100Ser with DTT (blue) and without DTT (red). Changes occur around residue Cys111 due to disulfide formation, and at Ala63 (see text). .
Biological significance of DsrC
In order to determine the importance of DsrC for oxidative sulfur metabolism in Ach. vinosum we attempted an in frame deletion of the gene. This method has repeatedly been used for the specific inactivation of individual dsr genes in this purple sulfur bacterium.10,22 However, the Ach. vinosum mutant lacking dsrC turned out to be genetically unstable and could neither be stably maintained in liquid culture on media containing malate as electron donor and carbon source nor be kept in the presence of reduced sulfur compounds. These findings indicate that DsrC is indispensable in Ach. vinosum even in the absence of reduced sulfur compounds. It should be noted that dsrC, which resides amidst other dsr genes, is constitutively transcribed from its own promoter located in the dsrF coding region.3 As a consequence, formation of DsrC is not turned off in Ach. vinosum strain 21D carrying an omega interposon in the dsrB gene located upstream of dsrC.2,3 In contrast to the mutant carrying an in frame deletion of dsrC the latter strain is viable and genetically stable. Our interpretation is furthermore supported by the observation that an Ach. vinsoum mutant lacking dsrE, dsrF and dsrH exhibits the same genetic instability as the dsrC deletion strain, most probably due to removal of the dsrC promoter in dsrF. In contrast, a strain lacking solely dsrE is viable and genetically stable albeit it is completely unable to oxidize intracellular sulfur (Frauke Grimm & Christiane Dahl, unpublished). In summary, our analyses of mutant strains underline the eminent importance of DsrC in central metabolic pathways of Ach. vinosum but on their own do not allow to pin point a function restricted to oxidative sulfur metabolism.
DsrC interactions with DsrEFH
An important role and specific function of DsrC in oxidative sulfur metabolism is indicated by the co-purification of Ach. vinosum DsrC with DsrAB, DsrKJ, and DsrE, DsrF and DsrH, all of which are encoded in immediate vicinity.2 The proteins DsrE, DsrF and DsrH form an α2β2γ2-structured heterohexamer and share sequence similarity with TusB (formerly YheL), TusC (formerly YheM) and TusD (formerly YheN) from E. coli, respectively.3,15 Conserved Cys78 of A. vinosum DsrE corresponds to the active cysteines of the structurally characterized E. coli TusD23 and also the related homo-oligomeric E. coli protein YchN.24 Furthermore, DsrC exhibits sequence similarities with the E. coli protein TusE (formerly YccK). It has been shown that within a sulfur relay system involved in thiouridine biosynthesis the TusBCD complex directly accepts a sulfur atom from the protein TusA in a TusE-dependent manner and that TusBCD is a common interaction partner of TusE in E. col.15 Together all these findings suggested that Ach. vinosum DsrC might specifically interact with DsrEFH. We assessed that possibility by using a band-shift technique under non-denaturing conditions. The native proteins were preincubated in binding buffer. After dilution with sample buffer, proteins were separated by native polyacrylamide electrophoresis. Indeed, two additional shifted bands were clearly identified after incubation of wild-type DsrC and DsrEFH. These bands migrated more slowly than DsrEFH alone. Although it is likely that the slower migrating bands are due to formation of a higher molecular weight complex, we can currently not completely exclude that conformational changes of DsrEFH induced by DsrC led to altered migration of DsrEFH in polyacrylamide gels. The amount of slower migrating DsrEFH was dependent on the amount of DsrC added. As shown in Fig. 7a titration of 100 pmol DsrEFH with DsrC in the range of 20–200 pmol steadily increased the intensity of the additional bands with the faster migrating of these two appearing first. The intensity of the additional bands even increased at optimum concentrations of the reducing agent TCEP (25 µM) at which the original DsrEFH band was completely transformed into the slower migrating forms (Fig. 7b).
Fig. 7. The ability of DsrC to interact with DsrEFH was determined by band-shift assays.
(a) 100 pmol of DsrEFH were incubated with increasing amounts of wild type DsrC monomer in a total volume of 60 µl under the conditions specified in materials and Methods. In this series of experiments TCEP was not added. Increasing amounts of DsrC produced an additional band migrating between DsrEFH and the wt DsrC monomer. A molar ratio DsrE2F2H2 : DsrC of 2 : 1 and lower led to formation of a second distinct retarded band. Both retarded bands increased in intensity upon increase of DsrC concentration. Intensity of the original DsrEFH band decreased concomitantly. (b) Effect of reducing agent (TCEP) on formation of additional bands in band shift assays. Upon addition of 25 µM TCEP 100 pmol DsrE2F2H2 are completely retrieved in two shifted bands in the presence of 200 pmol monomeric DsrC. Higher concentrations of TCEP decrease shifted band intensities. (c) Band shift assay of DsrEFH with wild type and mutant DsrC proteins. 100 pmol DsrEFH were incubated with 200 pmol of the respective DsrC protein in the presence of 25 µM TCEP as described in Materials and Methods. Lane 1, shifted bands are clearly visible for wt DsrC and DsrC-Cys100Ser but not present in samples with DsrC-Cys111Ser and DsrC-Cys100Ser/Cys111Ser.
