SUMMARY
The cyanobacterial lectin Cyanovirin-N (CV-N) exhibits antiviral activity against HIV at a low nanomolar concentration by interacting with high-mannose oligosaccharides on the virus surface envelope glycoprotein gp120. Atomic structures of wild-type CV-N revealed a monomer in solution and a domain-swapped dimer in the crystal, with the monomer comprising two independent carbohydrate binding sites that individually bind with micromolar affinity to di- and trimannoses. In the mutant CVNmutDB, the binding site on domain B was abolished and the protein was found to be completely inactive against HIV. We determined the solution NMR and crystal structures of this variant and characterized its sugar binding properties.
INTRODUCTION
Interactions between carbohydrates and proteins are involved in a broad spectrum of physiological processes (Lis and Sharon, 1998) and understanding the underlying mechanism inherent to lectin-carbohydrate recognition is of considerable interest. Proteins involved in carbohydrate recognition typically have multiple binding sites and carbohydrate ligands are usually presented in clusters. Therefore, each protein may bind more than one carbohydrate epitope simultaneously (Lee, 1992; Bundle et al., 1992). Results using model systems revealed that (i) polyvalent display of carbohydrates produces high avidities, although the affinities of monovalent, small sugars for their protein receptors are weak (Kiessling and Pohl, 1996; Sigal et al., 1996; Adler et al., 1995; Sabesan et al., 1991; DeFrees et al., 1996; Spengard et al., 1996; Meunier and Roy, 1996) and (ii) increased specificity may results from carbohydrates being presented in a polyvalent format (Lees et al., 1994; Mortell et al., 1996). Approaches aimed at understanding the molecular mechanisms and structural basis of high-affinity oligosaccharide recognition, as well as the energetic principles that govern these important interactions, ideally combine structural information derived from NMR or crystallography with thermodynamic data provided by isothermal titration calorimetry. Over the last couple of years we have embarked on such studies for the cyanobacterial lectin CV-N in order to gain insight into its potent HIV-inactivating properties. Although the precise mechanism and delineation of details in the individual steps of CV-N’s mode of action are still largely unknown, viral inactivation of HIV is intimately connected with recognition of N-linked high-mannose oligosaccharides (Man-8 and Man-9) on gp120 (O’Keefe et al., 2000; Bolmstedt et al., 2001; Shenoy et al., 2001). CV-N binds tightly to wild type gp120 but not to deglycosylated or non-glycosylated recombinant gp120 (Boyd et al., 1997; O’Keefe et al., 2000) and specifically recognizes Manα(1→2)Manα moieties at the termini of the D1 and D3 arms of Man-8 or Man-9 (Bolmstedt et al., 2001; Barrientos et. al., 2005).
CV-N is a 101 amino acid protein that was originally isolated from an aqueous extract of the cyanobacterium Nostoc ellipsosporum (Boyd et al., 1997; Patterson et al., 1993). A number of atomic structures of wild type CV-N, mutants and complexes have been solved by NMR and crystallography, revealing either monomeric (Bewley et al., 1998) or domain-swapped dimeric structures (Yang et al., 1999; Barrientos et al., 2002a). In solution, the monomeric, bilobal structure exhibits pseudo-symmetry and the two pseudo-symmetric domains AM and BM consist of residues [1–38/90–101] and [39–89], respectively (Figure 1A, Bewley et al., 1998). In the crystal or, under specific conditions, a domain-swapped dimeric structure is observed (Yang et al., 1999; Barrientos et al., 2002a) and the domains AD, A’D, BD and B’D in the domain-swapped dimer comprise residues [1–38 and 90’–101’], [1’–38’ and 90–101], [39’–50’ and 51–89] and [39–50 and 51’–89’], respectively (Figure 1A, Yang et al., 1999). For both monomeric and domain-swapped dimeric CV-N to coexist in solution under identical conditions, the free energies of folding for both quaternary states must be comparable and the kinetic barrier between the monomer and dimer was found to be significant (Barrientos et al., 2002a). Mutation of residues in the hinge region allowed us to change the free energy difference between monomer and dimer: Substituting the pivotal proline in the hinge region to glycine resulted in stabilization of the monomeric P51G mutant by about 5.2 kcal/mol compared to wild-type, with a less dramatic increase in stability for the dimer (Barrientos et al., 2002a). In addition, the energetic difference between the monomer and domain-swapped dimer is tipped more towards monomer for this mutant. Another interesting hinge region mutant was S52P, which yielded predominantly dimeric protein due to drastic destabilization of the monomer. Therefore, the removal or addition of a proline residue in the hinge region has a significant influence on the stability of the monomeric protein and the predominant quaternary state (Barrientos et al., 2002a). In addition, the single proline to glycine change dramatically reduced conformational and oligomeric heterogeneity during protein purification and folding.
Figure 1. NMR solution and X-ray structures of wild type and mutant CV-N.
(A) Ribbon representations of the monomeric solution structure of wild type CV-N [PDB accession code 2EZM] and the crystal structure of the domain-swapped dimer [PDB accession code 3EZM]. Residue positions changed by mutagenesis in CVNmutDB are indicated by blue spheres. The different polypeptide chains in the domain-swapped dimer are colored green and pink. N- and C-termini of the chains are marked by N and C, respectively and the pseudo-symmetric domains are labeled AM and BM in the monomer and AD, BD, A’D and B’D in the domain-swapped dimer. (B) Amino acid sequences of [P51G]CV-N and CVNmutDB. Residues belonging to domains A and B are labeled green and pink, respectively. Disulfide bonds are indicated by brackets and residues involved in protein-carbohydrate interactions are underlined in the [P51G]CV-N sequence. Identical amino acids in the aligned sequence repeats are marked by dots. Residues that were mutated in CVNmutDB are colored blue. (C) Ribbon superposition of the NMR solution structures of CVNmutDB (green) and wild type CV-N (pink).
One sugar binding site was identified in each domain by NMR titration experiments (Bewley and Otero-Quintero, 2001; Barrientos et al., 2002b; Shenoy et al., 2002): a semicircular cleft in domain AM and a deeper pocket in domain BM, with both sites separated by ~40 Å. The two sites are chemically and topologically distinct, despite considerable sequence similarity (Figure 1B). Each site recognizes α(1→2) linked di-mannoses, either as individual oligosaccharides or as part of the terminal arms of the branched Man-8 and Man-9 structures. Domain A exhibits a slight preference for the tri-mannose and domain B for the dimannose units, respectively (Bewley et al., 2002; Botos et al., 2002a; Sandström et al., 2004; Shenoy et al, 2002). In all cases, the reported binding constants are in the micromolar range (Bewley and Otero-Quintero, 2001; Bewley et al., 2002; Shenoy et al., 2002). Although considerable information is available on the interaction of CV-N with high-mannose sugars, it is not clear whether a single sugar-protein contact is sufficient for the anti-viral activity of CV-N. Indeed, the high affinity towards carbohydrate epitopes on viral glycoproteins, measured by ELISA, is in stark contrast to the micromolar solution binding constants for small sugars. Unfortunately, solution binding experiments with large, branched oligomannoses and wild type CV-N result in precipitation of the sugar-protein complexes due to multisite/multivalent crosslinking (Shenoy et al., 2002), preventing the extraction of accurate binding data for these high-mannose sugars.
