Abstract
Greater understanding of metastasis is required to improve cancer treatment outcomes. Recently, changes in expression of the scaffold protein HEF1/CAS-L/NEDD9 were found to be a potent pro-metastatic stimulus in melanoma and other cancers. Mechanistic studies suggest diverse cellular roles of HEF1 and highlight its importance in the response to extracellular cues that drive invasion and metastasis. As a metastatic “hub” for signaling in cancer, HEF1 may provide a useful target for drug discovery efforts.
Introduction
In recent years, our understanding of the metastatic process has evolved significantly. It is now appreciated that the formation of distant metastases requires functionally distinct events, including 1) invasion of a tumor through basement membrane and stroma, 2) tumor cell intravasation, 3) survival of a tumor in the blood stream, 4) (frequently, although not invariably) homing of the tumor to a specific site, and 5) extravasation and colony formation. This complexity either implies that the combined action of multiple proteins is necessary for metastasis, or that proteins which can promote metastasis are multifunctional.
The identification of tumor-encoded genes whose elevated expression marks transition to metastasis and which are required for establishment of tumors at distant sites will yield important diagnostic and therapeutic benefits. Many research groups have sought to define such genes, and in the past year, signaling activity and overexpression of HEF1/Cas-L/NEDD9 (1–3) (hereafter designated HEF1) have been shown to be required for invasion by glioblastomas (4) and strongly linked to promotion of melanoma metastasis (5). Significantly, extensive characterization of HEF1 over the past decade has revealed a multifaceted mode of action that suggests changes in HEF1 status may be fundamentally important for multiple aspects of the metastatic program. In this review, we will first discuss the signaling activities that most likely underlie the ability of HEF1 to influence metastasis, and then outline the cellular mechanisms influencing HEF1 expression and “activation” during metastatic transition. Finally, we will discuss intriguing recent findings that imply a broader role for HEF1 in the pathogenesis of aggressive non-solid tumors, and strategies for therapeutically targeting HEF1. We note that roles for HEF1 in additional cellular processes and diseases have recently been comprehensively reviewed (6, 7).
Biological activities of HEF1 that favor metastasis
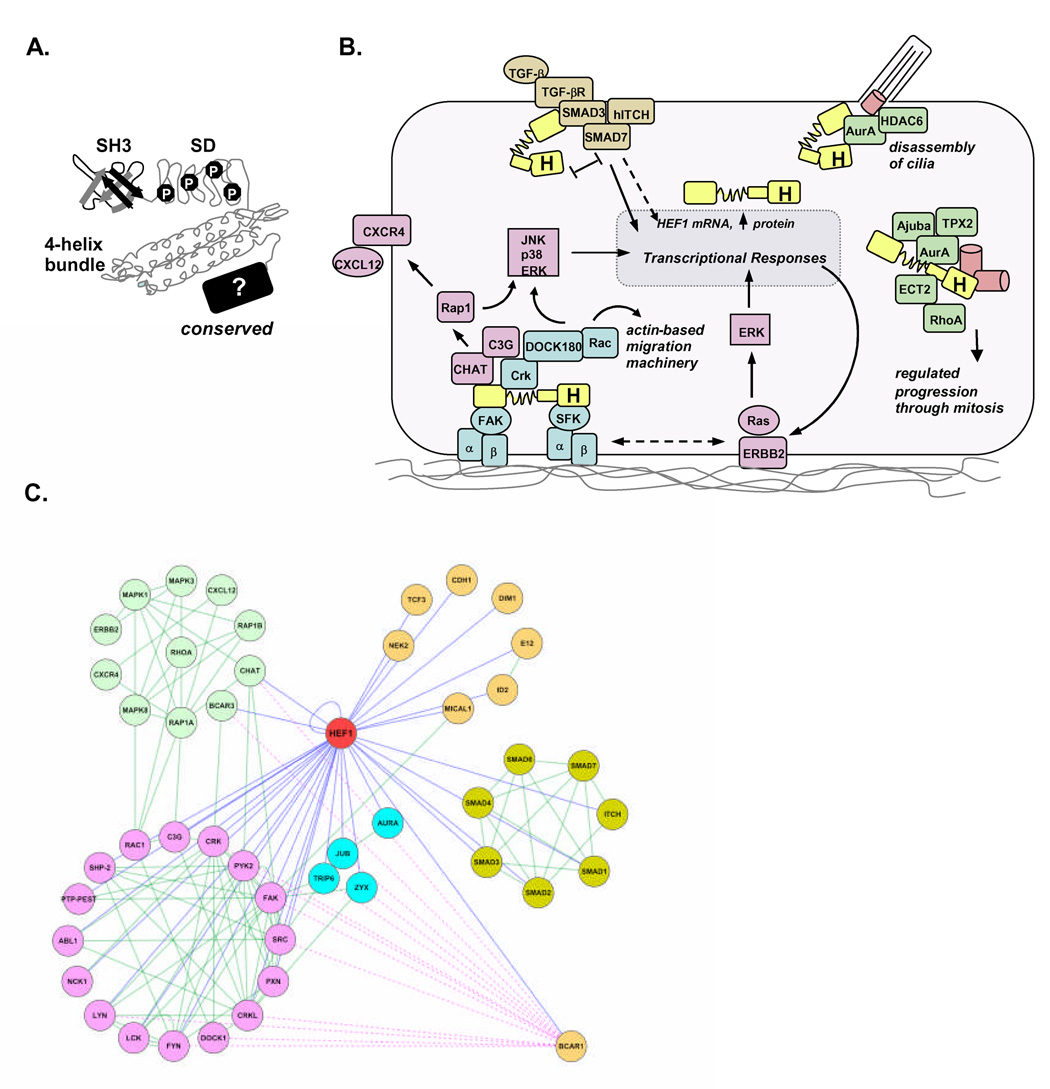
The HEF1 gene is conserved in all vertebrates, and localizes at chromosome 6p25-24 in humans, and chromosome 13 A3.3 in mice. The HEF1 protein is predominantly cytoplasmic, and concentrates at focal adhesions during interphase in adherent cells, and at centrosomes and other parts of the mitotic apparatus during G2/M. HEF1 lacks any known enzymatic function, but contains many functional modules for protein interaction, leading to its classification as a scaffolding protein (2, 3, 7, 8). Validated interaction