Complex formation between DsrC and DsrEFH was also assessed using NMR spectroscopy. Peaks in the 1H-15N-HSQC spectrum recorded on a 1:1 mixture of 13C,15N-DsrC and unlabeled DsrEFH had substantially broader linewidths overall compared to the same experiment recorded on DsrC alone (Fig. 6), aside from mobile amides near the N-terminus and in Asn and Gln residues. This is consistent with an increased rotational correlation time in DsrC due to formation of a high molecular weight complex with DsrEFH. Chemical shifts of most peaks did not change appreciably, but approximately 15 backbone amide peaks disappeared, suggesting the presence of a specific interface with DsrEFH rather than non-specific binding or aggregation.
In a final set of experiment we tested whether the interaction of DsrEFH and DsrC was dependent on the presence of the conserved DsrC cysteine residues (Fig. 7c). It clearly appeared that Cys111 is absolutely required for the interaction, as interacting bands are not formed when Cys111 alone or both Cys111 and Cys100 are mutated to Ser. The interaction is not prevented by removal of Cys100.
Discussion
Conserved residues in the unstructured carboxy-terminus suggest functional importance
Carboxy-terminal residues in all DsrC sequences are highly conserved, including an invariant cysteine (Cys111 in Ach. vinosum DsrC) at the penultimate position, suggesting some functional importance for these residues18. In the NMR structure of P. aerophilum DsrC, residues in the carboxy-terminus from 104 to 111 were found to be disordered, though the presence of a carboxy-terminal hexa-His tag meant that the disorder could have been caused by the disruption of the native structure by the hexa-His tag. The Ach. vinosum DsrC construct had a hexa-His tag at the amino-terminus instead, yet similar disorder was observed in residues 106–112, suggesting the lack of structure in these conserved residues is real. Disordered structure in conserved residues implies that they become structured during interactions with other proteins, otherwise there would be no selective pressure to maintain specific residues at each position. Invariant conservation of the penultimate cysteine residue (Cys111) in this tail implies that it is essential to the function of DsrC. Moreover, conservation of another cysteine (Cys100) in all dsrAB-containing organisms argues for its importance as well. This cysteine is occluded by the carboxy-terminal arm at its point of exit from the globular portion of the protein. In the crystal structure of Agl. fulgidus DsrC, the carboxy-terminal arm adopts a conformation which places the penultimate cysteine sulfur atom almost within bonding distance of the other cysteine sulfur; as the last ten amino acids are identical in Ach. vinosum DsrC, the same conformation should be possible in this protein as well. The localization of conserved residues from elsewhere in the sequence around these conserved structural characteristics also suggests that the carboxy-terminal arm and its surroundings are the “active site” of DsrC.
In native DsrC in oxidizing conditions, a kinetic dimer, then a thermodynamic monomeric disulfide forms
Our NMR spectroscopic studies of native DsrC and DsrC mutants show that an intermolecular disulfide bond between Cys111 residues in two DsrC monomers forms rapidly in oxidizing conditions. Cysteine 111 is the penultimate residue on the disordered carboxy-terminal arm of DsrC, and disulfide bond formation between unhindered thiols in these conditions is not unexpected. This intermolecular disulfide is resolved over time by displacement of one Cys111 sulfur by the Cys100 sulfur in the other molecule, yielding an intramolecular disulfide between Cys100 and Cys111. Mutation of Cys100 to serine halts breakage of the intermolecular Cys111-Cys111 disulfide-linked dimer. The intramolecular disulfide that forms in native DsrC can be reduced and subsequently reoxidized again in the same fashion. The oxidized intramolecular Cys100-Cys111 disulfide form of DsrC appears to be less structured or fluxional as many peaks in the 1H-15N-HSQC spectrum acquire a scratchy appearance with irregular line shapes or weaker peak intensities (Fig. 5). The reduced form can easily be regenerated by treatment with DTT or TCEP.
Evidence for both stages of disulfide bond formation was also seen in studies monitored by SDS-PAGE as well, and blocking of free Cys100 thiols by iodoacetamide following formation of the intermolecular disulfide dimer prevented subsequent resolution to the intramolecular Cys100-Cys111 disulfide monomer. The viability of such a disulfide is also suggested by the close proximity of Cys100 and Cys111 sulfur atoms in some molecules of the asymmetric unit of the Agl. fulgidus DsrC crystal structure.19 It should, however, be noted that the distance of these two cysteines is bigger than a typical disulfide S-S bond distance. The intramolecular Cys100-Cys111 disulfide must be the thermodynamically stable oxidation state of DsrC, suggesting the possibility that it is not incidental but actually physiologically important for DsrC function in sulfur oxidizers and sulfate reducers as part of a redox cycle that is not necessary in bacteria that only use DsrC (TusE) to transfer sulfur for ribonucleotide modification and therefore lack Cys100.
Presence of a 7-residue insertion in S-oxidizer DsrC sequences and TusE sequences is correlated with the presence of DsrEFH or TusBCD
While the substantial overall similarity in all DsrC/TusE sequences is notable, in one region DsrC from dsrAB-containing sulfur oxidizing bacteria is more similar to TusE than to DsrC from sulfate-reducing bacteria. In particular, DsrC from these groups share a 7–8 residue insertion around helices 3 and 4. Residues flanking the insertion are also conserved within the two groups and are different in sulfate-reducing bacteria (Fig. 1). A more distant residue in the sequence, Asp48, displays a similar pattern of conservation (Asp or Glu) and is located on helix 2 where it abuts residues in the insertion sequence in lysine-rich helix 3. Because both groups of organisms contain dsrEFH or tusBCD genes, the presence of the insertion itself and the sequence similarity within the insertion may reflect a common mode of interaction between DsrC and DsrEFH in Ach. vinosum and other sulfur-oxidizers with dsrAB, and between TusE and TusBCD in E. coli and other organisms that contain tusBCD and tusE but lack dsrAB. Additional work will be needed to determine whether residues in the insertion constitute a DsrEFH interaction motif, or whether the insertion serves a different purpose. As was noted above, the structure adopted by the insertion sequence and surrounding residues resembles the DNA-binding domains of some bacterial transcriptional regulatory proteins; the significance, if any, of this resemblance is unclear.