The CVNmutDB mutant was created to eliminate the binding pocket in domain B, resulting in a protein with only one carbohydrate recognition site (Figure 1B). This mutant is complementary to mutants in which the binding site on domain A was removed (Chang and Bewley, 2002; Barrientos et al., 2006) and a crystal structure of such a binding site variant that also includes the P51G hinge mutation was reported recently (Fromme et al., 2007). Here, we report the solution NMR and X-ray crystal structures of CVNmutDB and the interaction of this mutant protein with the sugars Man-2 and Man-3. The solution NMR structure shows a monomer and is very similar to that of wild type CV-N (Figure 1C), exhibiting only local differences around the mutation sites. Surprisingly, CVNmutDB in the crystal is also monomeric, like the recently reported mutant of binding site A (Fromme et al., 2007) and no domain-swapping is observed for this variant, although two monomers are found in the asymmetric unit.
Since CVNmutDB exhibits only a single carbohydrate binding site on domain A, equilibrium binding to several sugars was monitored by ITC and NMR. A dissociation constant of 3.4 µM was obtained for Man-3 binding, very similar to that observed for Man-9 (4.3 µM; Barrientos et al., 2006). The binding to di-mannose is very weak (Kd = 750 µM) and both ITC and NMR titration yielded similar binding constants.
Our results presented here demonstrate that: (i) the three-dimensional structures of CVNmutDB in solution (NMR) and the crystal are very similar and the protein no longer undergoes domain-swapping, (ii) abolishing the sugar binding site on domain BM results in micromolar affinities for mannose sugars, even large branched ones, involving the single binding site on domain A, and (iii) cross-linking, domain-swapping and the previously noted loss of antiviral activity (Barrientos et al., 2006) are most likely functionally linked.
RESULTS AND DISCUSSION
NMR solution structure
The solution structure of CVNmutDB was determined by NMR spectroscopy using uniformly 15N- and 13C/15N-labeled samples and established protocols commonly used in our laboratory. A set of 50 simulated annealing refined structures was calculated with 2076 interproton distance restraints and a final 20 conformer ensemble was generated using NH RDC refinement. All structures are well defined, satisfy all experimental constraints, display excellent covalent geometry and exhibit atomic r.m.s.d. deviations of 0.21 ± 0.05 and 0.60 ± 0.05 Å with respect to the mean coordinate positions for the backbone (N, Cα, C’) and all heavy atoms, respectively. 78.3% of residues lie in the most favorable region of the Ramachandran plot (Ramachandran and Sasisekharan, 1968). A summary of the experimental constraints as well as pertinent structural statistics for the refined ensemble of 20 conformers are provided in Table 1.
Table 1.
Structural statistics for the final 20 conformer NMR ensemble
| r.m.s. deviations from experimental distance restraints (Å) | |
| Total interproton (2076) | 0.006 ± 0.0008 |
| Sequential (|i – j| = 1) | 911 |
| Short range (1 < |i – j| < 5) | 295 |
| Long range (|i – j| ≥ 5) | 870 |
| RDC NH constraints | 62 |
| r.m.s. deviations from experimental dihedral restraints (deg.) | |
| Total dihedral angles (126) | 0.1 ± 0.05 |
| Deviations from idealized covalent geometry | |
| Bonds (Å) | 0.001 ± 0.0002 |
| Angles (deg.) | 0.28 ± 0.006 |
| Impropers (deg.) | 0.12 ±0.01 |
| Structural quality | |
| ELJ (kcal/mol) | 13.8 |
| PROCHECK | |
| Residues in most favorable regions | 78.3% |
| Residues in additional allowed regions | 9.1% |
| Residues in generously allowed regions | 0% |
| Residues in disallowed regions | 0% |
| Coordinate precision (Å) | |
| Backbone (N, Cα, C’) | 0.21 ±0.05 |
| All non-hydrogen atoms | 0.60 ±0.05 |
| RDC deviation (Hz) | 0.64 ±0.1 |
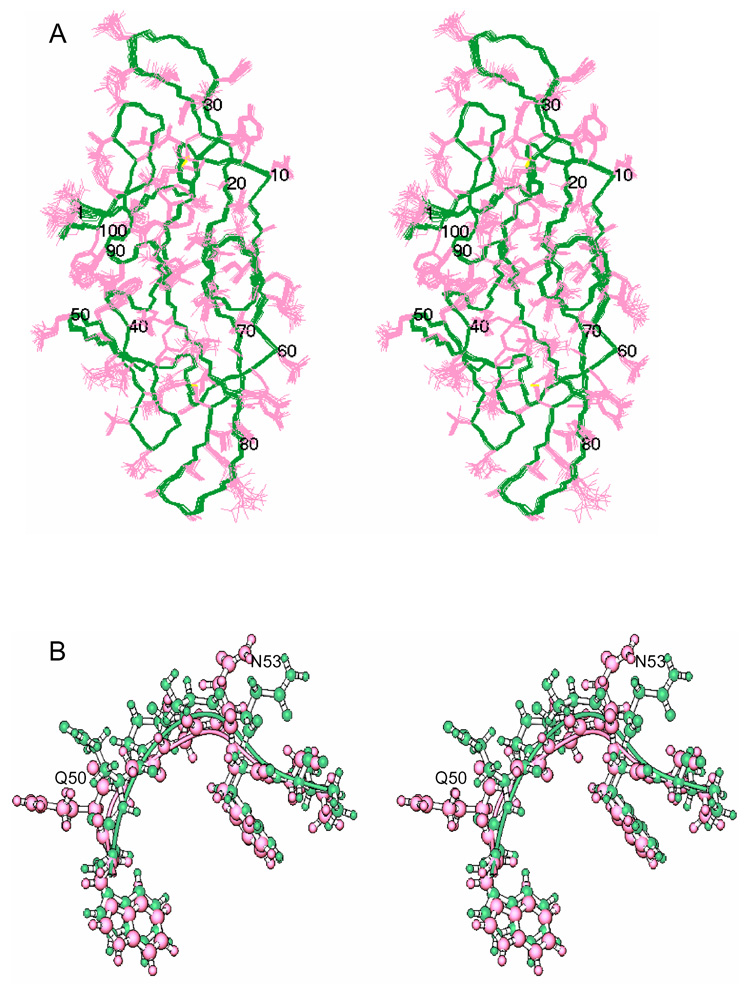
A ribbon representation of the overall solution structure of CVNmutDB is displayed in Figure 1C. Like wild type CV-N, the mutant structure exhibits two pseudo-symmetric domains and the architecture of both proteins as well as the secondary structure elements are very similar. The latter comprise two anti-parallel triple-stranded β-sheets (sheet 1: residues 8–13 (β1), 17–23 (β2) and 29–35 (β3); sheet 2: residues 58–64 (β6), 68–74 (β7) and 80–86 (β8)), two β-hairpins (residues 41–42 (β4) and 47–48 (β5); 91–94 (β9) and 97–100 (β10)) and four 310 helical turns (residues 4–7(α1), 87–89(α2), 36–38(α3) and 55–57(α4)). A stereoview of the refined 20-conformer ensemble is provided in Figure 2A. The similarity between the wild type CV-N structure [PDB accession code 2EZM] and the minimized mean structure of CVNmutDB is reflected in a r.m.s.d. value of 1.04 Å for the backbone atoms (N, Cα, C’). A distinct difference in structure is, however, observed around the P51G mutation site. Different orientations of the Q50 and N53 side chains are clearly present, while the aromatic rings of W49 and F54 are essentially in identical positions (Figure 2B).