sequences include (Figure 1A) an SH3 domain, at least 15 SH2 domain-binding sites, and an evolutionarily well-conserved carboxy-terminal domain of unknown structure. Proteins that functionally and/or physically interact with HEF1 (Figures 1B, 1C) include many with direct roles in promoting tumor invasion, as discussed below. Vertebrate HEF1 has two paralogs, p130Cas/BCAR1 and Efs/Sin, which conserve domain structure and many but not all functional interactions (6, 8): together, HEF1, p130Cas, and Efs are referred to as members of the Cas family.
A. Schematic of structure of the HEF1/NEDD9/CasL protein. Human HEF1 is 834 amino acids; key functional domains include an amino-terminal SH3 domains, which binds FAK; a “substrate domain” (SD) containing multiple embedded SH2 binding sites, analogous to the mechanosensing domain of p130Cas recently described by Sawada et al.; a likely four-helix bundle, based on molecular modeling of primary sequence of HEF1 against the crystal coordinates for the conserved region of p130Cas (results not shown); and an evolutionarily conserved C-terminal domain, which binds Src family kinases and other proteins (indicated with a “?” as details of structure remain unknown). B. HEF1 (indicated in bright yellow, with “H”) is a component of the integrin dependent Src-FAK-Crk migration signaling cascade; influences cellular homing through CHAT/CXCR4; engages in crosstalk with the Ras pathway; is an intermediate in TGF-β-dependent signaling; activates the centrosomal Aurora-A/Ajuba/TPX2 machinery governing mitotic entry and cytokinesis; and activates Aurora-A/HDAC6 at the basal body to initiate ciliary disassembly. C. Curated online resources (based on experimentally well-validated protein interaction data) indicate numerous direct HEF1 interactions with cancer-related signaling pathways. Blue lines indicate protein interactions; many HEF1 partners also take part in extensive self-interactions within functional clusters that are likely augmented by increased HEF1 levels. Clusters shown in rose are particularly relevant to FAK/Src/integrin; in green, to CHAT/Rap1; in blue, to Aurora A; in gold, to TGF-β/SMADs; in yellow, all others. All HEF1-only interactions present in online databases are shown as blue lines; for contrast, common interactions of BCAR1/p130Cas are also shown as dashed pink lines. Interactions between all other proteins (including functional as well as physical interactions) are shown in green. For clarity, only interactions relevant to the discussion in this review are shown. Data on protein-protein interactions were collected in Cytoscape (http://cytoscape.org/), combining data from a Bionet plugin (http://err.bio.nyu.edu/cytoscape/bionetbuilder/), and EMBL String (string.embl.de/), with each retrieving information from several databases, including DIP (http://dip.doe-mbi.ucla.edu/), BIND (http://www.bind.ca/), KEGG (http://www.genome.jp/kegg/), Prolinks (http://mysql5.mbi.ucla.edu/cgi-bin/functionator/pronav), HPRD (http://www.hprd.org/), and The BioGrid (http://www.thebiogrid.org/).