Sequence conservation of two cysteines in all DsrAB-containing organisms (sulfate/sulfite-reducers and sulfur oxidizers) argues for a role of both in DsrC function
Other sequence positions in DsrC exhibit residue type conservation among both sulfur-oxidizing bacteria and sulfate-reducing prokaryotes that is not present in organisms lacking dsrAB. Most strikingly, sequences from these two groups contain, in addition to the penultimate cysteine (Cys111), an invariant cysteine (Cys100 in Ach. vinosum DsrC) that is not present in DsrC (TusE) sequences from bacteria lacking dsrAB. A similar concordance is found for Thr109 and Gly110, which are invariant in sulfate reducers and sulfur oxidizers, but are replaced with Val, Ala, or Ser (for Thr109) and Lys, Arg, or Asn (for Gly110) in bacteria lacking DsrAB. In the crystal structure of Agl. fulgidus DsrC, the equivalent to Gly110 (Gly113) adopts a backbone conformation that is not possible for other residues, thereby allowing the carboxy-terminal arm to bend back towards the protein and put the cysteine sulfur atoms in close proximity.19 While the invariant cysteine next to the carboxy-terminal residue found in both DsrC sequences from dsrAB-containing organisms and in TusE sequences from other bacteria is certain to be significant, conservation of the second cysteine in all dsrAB-containing organisms is also likely to be meaningful. Thus, while TusE appears to function as a carrier of sulfur atoms via a persulfide at the penultimate cysteine, the second conserved cysteine in DsrC argues for the possible (additional) involvement of both conserved cysteines in an intramolecular disulfide bond. The observation that such an intramolecular disulfide forms easily in vitro does not prove its relevance, but together with other evidence, such as the conformational implications of the conserved residue Gly110, supports the idea that capability for intramolecular disulfide formation is built in to the sequence and structure, and is therefore functionally significant.
Proposed models of DsrC & DsrEFH function in sulfur-oxidizing bacteria
Observations of thiol-disulfide interconversion in dimeric and monomeric forms of DsrC, together with the identification of a DsrC-DsrEFH interaction suggest possible models for the function of these proteins in sulfur oxidizing bacteria.
The apparent supercomplex between DsrC, DsrEFH, DsrAB and membrane-bound DsrKMJOP2, together with the similarity between the DsrK component of the membrane complex and heterodisulfide reductase, suggests that perhaps DsrC is the “bacterial heterodisulfide”,5,6 analogous to the archaeal heterodisulfide formed from oxidized coenzyme M and coenzyme B during methanogenesis.25 Assuming that DsrAB operates in a reverse role from its function in sulfate-reducing prokaryotes, and that DsrC interacts transiently with DsrAB, DsrC could function as a carrier of electrons generated from DsrAB-catalyzed oxidation of sulfide to the membrane complex DsrKMJOP. DsrAB would reduce DsrC during sulfide oxidation; DsrKMJOP would oxidize it. However, a specific role for DsrEFH in sulfur oxidizers in such scenario is not really obvious. One possibility could be that it assists DsrKMJOP in some way, perhaps to accelerate interconversion of DsrC Cys111-Cys111 dimers and intramolecular Cys100-Cys111 disulfide (producing one reduced DsrC as well). Alternatively, DsrEFH could protect DsrC-DsrC intermolecular disulfide or DsrC intramolecular disulfide from reduction by the cytosol during transit from DsrMKJOP back to DsrAB.