Figure 2. NMR solution structure of CVNmutDB.
(A) Stereoview of the superposition of the final 20 conformer ensemble [PDB accession code 2rp3]. Backbone (N, Cα, C’) atoms are shown in green and all side-chain atoms in pink. The disulfide bridges are colored yellow. (B) Stereoview of the region around the P51G mutation. The structures of CVNmutDB and wild type CV-N are shown in ball and stick representation and colored green and pink, respectively. Note the different positions of the Q50 and N53 side chains.
Crystal structure
Previous crystallographic investigations yielded trigonal (Yang et al., 1999; Botos et al., 2002b) and tetragonal (Barrientos et al., 2002a; Botos et al., 2002a) crystals that showed domain-swapped dimer structures. More recently, monoclinic crystals of a mutant CV-N were obtained that revealed a monomer structure (Fromme et al., 2007). We obtained orthorhombic crystals (1.36 Å resolution) belonging to space group P212121 with cell constants a =34.16, b = 58.02, c = 87.79 Å that also showed monomeric protein, albeit with two molecules in the asymmetric unit (m1, m2) (Figure 3A). That indeed the X-ray data was incompatible with the presence of a domain swapped dimer was ascertained by omitting the region W49 - I55 in the starting model used for refinement. After the first rigid body refinement step, the composite omit electron density map clearly showed strong density for this region (Figure 3B) which connected the two separate starting segments in a monomeric arrangement. Therefore, the X-ray structure clearly does not comprise a dimer. A detailed comparison of the ϕ,ψ backbone dihedral angles for residues 49 to 54 between the present monomeric solution NMR and X-ray structures that form a helical loop shows that all angles are similar (see Table S1; available online). Their values are W49NMR (−82/−177), W49X-ray (−82/179), Q50NMR (46/35), Q50X-ray (59/54), G51NMR (−135/−174), G51X-ray (179/−172), S52NMR (−162/173), S52X-ray (−151/173), N53NMR (66/39), N53X-ray (69/32), F54NMR (−94/−44), F54X-ray (−60/−28). Within the crystal lattice, all protein molecules are tightly packed with no significant disordered solvent channels between them. This results in a highly ordered protein array and a large number of ordered water molecules, possibly contributing to the high resolution diffraction characteristics of these crystals. To the best of our knowledge, this is the highest resolution crystal structure solved for any CVN to date. All pertinent crystallographic statistics for the X-ray structure are reported in Table 2. Both independent monomers are very similar with r.m.s.d. values of 0.33 Å and 0.98 Å for the backbone and all atoms, respectively (see Table S2; available online). Interestingly, an unusual rotamer for K48 in one of the monomers (m1) was observed. H-bonding between the ε-amino group of this side chain and the backbone carbonyl groups of W49, Q50 and the side chain of N30 from a neighbor monomer within the unit cell most likely caused this unusual conformation. Indeed, the side chain of the equivalent K48 in the other monomer (m2) exhibits the common rotameric conformer and extends into the solvent.
Figure 3. Crystal structure of CVNmutDB.
(A) Stereoview illustrating the relative orientation of the two monomers in the asymmetric unit. Each β-sheet is numbered. (B) View of the composite omit electron density map contoured at 1.7 σ for the region W49 to F54. This region is the hinge in the domain-swapped structure of wild type CV-N and exhibits a loop conformation in the present monomeric structure.
Table 2.
Crystallographic data and refinement statistics
| Space group | P212121 |
|---|---|
| Unit cell | |
| a (Å) | 34.16 |
| b (Å) | 58.02 |
| c ( Å) | 87.79 |
| α ( °) | 90 |
| β ( °) | 90 |
| γ ( °) | 90 |
| Resolution | 1.36 Å |
| Completeness % (last shell) | 99.0 (94.7) |
| Total reflections | 167,871 |
| Unique reflections | 37,884 |
| Rmerge | 0.038 |
| Refinement statistics | |
| Resolution range (Å) | 31.83 – 1.36 |
| Number of reflections | 36,956 |
| R factor (last shell) | 16.5 (16.7) |
| Rfree (last shell) | 18.6 (18.7) |
| Number of residues | 202 |
| Number of waters | 448 |
| Average B factor (Å2) | |
| Main chain atoms | 7.5 |
| Side chain atoms | 9.9 |
| Solvent atoms | 21.55 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.0082 |
| Bond angles (deg.) | 1.52 |
The CVNmutDB crystal structure, like the NMR solution structure is very similar to that of wild type CV-N [PDB accession code 2EZM], with both monomers in the asymmetric unit exhibiting r.m.s.d. values of 0.81 Å and 0.83 Å for the backbone atoms (N, Cα, C’). These values are comparable to and only slightly lower than the r.m.s.d. value that was obtained between the two NMR solution structures (1.04 Å). Again, only small conformational changes around the P51G site of mutation are seen. Comparison of the present mutant solution NMR and the X-ray structures also yield similar r.m.s.d. values of 0.93 Å and 0.97 Å for both monomers. Therefore, the mutant and wild type structures are all within 0.92±0.12 Å r.m.s.d. for the backbone, irrespective of the methodology used.
Within the asymmetric unit the two monomers are oriented with an angle of ~64° between the long axes of the two domains (AB’, A’B), using the Sγ atoms of the cysteines in the two disulfide bonds (i.e., Cys8/Cys58’/Cys58/Cys8’) for relative axis angle determination. This spatial arrangement is reminiscent of the orientation previously observed between the two halves of the domain-swapped dimer structures in the tetragonal crystal (P41212) [PDB accession code 1L5B] (Barrientos et al., 2002a) and very different from that of the monoclinic mutant monomer structure [PDB accession code 2Z21] (Fromme et al., 2007) (Figure 4A). The strikingly similarity in orientation for the pseudo-monomers in the domain-swapped dimer [PDB accession code 1L5B] and the current momomer arrangement is easily appreciated from the illustration provided in Figure 4B. Analysis of the protein-protein interface reveals that approximately the same regions of the structures are involved. D35, N37 and S16 of one unit (monomer or chain) interact with K99 and Y100 of the other unit (Figure 4C). In particular, the side chain of N37 of CVNmutDB in monomer 2 that hydrogen bonds with the amide backbone nitrogen of Y100 in monomer 1 shows a similar contact in the domain-swapped dimer, although the distance is slightly larger. A detailed analysis of the interfaces reveals that overall, CVNmutDB buries less surface area (724 Å2) than the tetragonal domain-swapped dimer (1162 Å2) (Table S3). Interestingly, the triclinic domain-swapped dimer also buries less surface area than the other dimer structures (732 Å2). This is caused by a notable difference in orientation between the two dimeric halves in these two dimers. The other monomer structure, [PDB accession code 2Z21] exhibits more close contacts between the two monomers caused by the almost parallel arrangement of the molecules, resulting in an overall buried surface area of 1084 Å2.