Significantly elevated levels of HEF1 mRNA (10–70 fold) and protein exist in >35% of the metastatic melanomas produced by “escaper” tumors in Tyr-rtTA+;Tet-RAS+;Ink4a/Arf−/− mutant mice, as well as in a similar percentage of human metastatic melanomas (5). Importantly, elevated HEF1 protein expression was essential for the metastatic properties of the involved tumors both in vitro and in vivo (5). As HEF1 is a scaffolding protein, its action involves regulated assembly of protein complexes. Consequently, the effect of altered HEF1 expression is dependent on the relative stoichiometry and availability of other complex constituents. Hence, moderate overexpression may drive the assembly of functional complexes, causing constitutive activation of downstream effector pathways. Conversely, excessive HEF1 over-expression may be equivalent to loss of HEF1 expression, if either condition induces complex disruption. It is perhaps significant that one study has now identified reduced HEF1 expression as part of a signature for metastatic breast cancers ((9), discussed further below). Notably, both overexpression and depletion of HEF1 cause mitotic defects in cultured cells (10, 11). While the exact mechanism of HEF1 action in metastasis requires further investigation, to date, studies of HEF1 overexpression, depletion, and genetic deletion have revealed the following metastasis-relevant properties:
1] HEF1 positively regulates the Src-FAK-Crk “migratory switch”
The initial reports identifying HEF1 established that this protein interacts directly with FAK, Src, and Crk (2, 3). FAK is commonly constitutively activated in melanomas, and an important target of cancer drug development. The consensus of work by many groups suggests a mechanism in which cell attachment triggers the interaction of Src, HEF1, and FAK: overexpression or mutational activation of one of these proteins can also drive complex formation. These interactions enhance the activation of Src and FAK, and lead to extensive tyrosine phosphorylation of HEF1, creating binding sites for effector proteins with SH2 domains: the most important of these are Crk and Crk-L. A Cas-Crk complex has been described as a “master switch” for cell migration; through Crk-L and DOCK180, HEF1 activates Rac and other components of the cell migration machinery (reviewed in (8)).
2] HEF1 activates machinery for tumor growth, invasion and homing
Tumor progression depends on the activation of essential effector kinases that mediate proliferation and survival in an expanding hypoxic tumor mass. Tumor invasion depends on the increased expression and activity of extracellular proteases that degrade or remodel basement membranes to allow cellular movement, while activating latent growth factors that promote tumor progression. Related to this, HEF1 overexpression may induce molecules involved in tissue remodeling and invasion (12). HEF1 overexpression also induces the activation of ERK, p38, and JNK kinases through interactions with intermediary signaling effectors (reviewed in (6)). For example, overexpressed HEF1 transcriptionally induces ErbB2/HER2/neu, a growth factor receptor at the top of the Ras > Raf > MEK > ERK pathway, while the HEF1 interactor FAK directly binds Shc, which influences activation of Ras through its binding partners EGFR and Grb2.
HEF1 also binds proteins of the AND-34/CHAT family (13), which activate JNK and ERK by signaling through Rap1 (14). Activated Rap1 is also an important intermediate in “inside-out” cell signaling, in which internally-derived signals activate integrin-ligand binding in response to upstream cues that typically involve chemokine stimulation. This process, much studied in consideration of the migration and “homing” of lymphoid cells, is now appreciated as playing a role in tumor invasion and targeting of metastases. Lymphoid cells from HEF1 null mice, or with depleted HEF1, are greatly impaired for chemokine response, migration, and homing, accompanied by failed activation of CHAT-H and Rap1 (14, 15). Critically, these defects involve the CXCR4-CXCL12 targeting system, which is important not only for targeting of lymphocytes to secondary organs, but is also a major contributor to tumor metastasis (16). By inference, CXCR4 signaling is expected to be hyperactivated in cells with overexpressed HEF1.