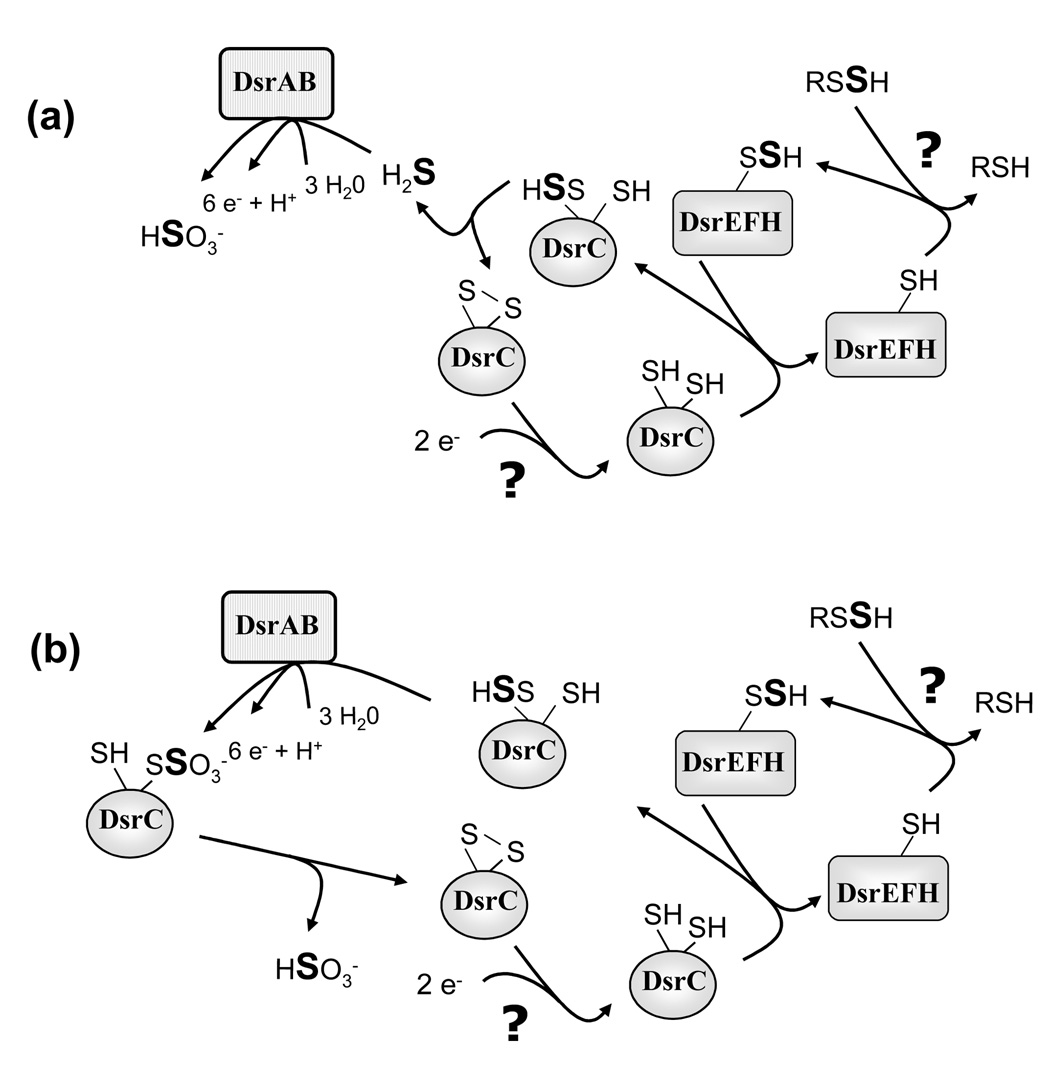
The possibilities discussed above do not take into account that DsrEFH and DsrC might well be proteins involved in a sulfur relay system just as it is firmly established for the related proteins in E. coli.15 Purple sulfur bacteria of the family Chromatiaceae like Ach. vinosum produce sulfur stored in periplasmic sulfur globules as an obligate intermediate during the oxidation of sulfide and thiosulfate.3 Currently, a model is promoted that implies transport of sulfur from the periplasmic sulfur globules to the cytoplasm via a perthiolic carrier molecule (RSSH in Fig. 8).4–6 As illustrated in Fig. 8 DsrEFH could act as the cytoplasmic acceptor for the persulfide sulfur via its conserved probable active site cysteine (Cys78 of DsrE). In analogy to the related E. coli proteins, DsrC may accept a sulfur atom from DsrE. Now, two possibilities are feasible: (1) The persulfide sulfur could be reductively released as a substrate for sulfite reductase by formation of a disulfide between the two conserved cysteine residues of DsrC (note the difference to E. coli TusE which has only one conserved carboxy-terminal cysteine). Sulfide is a likely substrate for sulfite reductase operating in the oxidizing direction as it has been established as a product of sulfite reductases from sulfate-reducing prokaryotes. Binding to of DsrC to DsrAB could stimulate reductive sulfide release by forcing the carboxy-terminal arm to adopt a conformation placing the Cys100 and Cys111 thiols in close proximity, such as is seen in the Agl. fulgidus DsrC X-ray structure. This would enable the Cys100 thiol to displace SH− from the Cys111 persulfide, resulting in an intramolecular Cys100-Cys111 disulfide. (2) DsrC could act as an even more direct substrate-donating molecule to sulfite reductase by presenting sulfur bound to its flexible carboxy-terminal arm immediately to the active site of DsrAB. Tight complex formation between DsrAB and DsrC as would be necessary for such a reaction has already been described for several sulfate reducers.12,13 DsrC would then leave sulfite reductase carrying a sulfonate group. From there, sulfite could be reductively released as the final product of the reaction sequence by formation of a Cys100-Cys111 disulfide. Such a scenario is reasonable as the long known reaction of disulfide bonds with sulfite according to the equation RS-SR + SO32− ↔ RS− + RS-SO32− 26,27 is fully reversible at pH values above 7.0–7.5 when the concentration of RSH is higher than that of RS−. Cysteine 100 occupies a positively charged region of DsrC created by nearby conserved basic residues (Lys97, Arg101, and Lys107) that could stabilize a negatively charged thiolate or thiosulfonate anion during sulfur oxidation in the DsrAB-bound state. DsrC would finally have to be restored as an acceptor for the next sulfur atom in a so far unknown manner involving a disulfide reducing enzyme. DsrL, a protein exhibiting NADH:acceptor oxidoreductase reductase activity in vitro and carrying a thioredoxin–like Cys-X2-Cys motif,2 (and Y. Lübbe and C. Dahl, unpublished) is a possible candidate for this reaction. In the models depicted in Fig. 8, the specific presence of DsrEFH only in sulfur oxidizers is evident: it is necessary for delivery of the sulfur originally deposited in periplasmic (as in Chromatiaceae or chemotrophic sulfur oxidizers like Beggiatoa) or extracellular sulfur globules (as in Ectothiorhodospiraceae or green sulfur bacteria) to DsrC and finally to sulfite reductase. In sulfate-reducing prokaryotes DsrEFH is obsolete, disulfidic DsrC could directly react with sulfite and could then present oxidized substrate to sulfite reductase. A disadvantage of the model discussed here is that it currently cannot explain how electrons gained by the action of sulfite reductase in sulfur oxidizers are further transmitted into membrane bound electron-transport chains.