Figure 4. Comparison between different crystal structures of CV-N.
(A) Stereoview of backbone superpositions of the two monomeric units of CVNmutDB (green) [PDB accession code 3CZZ], the M4-P51G mutant (black) [PDB accession code 2Z21] and the domain-swapped X-ray structure of wild type CV-N (pink) [PDB accession code 15LB]. All structures are displayed in the same orientation after best fit superpositions of one monomer unit each and the pseudo-monomer AB’ unit of the domain-swapped dimer. (B) Side by side view of the two protein molecules in the asymmetric unit of CVNmutDB (green) and the domain-swapped wild type CV-N (pink) [PDB accession code 15LB], illustrating the similarity in relative orientation. (C) Protein-protein interfaces for the two monomers of CVNmutDB and the pseudo-monomers of wild-type domain-swapped dimer [PDB accession code 1L5B]. Similar interactions are present, either between amino acids belonging to different monomers (CVNmutDB) or different chains (wild type CV-N).
Carbohydrate Binding
Binding studies using isothermal titration calorimetry (ITC) and NMR with monomeric wild type CV-N identified the termini of the D1 and D3 arms of Man-9 as the consensus binding epitopes on the sugar and delineated two carbohydrate binding sites on the protein, one each on domains AM and BM (Bewley and Otero-Quintero, 2001; Barrientos et al., 2002b; Bewley et al., 2002; Shenoy et al., 2002). We previously showed by NMR that in CVNmutDB only domain A was capable of interacting with sugars (Barrientos et al., 2006), and a true equilibrium binding constant for high mannose sugars (Man-9) was obtained, without the complications inherent to multivalent binding. Here, we carried out ITC titrations with Man-3 and Man-2, the D1 and D3 arms of Man-9, respectively. The observed binding isotherms are provided in Figure 5. Analysis of the binding isotherm for Man-3 (Figure 5A) yielded a ΔG value of −7.57 kcal/mol, ΔH= − 8.22 kcal/mol, with a binding entropy TΔS of − 0.65 kcal/mol. The derived equilibrium dissociation constant was Kd= 3.4 ± 0.05 µM. For comparison, a ΔG value of −7.4 kcal/mol, ΔH= −11.1 kcal/mol and an equilibrium dissociation constant Kd of 4.3 ± 0.3 µM were reported for Man-9, (Barrientos et al., 2006). It is noteworthy that the binding parameters for Man-3 and Man-9 are very similar, indicative of the fact that only a single binding site is available for the short (Man-3) as well as the high-mannose (Man-9) oligosaccharide. For both sugars, binding was driven by enthalpic contributions (with negative ΔH values of −8.22 kcal/mol and −11.1 kcal/mol for Man-3 and Man-9, respectively), with an unfavorable entropic contribution. A significantly larger negative TΔS (−3.68 kcal/mol) value was obtained for Man-9 compared to Man-3 (−0.65 kcal/mol), consistent with bigger losses in rotational, translational, and conformational freedom for the larger oligosaccharide upon complex formation.
Figure 5. Titration of CVNmutDB with mannose sugars.
(A) Isothermal titration of CVNmutDB with Man-3. The titration data for 25 automated injections are shown in the top panel and the total heat released as a function of the molar ratio of Man-3 versus CVNmutDB is displayed in the bottom panel (black). For comparison, previous titration data obtained for CVNmutDB with Man-9 are shown by a dotted line (Barrientos et al., 2006). (B) Isothermal titration of CVNmutDB with Man-2. The titration data for 20 automated injections are shown in the top panel and the total heat released as a function of the molar ratio of Man-2 versus CVNmutDB is displayed in the bottom panel. The continuous line represents the nonlinear least-squares best fit to the experimental data using a one site model. (C) NMR titration of CVNmutDB with Man-2. The top right hand panel shows the 1H-15N HSQC chemical shift titration curve (bound – free state) versus ligand/ protein molar ratio. The chemical shift difference is defined as: Δδ = [(Δδ HN) 2 + (ΔδN × 0.17) 2]1/2 The bottom panel shows the superposition of 1H-15N HSQC spectra without and in the presence of 12 equivalents of Man-2 in black and magenta, respectively. Selected resonances that experience large shifts upon addition of the disaccharide are labeled with residue name and number in an expanded view in the two top left panels.
Very weak binding was observed for di-mannose interacting with CNVmutDB. Such weak binding precludes the measurement of a complete binding isotherm by ITC, although sufficient data was available for fitting the binding curve (Figure 5B). Analysis of the titration yielded an equilibrium dissociation constant of Kd = 757 ± 80 µM, a ΔG value of −4.28 kcal/mol, a favorable enthalpic contribution, ΔH= −2.98 kcal/mol and a binding entropy of TΔS = 1.3 kcal/mol.
Since NMR is ideally suited to follow ligand binding in titration experiments, we also determined the binding constant for Man-2 by NMR. In particular, very low affinity interactions that are difficult to quantitatively assess by ITC are still amenable to NMR titration. The results of such an experiment for the Man-2 titration of CVNmutB are shown in Figure 5C with the HSQC titration data displayed in the bottom panel, with an expanded view of two selected regions in the top two left panels, and a binding isotherm derived from the chemical shift changes in the top right hand panel. The equilibrium dissociation constant (Kd) for Man-2 binding was determined from the titration shifts (chemical shift Δδ (ppm) versus the ratio of ligand to protein concentration) using several resonances. The extracted average Kd was 745 ± 33 µM. This value is in excellent agreement with the value extracted from the ITC binding curve (see above). A summary of all thermodynamic parameters determined for Man-2 and Man-3 is provided in Table 3.
Table 3.