Fig. 8. Models of DsrC function during sulfur oxidation in A. vinosum integrating probable sulfur transfer functions of DsrEFH and DsrC.
(a) Release of free sulfide as a substrate for sulfite reductase is suggested. (b) DsrC is proposed to act as a substrate-donor for sulfite reductase and to release sulfite via disulfide formation. See discussion for detailed explanation.
Materials and Methods
Cloning of dsrC and site-directed mutagenesis
For PCR amplification of dsrC with chromosomal Ach. vinosum DNA as the template, CEXf (5′-AGGAAGATTCATATGGCCGACACGAT-3′) and CEXr (5′-CCGGACGCGGATCCGCTTAGACGCA-3′) were used as primers (underlined portions of the two primers indicate restriction sites of NdeI and BamHI, respectively). After digestion with NdeI plus BamHI, the gene was cloned into pET15b (Novagen) resulting in plasmid pETCEX and the protein was overproduced with an amino-terminal His tag in E. coli BL21 (DE3). Point mutations were introduced into dsrC by gene splicing by overlap extension28 using standard PCR with Pfu DNA polymerase (Fermentas, St. Leon-Rot) and pETCEX as the template. For the Cys100Ser exchange two fragments were amplified with the following primers: for the first fragment T7ProPr (5’-TAATACGACTCACTATAGGG-3’) and CEXSer100rev (5’-GAAACGGGACGCCTGCTT-3’), for the second fragment CEXSer100for (5’-AAGCAGGCGTCCCGTTCC-3’) and CEXrev (5’-GTCTTCAAGAATTCTCATGTT-3’). Both fragments were used as templates for amplification of the complete dsrC gene carrying the desired point mutation. In this step T7ProPr and CEXrev served as primers. The resulting fragment was restricted with NdeI and BamHI and cloned into pET15b resulting in plasmid pETCEXSer100. The Cys111Ser exchange was done accordingly using primers CEXSer111rev (5’-ATCCGCTTAGACGGAGCCGGTCGG-3’) and CEXSer111for (5’-CCGACCGGCTCCGTCTAAGCGGAT-3’). For the construction of the double mutation, plasmid pETCEXSer100 was used as the template for introduction of the Cys111Ser mutation using primers CEXSer111 rev and CEXSer111for. The sequences of all mutated dsrC genes were verified by nucleotide sequencing.
Construction of an in frame deletion of dsrC in A. vinosum
The in frame deletion of dsrC was achieved by utilizing the gene splicing by overlap extension PCR,28 using the following primers: CXbaf1 (5’-TCAACGAGTCTAGATATCAGCATCAGGCGTC-3’), Crev1 (5’-CTCGTCCGCTTAGACATCGACTTCGATCGT-3’), Cfor1 (5’-ACGATCGAAGTCGATGTCTAAGCGGACGAG—3’) and CXbar1 (5’-CGCACTTGTCTAGAGCGTCCATGTAGACGC-3’). An XbaI restriction site was introduced to the 5’ and 3’ ends of the final deletion framing PCR fragment. The PCR amplicons were cloned into the XbaI site of the mobilizable suicide vector pK16mobsacB.29 The resulting plasmid pK18mobsacBΔC was transferred into A. vinosum Rif50 and the gene exchange was achieved and verified as described by Lübbe.22 Growth of A. vinosum strain was examined in batch culture as described by Prange.30
Preparations of samples for NMR
Competent cells (Novagen) were transformed by 42 °C heat shock and plated on LB agar media containing 50 µg/mL carbenecillin. Colonies from plates were used to inoculate two 4 mL volumes of M9 minimal media made with 15NH4Cl and U-13C-glucose (2g/L). The medium also contained 10 µM Fe3+ and trace metals. The 4 mL cultures were grown overnight at 37 °C with shaking and then used to inoculate two 40 mL starter cultures. These cultures were allowed to grow to mid-log phase, then each was added to 450 mL media in 2 L flasks. These were grown until OD600 = 0.6, then 130 mg IPTG was added to each flask (final concentration ca. 1.1 mM IPTG) and the temperature was lowered to 30 °C for 8 hours until cells were harvested by centrifugation and frozen. The wet weight yield of cells totaled 4.2 g. Cells were thawed and resuspended in Ni2+ column loading buffer (50 mM sodium phosphate pH 8.0, 500 mM NaCl, 10 mM imidazole). The cell suspension was passed several times through a French press. Lysed cells were centrifuged 12 minutes at 19000 × g to remove insoluble cell material. Protamine sulfate (12.5 mg in 1 mL buffer) was added to precipitate nucleic acids. The supernatant was subjected to ultracentrifugation for 1 hour (308000 ×g at rmax), then loaded onto a 10 mL bed volume Ni2+-NTA (Qiagen) gravity column equilibrated with loading buffer. Bound protein was washed with 50 mL load buffer, then removed with elute buffer containing 400 mL imidazole. Upon elution, β-mercaptoethanol was added to 2 mM to each fraction. Fractions containing protein were concentrated from 10 mL to 2.5 mL in a centriprep-3 concentrator, then exchanged into NMR buffer (50 mM Tris pH 7.6 @ 25 °C, 500 mM NaCl, 10 mM DTT) on a PD-10 column. The protein in NMR buffer was concentrated further to 1 mM in a centricon-3 device, and D2O (10% v/v) and NaN3 (0.02% w/v) were added. NMR samples (250 µL) were put in Shigemi NMR tubes, and remaining unused DsrC protein in NMR buffer (ca. 1 mL) was stored in a 15 mL plastic conical tube. NMR samples of DsrC mutants Cys100Ser, Cys111Ser, and Cys100Ser/Cys111Ser were prepared using the same protocol as for native DsrC. The DsrC+DsrEFH complex was prepared for NMR by combining approximately equimolar amounts of 13C,15N-labeled DsrC and unlabeled DsrEFH, then concentrating the mixture 10-fold in a Centricon-30 ultrafiltration device to separate the majority of any excess or unbound DsrC (12kDa) from the complex (ca. 90 kDa). The resulting sample (ca. 0.05 mM) was free of precipitate.
NMR Spectroscopy
NMR experiments were conducted at the Environmental Molecular Sciences Laboratory of Pacific Northwest National Laboratory, Richland WA. Backbone and side chain correlation experiments were collected on Varian Inova 600 and Unity 600 instruments. 3D-NOESY experiments were collected on a Varian Inova 800. The 4D-NOESY experiment was collected on a Varian Inova 600. Data was collected at 25 °C using standard triple-resonance pulse sequences.31 The following experiments were recorded: 1H-15N-HSQC, 1H-13C-HSQC, HNCACB, CBCACONNH, HNCO, CCC-TOCSY-NNH, HCC-TOCSY-NNH, HCCH-TOCSY, 3D-15N-NOESY (150 msec mixing time), 3D-simultaneous-13C-15N-NOESY (125 msec mixing time), and 4D-13C-13C-HMQC-NOESY-HMQC (125 msec mixing time), HNHA, and aromatic ring side chain correlation experiments (HBCBCG-CDHD/-CEHE-aro). All pulse sequences were from Lewis Kay (University of Toronto). Amide proton exchange was monitored by acquiring 1H-15N-HSQC spectra following dissolution of a lyophilized protein sample in D2O. Stereospecific Leu and Val side chain assignments were obtained from a 1H-13C-HSQC experiment recorded on the sample prepared from cells grown on 5% U-13C-glucose / 95% unenriched glucose.32 The 1H-15N-HSQC spectra recorded on the DsrC+DsrEFH complex were collected with shortened delays (1/2JHN = 2.3 ms) and long exeriment times (~20 hrs) to overcome losses due to relaxation. The raw data and pulse sequences used have been deposited in the BioMagResBank (Madison, WI). Data were processed with Felix (MSI) and analyzed with Felix and Sparky (http://www.cgl.ucsf.edu/home/sparky).
Calculation of structural ensemble and analysis of the ensemble
NOESY cross peaks were picked using restricted peak picking in Sparky and were automatically assigned and given upper distance bounds by AutoStructure.33 These NOE distance restraints had uniform lower bounds of 1.8 Å and upper bounds of either 2.8, 3.2. 4.0 or 5.0 Å. Amide 1H-15N-HSQC cross peaks still present 30 min. after dissolution of a lyophilized sample in D2O were used to derive hydrogen bond restraints with bounds of 1.8–2.5 Å for the HN-O distance and 2.8–3.5 Å for the N-O distance, provided preliminary structural ensembles clearly indicated the correct acceptor atom. Dihedral restraints for phi were derived from the HNHA experiment34 and TALOS 35 and had bounds of −55 ± 30 degrees for helical residues with J < 5 Hz and −120 ± 50 degrees for extended residues with J > 7.5 Hz. Dihedral restraints for psi had values of −47 ± 30 degrees for helical residues and 140 ± 50 degrees for extended residues. Psi restraints derived from TALOS were added only for residues in helices and beta sheets and only after consideration of amide-alpha NOE ratios (HNi-HAi vs. HNi-HAi-1), alpha carbon chemical shifts, and the evident secondary structure propensities in preliminary ensembles of structures.
Preliminary structures were calculated with AutoStructure33 and the resulting list of assigned NOEs was manually edited during refinement to remove non-restraining restraints and correct errors caused by misassigned chemical shifts. The resulting distance restraints, together with the manually derived dihedral restraints, were used to generate a set of 30 structures with NIH-Xplor36 using distance geometry and simulated annealing. The routines dg_sub_embed, dg_full_embed, and dgsa were used as provided except that in dgsa, an inital temperature of 2000K was used with 30000 high temperature steps and 200000 cooling steps. Sum averaging was used for methyl groups and methylene proton pairs. A final refinement in explicit water was performed in CNS.37 Twenty (out of 30 total) structures were selected to form the final ensemble on the basis of minimal restraint violations and energies. A structure validation report is available at www.nesg.org. A. vinosum DsrC is target OP4 of the Northeast Structural Genomics Consortium.