Overall thermodynamic parameters from ITC binding data
| Enthalpy ΔH (kcal/mol) | Entropy TΔS (kcal/mol) | Free energy ΔG (kcal/mol) | Affinity Kd (µM) | |
|---|---|---|---|---|
| CVN mutDB- Man 2 | −2.98 ± 0.02 | 1.3 ± 0.05 | −4.28 ± 0.04 | 757 ± 80 |
| CVN mutDB- Man 3 | −8.22 ± 0.03 | − 0.65 ± 0.03 | −7.57 ± 0.08 | 3.4 ± 0.05 |
| CVN mutDB- Man 9(a) | −11.12 ± 0.1 | −3.68 ± 0.06 | −7.43 ± 0.04 | 4.3 ± 0.3 |
Evaluation of the solution and crystal structures using RDCs
It is frequently observed that crystal structures agree better with experimentally measured RDC values than the corresponding NMR structures, probably because of superior local backbone geometries in the X-ray structures. Reasons for a poor agreement between measured and predicted RDCs (linear correlation coefficients < 0.9) are associated with the use of an incorrect or distant model, either for local parts of the structure or globally. Local areas of disagreement are usually easily discerned since they manifest themselves by a few outliers in the correlation whose sequence identities cluster to particular stretches of the polypeptide chain. Other causes for poor correlations may arise from large motions that leads to averaging of the RDCs. The agreement between predicted and calculated values are generally expressed with a quality Q factor (Cornilescu et al., 1998) and "good" models usually exhibit Q factors below 0.5. The present work represents an ideal test case for examining this issue. We compared measured and predicted RDC values for two solution NMR model structures, the highly refined structure of wild type CV-N and the current NMR mutant structure calculated without RDCs. In addition, the present two X-ray structure models were also used as the input models for RDC prediction. For all models we carried out a best fit of the alignment tensors to the coordinates of the structures by singular value decomposition (SVD) and compared the measured RDC values with those predicted based on the NH vector orientations in the models. To ensure that all models are oriented essentially identically in the same molecular frame prior to any calculations, a best fit superposition of all structures was carried out. The results of the RDC comparisons for all our structures are provided in Figure 6. Comparing predicted and measured values using the wild type CV-N NMR structure as the model shows good agreement with a correlation coefficient of 0.91 (Figure 6A). The major outliers in the correlation are associated with residues in the loop region around the P51G mutation site that clearly exhibits a slightly different backbone conformation as shown above (see also Figure 2B). In particular, Q50, S52 and N53 lie outside the error range. The other two residues that exhibit a poorer fit are S38 and Q79, with S38 located in the loop close to residues A41 and A42, two of the amino acid changes that were introduced into the β-hairpin in the mutant (residues 41–42 (β4) and 47–48 (β5)). Q79 is close to the other two amino acids that were changed, A76 and G78. Eliminating the three outliers (Q50, S52 and N53) that are associated with the P51G mutation from the correlation results in an increase of the linear correlation coefficient from 0.91 to 0.95 and the r.m.s.d between observed and predicted RDCs is reduced from 4.02 to 3.03 Hz (Table 4). Carrying out the analysis for each of the X-ray monomers as input model (Figure 6B) yield r.m.s.d. values for 1DNH of 3.42 Hz and 3.65 Hz for monomer 1 and 2, respectively. A linear correlation coefficient of 0.94 was obtained for both independent X-ray models, suggesting that they are essentially identical with regard to their quality as input models. Using the mutant solution structure determined without RDC refinement yields a 1DNH r.m.s.d. value of 4.28 Hz and a linear correlation coefficient of 0.91 (Figure 6C). Naturally, the final CVNmutDB NMR structure that was calculated with RDCs results in excellent agreement (correlation coefficient of 0.99) between calculated and predicted RDCs values. All values for the RDC predictions are summarized in Table 4. It is interesting that both, the highly refined CV-N wild type solution NMR structure as well as the X-ray models of the identical protein exhibit comparable quality when used as input models, suggesting that no major difference exists between NMR and X-ray models when evaluated by RDC values.
Figure 6. Comparison between experimental and predicted residual dipolar couplings (RDCs) calculated for different models.
(A) Correlation between 1DobsNH measured for CVNmutDB and 1DcalcNH predicted using the wild type CV-N solution NMR. The identity of data points exhibiting the largest deviations are indicated by residue type and number. (B) Correlation between 1DobsNH measured for CVNmutDB and 1DcalcNH predicted using the two monomers in the crystal structure of CVNmutDB. Data for monomer 1 and 2 are shown in green and pink, respectively. (C) Correlation between 1DobsNH measured for CVNmutDB and 1DcalcNH predicted using the CVNmutDB solution NMR structure before RDC refinement.
Table 4.
Alignment tensor parameters and correlation between predicted and measured RDCs for different model structures
| Model | D(a) (Hz) | R(b) | Q(c) | RMSD(d) (Hz) | Corr. Coef. | Euler angles(e) (°) | ||
|---|---|---|---|---|---|---|---|---|
| α1 | α2 | α3 | ||||||
| CVNmutDB (NMR) w/o RDC refinement | −11.54 | 0.37 | 0.39 | 4.28 | 0.91 | 38.95 | −50.38 | −80.8 |
| CVNmutDB (X-ray) monomer 1 | −10.96 | 0.56 | 0.32 | 3.42 | 0.94 | 36.65 | −48.95 | −75.76 |
| CVNmutDB (X-ray) monomer 2 | −10.62 | 0.56 | 0.34 | 3.65 | 0.94 | 35.88 | −48.47 | −74.51 |
| wild type CV-N (NMR, PDB code 2EZM) | −11.09 | 0.52 | 0.37 | 4.02 | 0.91 | 35.37 | −49.65 | −73.52 |
| wild type CV-N (NMR, PDB code 2EZM ) 3 outliers removed | −11.62 | 0.55 | 0.26 | 3.03 | 0.95 | 34.2 | −48.6 | −72.8 |
D, R, magnitude and rhombicity of the alignment tensor.
Quality factor.
Root mean square deviation between measured and calculated RDCs.
α1, α2, α3, Euler angles describing the orientation of the alignment tensor in the molecular frame.
CONCLUSION
We have determined the NMR solution and X-ray crystal structures of CVNmutDB, a mutant that is devoid of the carbohydrate binding site on domain B. Under both conditions, the protein is monomeric and the structures are very similar. The CVNmutDB variant is the second mutant that does not exhibit a domain-swapped dimer in the crystal, in addition to the very recently determined crystal structure of another sugar binding site mutant (Fromme et al., 2007). In the latter, the binding site on domain A was destroyed and the sequence also contains the hinge-loop mutation P51G that renders the protein significantly more stable than wild type CV-N (Barrientos et al., 2002a). In addition to the increased stability, it is expected that the presence of a glycine in the loop influences the folding and dynamics of the protein (Nagi and Regan, 1997). The question therefore arises whether the abolishment of one sugar binding site or the proline substitution is responsible for preventing domain-swapping in the crystal. From all the available data we know that the relative proportion of monomer versus domain-swapped dimer in solution can be modulated by protein concentration, temperature, mutagenesis and solvent composition. However, up to now, we have not found conditions that resulted in diffraction quality crystals of the P51G variant of CV-N, although we were able to prepare dimeric [P51G]CV-N in solution (EM and AMG, unpublished results). In a complementary approach, we attempted to create a domain-swapped dimer in solution for CVNmutDB using the identical protocol that resulted in the [P51G]CV-N domain-swapped dimer in solution, however, only monomeric protein was seen for concentrations up to 6 mM (data not shown). In this respect it is worth mentioning that the thermodynamic stability of CVNmutDB is very similar to that of the [P51G]CV-N variant. The ΔG values for CVNmutDB and wild type CV-N are 9.6 and 9.8 kcal/mol, respectively. This suggests that folding speed or the smoothness of the folding landscape may play an important role in domain-swapped dimer formation and not overall thermodynamic stability. The smoothing of the folding landscape by the introduction of a glycine in a pivotal loop region in conjunction with the other amino acid changes clearly could prevent domain-swapping and dimer formation.