Sequence and structure analysis
All amino acid sequences were obtained from GenBank. Psi-Blast was used with default parameters to generate the protein sequence family.38 ClustalW was used to generate the multiple sequence alignment.39 ConSurf was used to visualize the structural distribution of conserved residues.40 The structural ensemble was analyzed with PROCHECK-NMR.41 Surface electrostatic features of the protein were examined using ABPS42 with PyMol.43 Structure similarity searches using Dali44 were conducted using the residues 3–108 of the first structure from the ensemble as a representative structure for similarity searching. Pairwise structure comparisons were done with DaliLite.45
Purification of recombinant Dsr proteins and band shift assay
DsrEFH and wild type and mutated DsrC proteins were overproduced in and purified from E. coli BL21 (DE3) as described earlier.2,16 Unless specified otherwise, combinations of recombinant proteins (100 pmol DsrEFH and varying amounts of wild type or mutated DsrC) were incubated for 1 h at room temperature in a final volume of 60 µl containing 10 μl buffer (5 mM HEPES, pH 7.8, 0.1 M KCL, 0.01% Tween 20), and 25 µM TCEP. A volume of 20 µl per lane of these reaction mixtures was mixed with sample loading buffer (4×: 100 mM MOPS, pH 6.8, 40% glycerol, 0.001% bromphenol blue), separated by native electrophoresis in 7.5 % polyacrylamide gels and stained with Coomassie Blue.
SDS-PAGE analysis and thiol blocking with iodoacetamide
SDS polyacrylamide gel electrophoresis was performed in 15% gels in the absence of reducing agent according to the standard protocol of Laemmli.46 Per lane, 0.5–1 µg protein were applied. Non-reducing sample buffer (2×) contained 100 mM Tris-HCl, pH 6.8, 4% SDS, 0.001% bromphenol blue and 20% glycerol. Thiol-blocking was achieved by incubation of protein samples in the presence of 10 mM iodoacetamide for 2 h at 30°C. Reduction of such samples was performed by addition of 2-mercaptoethanol to a final concentration of 20 mM. Samples were then mixed with an equivalent volume of non-reducing sample buffer and analyzed by SDS-PAGE as described above.
Acknowledgements
NMR spectra were acquired in the Environmental Molecular Sciences Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy Office of Biological and Environmental Research) located at Pacific Northwest National Laboratory and operated for DOE by Battelle (contract KP130103). This work was supported by grants Da 351/3–4 and Da 351/3–5 to CD from the Deutsche Forschungsgemeinschaft. JRC and MAK acknowledge support from the the NIH Protein Structure Initiative (Grant P50-GM62413-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Code:
The structure was deposited to the Protein Data Bank with the accession number 1YX3.
References
- 1.Schedel M, Vanselow M, Trüper HG. Siroheme sulfite reductase from Chromatium vinosum. Purification and investigation of some of its molecular and catalytic properties. Arch. Microbiol. 1979;121:29–36. [Google Scholar]
- 2.Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, Deuster O, Brune DC. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 2005;187:1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pott AS, Dahl C. Sirohaem-sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology. 1998;144:1881–1894. doi: 10.1099/00221287-144-7-1881. [DOI] [PubMed] [Google Scholar]
- 4.Frigaard N-U, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 2008;54 doi: 10.1016/S0065-2911(08)00002-7. in press. [DOI] [PubMed] [Google Scholar]
- 5.Grimm F, Franz B, Dahl C. Thiosulfate and sulfur oxidation in purple sulfur bacteria. In: Dahl C, Friedrich CG, editors. Microbial Sulfur Metabolism. Berlin: Springer, Heidelberg; 2008. pp. 101–116. [Google Scholar]
- 6.Dahl C. Inorganic sulfur compounds as electron donors in purple sulfur bacteria. In: Hell R, Dahl C, Knaff DB, Leustek T, editors. Sulfur in Phototrophic Organisms. New York: Springer; 2008. pp. 289–317. [Google Scholar]
- 7.Matias PM, Pereira IAC, Soares CM, Carrondo MA. Sulphate respiration from hydrogen in Desulfovibrio bacteria: a structural biology overview. Prog. Biophys. Mol. Biol. 2005;89:292–329. doi: 10.1016/j.pbiomolbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Karkhoff-Schweizer RR, Bruschi M, Voordouw G. Expression of the γ-subunit gene of desulfoviridin-type dissimilatory sulfite reductase and of the α- and β-subunit genes is not coordinately regulated. Eur. J. Biochem. 1993;211:501–507. doi: 10.1111/j.1432-1033.1993.tb17576.x. [DOI] [PubMed] [Google Scholar]
- 9.Mussmann M, Richter M, Lombardot T, Meyerdierks A, Kuever J, Kube M, Glöckner FO, Amann R. Clustered genes related to sulfate respiration in uncultured prokaryotes support the theory of their concomittant horizontal transfer. J. Bacteriol. 2005;187:7126–7137. doi: 10.1128/JB.187.20.7126-7137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander J, Engels-Schwarzlose S, Dahl C. Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 2006;186:357–366. doi: 10.1007/s00203-006-0156-y. [DOI] [PubMed] [Google Scholar]
- 11.Loy A, Duller S, Wagner M. Evolution and ecology of microbes dissimilating sulfur compounds: insights from siroheme sulfite reductase. In: Dahl C, Friedrich CG, editors. Microbial Sulfur Metabolism. Berlin: Springer, Heidelberg; 2008. pp. 46–59. [Google Scholar]