Domain swapping is frequently observed in X-ray structures of proteins and is believed to play an important role in oligomerization and aggregation phenomenon (Liu and Eisenberg 2002, Newcomer, 2002). Multimers are the easiest way for proteins to acquire multiple binding sites. We previously showed that cross-linking and multi-site/multivalent binding is intimately connected with CV-N’s carbohydrate mediated gp120 binding and the associated HIV-inactivating activity (Barrientos et al., 2006). The observation that the two variants for which no domain-swapping has been observed are devoid of anti-viral activity further substantiates this notion.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Protein was expressed from a synthetic gene for CVNmutDB using pET26b(+) (Novagen; Madison, WI) and E. coli BL-21(DE3) as vector and host strain (Barrientos et al., 2006), respectively (the amino acid sequence is displayed in Figure 1B). Uniformly 15N and 15N/13C -labeling was achieved utilizing 15NH4Cl (1g/L) and 13C6-D-glucose (2g/L) (Cambridge Isotope Laboratories, Inc; Andover, MA) as sole nitrogen and carbon sources. The expressed protein was isolated from the periplasmic fraction of E. coli cells by twice heating (62°C) and cooling (0°C) the cell suspension in PBS buffer (pH 7.4). After removal of insoluble material by centrifugation, the supernatant containing soluble CVNmutDB was fractionated by gel filtration on Superdex-75 (HiLoad 2.6 cm × 60 cm, Amersham Biosciences, Piscataway, NJ), equilibrated in 20 mM sodium phosphate buffer (pH 6.0). CVNmutDB protein was isolated as pure, folded, monomeric protein as verified by light scattering (data not shown). The purity and identity of the protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and mass spectrometry (measured molecular mass of 10,683 Da).
NMR spectroscopy
NMR spectra were recorded at 30 °C on Bruker DRX600 spectrometers, equipped with 5 mm, triple resonance, three axes gradient probes or z-axis gradient cryoprobes. Spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRview (Johnson, 2004). Samples contained 1.5 mM protein in 20 mM sodium phosphate buffer (pH 6.0). For chemical shift assignments, a series of heteronuclear, multi-dimensional experiments, routinely used in our laboratory were employed (Bax and Grzesiek, 1993; Fesik and Zuiderweg, 1990; Grzesiek et al., 1993; Logan et al., 1993). Complete 1H, 15N, and 13C backbone and side chain resonance assignments were obtained using the following heteronuclear 2D and 3D experiments: 1H-15N HSQC, HNCACB, CBCA(CO)NH, HCCH-TOCSY, as well as a 3D 13C/15N NOESY experiment. NOE spectra to derive inter-proton distance constraints included 3D 13C-edited and 15N-edited NOE experiments (Muhandiram et al., 1993; Talluri and Wagner, 1996; Mori et al., 1995), recorded with mixing times of 100 ms and 120 ms. Residual HN dipolar couplings (1DNH) were measured using in-phase/anti-phase 15N-1H HSQC experiments (Ottiger et al., 1998) on protein samples in magnetically oriented (10 mg/ml of pf1 phage) and isotropic solution (without phage) at 30 °C on a sample containing 150 µM CVNMutDB in 20 mM sodium phosphate buffer (pH 6.0). Only those NH resonances that exhibited no overlap were considered. A total of 62 residual dipolar couplings were used.
NMR Solution structure determination
All NOE cross peaks were picked using NMRView (Johnson, 2004) and inspected/sorted manually for accuracy. NOE cross peaks were assigned using the ATNOS algorithm (Herrmann et al., 2002) of CYANA (Güntert et al., 1997) in an automated fashion. The input for ATNOS consisted of the amino acid sequence of the protein, chemical shift lists from sequence-specific resonance assignment and NOEs from 15N and 13C-edited 3D-NOESY spectra. Throughout all calculations, 126 backbone torsion angle constraints derived from TALOS (Cornilescu et al., 1999) were employed. CYANA calculations were performed for seven iterative cycles with an additional cycle for stereo specific chemical shift assignment. From the second cycle onwards, the intermediate protein structures were used as an additional guide for the interpretation of the NOESY spectra. To obtain the best structural quality with the lowest energy target function and no restraints violations, successive runs incorporated optimized input NOE peaks and chemical shifts assignments. Continued analysis of the raw NMR data during the process of automated protein structure determination allows for direct feedback between the structure, NOE assignments and experimental NOESY spectra in an iterative fashion (Herrmann et al., 2002).
The initial CYANA calculations were used to generate the interproton distance restraints (2076) that were incorporated into further simulated annealing refinement with CNS (Brünger et al., 1998). From the NOE-derived ensemble of 50 structures the 20 lowest energy structures were further refined against residual dipolar couplings (62) with the program Dynamo included in the NMRpipe package, and final structures were examined using PROCHECK-NMR (Laskowski et al., 1996).
The atomic coordinates and NMR constraints have been deposited in the RCSB Protein Data Bank under accession code 2rp3. All structure figures were generated using the program RIBBONS 2.0 (Carson, 1991) and Swiss-PdbViewer v3.7 (Guex and Peitsch, 1997).
Crystallization and X-ray Data Collection
Purified CVNmutDB protein at a concentration of 4.8 mM in 20 mM sodium phosphate buffer, 0.01NaN3 (pH 6.0), was crystallized by sitting drop vapor diffusion. The best crystals were obtained at room temperature with 22–26% polyethylene glycol 4000 monomethyl ether as the precipitant in 0.2M ammonium acetate, 20 mM sodium phosphate buffer, 0.01 NaN3 (pH 6.0). Crystal growth took about 4 days with crystals typically having dimensions of 0.10 × 0.10 × 0.50 mm. X-ray diffraction data were collected from a single flash-cooled crystal (−180°C) using a Rigaku FR-E generator with a Saturn 944 CCD detector and high flux VariMax optics. To 1.36 Å resolution, 167,871 total observations were reduced to yield 37,884 unique reflections (94.7 % complete) with an internal R factor (based on intensities) of 0.038. The data were processed and scaled with the d*TREK package (Pflugrath, 1999).
Crystal Structure Determination and Refinement
The crystal structure of CVNmutDB was solved by molecular replacement using the domain-swapped structure of wild type CVN [PDB accession code 1L5B] as search model with the program Phaser 1.3.1. (McCoy et al., 2005). The initial model included two independent segments of the chain comprising residues L1-K48 and E56-E101. The hinge-loop region (W49-I55) was omitted from the model. Following rigid body and simulated annealing refinement, the model was used to generate an electron density composite omit map using the program CNS (Brünger et al., 1998). The atomic model for the missing region was built using Coot (Emsley and Cowtan, 2004) and further refined by simulated annealing. The final refinement procedure included periodic examinations of composite omit and difference maps, as well as the introduction of water molecules. Analysis of the final structure was performed using WHAT CHECK (Hooft et al., 1996) and PROCHECK (Laskowski et al., 1993). 92% of all residues are found in the most favored region in the Ramachandran plot (Ramachandran and Sasisekharan, 1968) with no residues in the disallowed regions.
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession code 3CZZ. All structural figures were generated with RIBBONS (Carson, 1991).
Isothermal Titration Calorimetry
Calorimetric titrations were performed using a VP-ITC isothermal titration calorimeter (MicroCal, LLC; Northampton, MA). Titrations were carried out at 30°C and all solutions contained 50 mM sodium phosphate buffer, 0.2 M NaCl, 0.02 % NaN3 (pH 7.4).
44 µM CVNmutDB solution was placed in the calorimeter cell (~1.44 ml active volume), stirred at 310 rpm and 12 µl aliquots of 465 µM Man-3 were added at 2 min intervals from a 295 µl stirring syringe. A total of 25 injections were performed. For the di-mannose titration, 14 µl aliquots of 15 mM Man-2 were added to 420 µM CVNmutDB for a total of 20 injections. Binding isotherms were fit using the Origin 7.0 software using a standard one-site model. Values for the binding enthalpy, the apparent number of binding sites and affinities were obtained from the fit to the experimental data. Other thermodynamic quantities were calculated using the standard expressions: ΔG = −RT ln Ka ; ΔG = ΔH −TΔS
Concentrations of the oligosaccharides were determined by compositional analysis at the Complex Carbohydrate Research Center at the University of Georgia.
NMR Titrations
Titration experiments were performed using uniformly 15N-labeled CVNmutDB (0.7 mM) with di-mannose (Man-2) in 20 mM sodium phosphate buffer (pH 6.0), 0.01% sodium azide and 90% H2O/10% D2O. A series of 1H-15N HSQC spectra were recorded after addition of sugar aliquots from stock solutions of 50 mM Man-2 at ligand: protein molar ratios of 0, 0.45, 0.83, 1.08, 1.26, 1.71, 2.6, 3.5, 4.3, 6.0, 7.5, 12.
Analysis of the NMR titration binding isotherm was carried out for resonances in fast exchange. For the fast exchange regime the observed chemical shift during the titration is given by: Δδ = [PL]/[P] (δb - δf), where [P], [PL] are protein and ligand-protein complex concentrations, δb , δf are the chemical shift of protein in the completely free and fully bound state. The dissociation constant Kd, was obtained from the best fit of titration curve (chemical shift Δδ vs. molar ratio M) using KaleidaGraph software and the following equation:
where relative chemical shift was estimated as: Δδ = [(Δδ HN) 2 + (ΔδN × 0.17) 2]1/2
Supplementary Material
Supplemental Data include three tables and can be found with this article online at http://www.structure.org/cgi/content/full/16/8/1183/DC1/.
ACKNOWLEDGMENTS
We thank Laura Barrientos for kindly providing the CVNmutDB clone and guidance, In-Ja Beyon for continuous help setting-up NMR experiments, JunGoo Jee for assistance and useful discussions regarding CYANA and John Louis for generous advice, expertise and discussions. We are grateful to Corine Sandström for kindly supplying the trimannoside sample. This work was in part funded by a National Institutes of Health grant to A.M.G. (R01GM080642).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adler P, Wood SJ, Lee YC, Lee RT, Petri WA, Jr, Schnaar RL. High affinity binding of the Entamoeba histolytica lectin to polyvalent N-acetylgalactosaminides. J Biol Chem. 1995;270:5164–5171. doi: 10.1074/jbc.270.10.5164. [DOI] [PubMed] [Google Scholar]
- Barrientos LG, Louis JM, Botos I, Mori T, Han Z, O'Keefe BR, Boyd MR, Wlodawer A, Gronenborn AM. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002a;10:673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- Barrientos LG, Gronenborn AM. The domain-swapped dimer of cyanovirin-N contains two sets of oligosaccharide binding sites in solution. Biochem Biophys Res Commun. 2002b;298:598–602. doi: 10.1016/s0006-291x(02)02489-0. [DOI] [PubMed] [Google Scholar]
- Barrientos LG, Gronenborn AM. The highly specific carbohydrate-binding protein cyanovirin-N: structure, anti-HIV/Ebola activity and possibilities for therapy. Mini Rev Med Chem. 2005;5:21–31. doi: 10.2174/1389557053402783. [DOI] [PubMed] [Google Scholar]
- Barrientos LG, Matei E, Lasala F, Delgado R, Gronenborn AM. Dissecting carbohydrate-Cyanovirin-N binding by structure-guided mutagenesis: functional implications for viral entry inhibition. Protein Eng Des Sel. 2006;19:525–535. doi: 10.1093/protein/gzl040. [DOI] [PubMed] [Google Scholar]
- Bax A, Grzesiek S. Methodological Advances in Protein NMR. Accounts of Chemical Research. 1993;26:131–138. [Google Scholar]
- Bewley CA, Gustafson KR, Boyd MR, Covell DG, Bax A, Clore GM, Gronenborn AM. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat Struct Biol. 1998;5:571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Otero-Quintero S. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man(8) D1D3 and Man(9) with nanomolar affinity: implications for binding to the HIV envelope protein gp120. J Am Chem Soc. 2001;123:3892–3902. doi: 10.1021/ja004040e. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Kiyonaka S, Hamachi I. Site-specific discrimination by cyanovirin-N for alpha-linked trisaccharides comprising the three arms of Man(8) and Man(9) J Mol Biol. 2002;322:881–889. doi: 10.1016/s0022-2836(02)00842-2. [DOI] [PubMed] [Google Scholar]
- Bolmstedt AJ, O'Keefe BR, Shenoy SR, McMahon JB, Boyd MR. Cyanovirin-N defines a new class of antiviral agent targeting N-linked, high-mannose glycans in an oligosaccharide-specific manner. Mol Pharmacol. 2001;59:949–954. doi: 10.1124/mol.59.5.949. [DOI] [PubMed] [Google Scholar]
- Botos I, O'Keefe BR, Shenoy SR, Cartner LK, Ratner DM, Seeberger PH, Boyd MR, Wlodawer A. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J Biol Chem. 2002a;277:34336–34342. doi: 10.1074/jbc.M205909200. [DOI] [PubMed] [Google Scholar]