- 12.Pierik AJ, Duyvis MG, van Helvoort JMLM, Wolbert RBG, Hagen WR. The third subunit of desulfoviridin-type dissimilatory sulfite reductases. Eur. J. Biochem. 1992;205:111–115. doi: 10.1111/j.1432-1033.1992.tb16757.x. [DOI] [PubMed] [Google Scholar]
- 13.Steuber J, Arendsen AF, Hagen WR, Kroneck PMH. Molecular properties of the dissimilatory sulfite reductase from (Essex) and comparison with the enzyme from Desulfovibrio vulgaris (Hildenborough) Eur. J. Biochem. 1995;233:873–879. doi: 10.1111/j.1432-1033.1995.873_3.x. [DOI] [PubMed] [Google Scholar]
- 14.Dahl C, Kredich NM, Deutzmann R, Trüper HG. Dissimilatory sulphite reductase from Archaeoglobus fulgidus: physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. J. Gen. Microbiol. 1993;139:1817–1828. doi: 10.1099/00221287-139-8-1817. [DOI] [PubMed] [Google Scholar]
- 15.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Dahl C, Schulte A, Shin DH. Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of DsrEFH from Allochromatium vinosum. Acta Cryst. 2007;F63:890–892. doi: 10.1107/S1744309107041188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pires RH, Venceslau SS, Morais F, Teixeira M, Xavier AV, Pereira IAC. Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex - a membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry. 2006;45:249–262. doi: 10.1021/bi0515265. [DOI] [PubMed] [Google Scholar]
- 18.Cort JR, Mariappan SVS, Kim C-Y, Park MS, Peat TS, Waldo GS, Terwilliger TC, Kennedy MA. Solution structure of Pyrobaculum aerophilum DsrC, an archaeal homologue of the gamma subunit of dissimilatory sulfite reductase. Eur. J. Biochem. 2001;268:5842–5850. doi: 10.1046/j.0014-2956.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- 19.Mander GJ, Weiss MS, Hedderich R, Kahnt J, Ermler U, Warkentin E. X-ray structure of the γ-subunit of a dissimilatory sulfite reductase: fixed and flexible C-terminal arms. FEBS Lett. 2005;579:4600–4604. doi: 10.1016/j.febslet.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 20.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 21.Dover LG, Corsino PE, Daniels IR, Cocklin SL, Tatituri V, Besra GS, Futterer K. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 2004;340:1095–1105. doi: 10.1016/j.jmb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Lübbe YJ, Youn H-S, Timkovich R, Dahl C. Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c diamide synthase for sulfur oxidation. FEMS Microbiol. Lett. 2006;261:194–202. doi: 10.1111/j.1574-6968.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 23.Numata T, Fukai S, Ikeuchi Y, Suzuki T, Nureki O. Structural basis for sulfur relay to RNA mediated by heterohexameric TusBCD complex. Structure. 2006;14:357–366. doi: 10.1016/j.str.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Shin DH, Yokota H, Kim R, Kim S-H. Crystal structure of a conserved hypothetical protein from Escherichia coli. J. Struct. Funct. Genom. 2002;14:53–66. doi: 10.1023/a:1014450817696. [DOI] [PubMed] [Google Scholar]
- 25.Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter KO. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 1999;22:353–381. [Google Scholar]
- 26.Cecil R, Wake RG. Reactions of inter- and intra-chain disulphide bonds in proteins with sulphite. Biochem. J. 1962;82:401–406. doi: 10.1042/bj0820401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey JL, Cole RD. Studies on the reaction of sulfite with proteins. J. Biol. Chem. 1959;234:1733–1739. [PubMed] [Google Scholar]
- 28.Horton RM. PCR mediated recombination and mutagenesis: SOEing together tailor-made genes. Mol. Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 29.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 30.Prange A, Engelhardt H, Trüper HG, Dahl C. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT PCR. Arch. Microbiol. 2004;182:165–174. doi: 10.1007/s00203-004-0683-3. [DOI] [PubMed] [Google Scholar]
- 31.Cavanagh J, Fairbrother WJ, Palmer AGI, Skelton NJ. Protein NMR spectroscopy, principle and practice. San Diego: Academic Press; 1996. [Google Scholar]
- 32.Neri D, Szyperski T, Otting G, Senn H, Wütrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the dNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 33.Huang YJ, Moseley HN, Baran MC, Arrowsmith C, Powers R, Tejero R, Szyperski T, Montelione GT. An integrated platform for automated analysis of protein NMR structures. Meth. Enzymol. 2005;394:111–141. doi: 10.1016/S0076-6879(05)94005-6. [DOI] [PubMed] [Google Scholar]
- 34.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHα) coupling constants in15N-enriched proteins. J. Am. Chem. Soc. 1993;115:7772–7777. [Google Scholar]
- 35.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 36.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR moelcular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 37.Linge JP, Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR. 1999;13:51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 42.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delano WL. The PyMOL molecular graphics system. San Carlos, California, USA: DeLano Scientific; 2002. [Google Scholar]
- 44.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 46.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]