- Botos I, Mori T, Cartner LK, Boyd MR, Wlodawer A. Domain-swapped structure of a mutant of cyanovirin-N. Biochem Biophys Res Commun. 2002b;294:184–190. doi: 10.1016/S0006-291X(02)00455-2. [DOI] [PubMed] [Google Scholar]
- Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O'Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bundle DR, Young NM. Carbohydrate-protein interactions in antibodies and lectins. Curr Opin Struct Biol. 1992;2:666–673. [Google Scholar]
- Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- Chang LC, Bewley CA. Potent inhibition of HIV-1 fusion by cyanovirin-N requires only a single high affinity carbohydrate binding site: characterization of low affinity carbohydrate binding site knockout mutants. J Mol Biol. 2002;318:1–8. doi: 10.1016/S0022-2836(02)00045-1. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of Protein Structure from Anisotropic Carbonyl Chemical Shifts in a Dilute Liquid Crystalline Phase. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- DeFrees S, Phillips L, Guo L, Zalipsky S. Sialyl Lewis x Liposomes as a Multivalent Ligand and Inhibitor of E-Selectin Mediated Cellular Adhesion. J Am Chem Soc. 1996;118:6101–6104. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fesik SW, Zuiderweg ER. Heteronuclear three-dimensional NMR spectroscopy of isotopically labelled biological macromolecules. Q Rev Biophys. 1990;23:97–131. doi: 10.1017/s0033583500005515. [DOI] [PubMed] [Google Scholar]
- Fromme R, Katiliene Z, Giomarelli B, Bogani F, Mc Mahon J, Mori T, Fromme P, Ghirlanda G. A monovalent mutant of cyanovirin-N provides insight into the role of multiple interactions with gp120 for antiviral activity. Biochemistry. 2007;46:9199–9207. doi: 10.1021/bi700666m. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Vuister GW, Bax A. A simple and sensitive experiment for measurement of JCC couplings between backbone carbonyl and methyl carbons in isotopically enriched proteins. J Biomol NMR. 1993;3:487–493. doi: 10.1007/BF00176014. [DOI] [PubMed] [Google Scholar]
- Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. http://www.expasy.org/spdbv/ [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kiessling LL, Pohl NL. Strength in numbers: non-natural polyvalent carbohydrate derivatives. Chem Biol. 1996;3:71–77. doi: 10.1016/s1074-5521(96)90280-x. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lee YC. Biochemistry of carbohydrate-protein interaction. FASEB J. 1992;6:3193–3200. doi: 10.1096/fasebj.6.13.1397841. [DOI] [PubMed] [Google Scholar]
- Lees WJ, Spaltenstein A, Kingery-Wood JE, Whitesides GM. Polyacrylamides bearing pendant alpha-sialoside groups strongly inhibit agglutination of erythrocytes by influenza A virus: multivalency and steric stabilization of particulate biological systems. J Med Chem. 1994;37:3419–3433. doi: 10.1021/jm00046a027. [DOI] [PubMed] [Google Scholar]
- Lis H, Sharon N. Lectins: Carbohydrate-Specific Proteins That Mediate Cellular Recognition. Chem Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan TM, Olejniczak ET, Xu RX, Fesik SW. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Meunier SJ, Roy R. Polysialosides scaffolded on p-Tert-butylcalix[4]arene. Tetrahedron Letters. 1996;37:5469–5472. [Google Scholar]
- Mortell K, Weatherman R, Kiessling L. Recognition Specificity of Neoglycopolymers Prepared by Ring-Opening Metathesis Polymerization. J Am Chem Soc. 1996;118:2297–2298. [Google Scholar]
- Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Farrow NA, Xu GY, Smallcombe SH, Kay LE. A Gradient 13C NOESY-HSQC Experiment for Recording NOESY Spectra of 13C-Labeled Proteins Dissolved in H2O. Journal of Magnetic Resonance, Series B. 1993;102:317–321. [Google Scholar]
- Nagi AD, Regan L. An inverse correlation between loop length and stability in a four-helix-bundle protein. Fold Des. 1997;2:67–75. doi: 10.1016/S1359-0278(97)00007-2. [DOI] [PubMed] [Google Scholar]
- Newcomer ME. Protein folding and three-dimensional domain swapping: a strained relationship? Curr Opin Struct Biol. 2002;12:48–53. doi: 10.1016/s0959-440x(02)00288-9. [DOI] [PubMed] [Google Scholar]
- O'Keefe BR, Shenoy SR, Xie D, Zhang W, Muschik JM, Currens MJ, Chaiken I, Boyd MR. Analysis of the interaction between the HIV-inactivating protein cyanovirin-N and soluble forms of the envelope glycoproteins gp120 and gp41. Mol Pharmacol. 2000;58:982–992. doi: 10.1124/mol.58.5.982. [DOI] [PubMed] [Google Scholar]
- Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Patterson GML, Baker KK, Baldwin CL, Bolis CM, Caplan FR, Larsen LK, Levine IA, Moore RE, Nelson CS, Tschappat KD, et al. Antiviral activity of cultured blue-green algae (Cyanophyta) J Phycol. 1993;29:125–130. [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V. RPMS: Ramachandran plot for multiple structures. Adv Protein Chem. 1968;23:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Sabesan S, Bock K, Paulson JC. Conformational analysis of sialyloligosaccharides. Carbohydr Res. 1991;218:27–54. doi: 10.1016/0008-6215(91)84084-r. [DOI] [PubMed] [Google Scholar]
- Sandström C, Berteau O, Gemma E, Oscarson S, Kenne L, Gronenborn AM. Atomic mapping of the interactions between the antiviral agent cyanovirin-N and oligomannosides by saturation-transfer difference NMR. Biochemistry. 2004;43:13926–13931. doi: 10.1021/bi048676k. [DOI] [PubMed] [Google Scholar]
- Shenoy SR, O'Keefe BR, Bolmstedt AJ, Cartner LK, Boyd MR. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J Pharmacol Exp Ther. 2001;297:704–710. [PubMed] [Google Scholar]
- Shenoy SR, Barrientos LG, Ratner DM, O'Keefe BR, Seeberger PH, Gronenborn AM, Boyd MR. Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: calorimetric and NMR characterization. Chem Biol. 2002;9:1109–1118. doi: 10.1016/s1074-5521(02)00237-5. [DOI] [PubMed] [Google Scholar]
- Sigal GB, Mammen M, Dahmann G, Whitesides GM. Polyacrylamides Bearing Pendant R-Sialoside Groups Strongly Inhibit Agglutination of Erythrocytes by Influenza Virus: The Strong Inhibition Reflects Enhanced Binding through Cooperative Polyvalent Interactions. J Am Chem Soc. 1996;118:3789–3800. [Google Scholar]
- Spengard U, Schudok M, Schmidt W, Wolfgang S, Kretzschmar G, Kunz H. Multiple sialyl Lewis(x) N-glycopeptides: Effective ligands for E-selectin. Angew Chem Int Ed Engl. 1996;35:321–324. [Google Scholar]
- Talluri S, Wagner G. An optimized 3D NOESY-HSQC. J Magn Reson B. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- Yang F, Bewley CA, Louis JM, Gustafson KR, Boyd MR, Gronenborn AM, Clore GM, Wlodawer A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J Mol Biol. 1999;288:403–412. doi: 10.1006/jmbi.1999.2693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include three tables and can be found with this article online at http://www.structure.org/cgi/content/full/16/8/1183/DC1/.