3] HEF1 conditions TGF-β responses
One of the enigmas of tumor progression is how tumor interpretation of TGF-β signals modulates over time. Extrinsic TGF-β inhibits the growth of early tumors; however, TGF-β promotes the growth of later stage invasive tumors (17), downregulating E-cadherin and promoting mesenchymal transformation. Intriguingly, HEF1 binds directly to TGF-β pathway effectors and inhibitors, including multiple SMADs (e.g., (18), and discussed in (6)). Via these interactions, HEF1 induces negative feedback for aspects of TGF-β-dependent signaling. Intriguingly, the TGF-β pathway signaling molecule SMAD7 has very recently been shown to inhibit melanoma metastasis to bone (19); HEF1 overexpression would be predicted to limit SMAD7 activity, thus promoting metastasis.
4] HEF1 activates RhoA and Aurora A: counter-pressures for tumor growth
Given the extensive biology linking HEF1 to invasion signaling pathways, recent observations that this protein also regulates cell cycle progression through mitosis were unexpected (6, 10, 11). In MCF-7 cells and other epithelial cell lines, HEF1 functions at two discrete points during cytokinesis. At the centrosome, HEF1 interacts with and activates Aurora A kinase during mitotic entry. Overexpressed HEF1 hyper-activates Aurora A, inducing failure of cytokinesis. Separately, HEF1 positively regulates RhoA activation and elevated HEF1 expression leads to abnormally persistent RhoA activity throughout cytokinesis, preventing normal cellular reattachment to surrounding matrix, and providing a second stimulus for deficient cytokinesis. HEF1-overexpressing cells exhibiting defective cytokinesis then arrest in G1, and subsequently undergo apoptosis at high frequency, implying the triggering of cell division checkpoints.
The recognition that HEF1 overexpression triggers cell division checkpoints and apoptosis may be particularly important in understanding why HEF1 overexpression is associated with later (rather than early) stages of tumor progression. In the Tyr-rtTA+;Tet-RAS+;Ink4a/Arf−/− melanoma mouse model, both Rb- and p53-dependent cell division checkpoints have been disabled and concurrently constitutive Ras overexpression provides a strong stimulus towards continued proliferation. Such prior changes may be essential for cells to tolerate pro-apoptotic effects of sustained HEF1 overexpression. In this context, it is interesting that early metastatic melanomas are often characterized by genomic rearrangements and aneuploidy, and manifest a high level of apoptosis relative to pre-metastatic tumors.
5] HEF1 and Aurora A regulate ciliary disassembly
Cilia are small organelles that protrude from the surface of many mammalian cell types and act as cellular “antennas”, with growth factor receptors localized at cilia sensing extracellular cues to regulate cell growth. Defects in ciliary structural integrity or associated signaling induce numerous developmental syndromes, are a primary cause of polycystic kidney disease, and have in the past year been strongly linked to cancer development (reviewed in (20)). Very recently, HEF1 activation of Aurora A at the ciliary basal body was shown to trigger a ciliary resorption pathway involving the tubulin deacetylase HDAC6 as an effector protein (21). This unexpected finding suggests a totally new mechanism by which overexpression of HEF1 can influence the growth properties of metastatic cancers: much more work is necessary to understand the importance of this observation.
What causes HEF1 upregulation prior to melanoma metastasis?
In normal and transformed cells, HEF1 is dynamically regulated in response to both intracellular and extracellular signals. Molecular mechanisms important for control of HEF1 expression and scaffolding function include transcriptional activation, phosphorylation, and both proteasome- and caspase-mediated proteolysis (reviewed in detail in (6)). Although upregulation of the HEF1 mRNA in metastasis has been shown in some cases to arise from chromosomal amplification (5), this upregulation more commonly occurs at the level of mRNA transcription. Given that the HEF1 mRNA is known to be downregulated during nervous system development ((1), and discussed in (6)), it is interesting to speculate that HEF1 overexpression in melanoma may reflect aberrant reactivation of a developmental program.
A growing number of transcriptional pathways have been reported to regulate HEF1; intriguingly, some of these are particularly relevant to metastasis. For example, hypoxia is now appreciated as creating conditions conducive for metastasis, and hypoxia is emerging as a potentially important inducer of HEF1 mRNA expression (22, 23). TGF-β strongly induces transcription of HEF1 (24): together with the role for HEF1 in negative regulation of TGF-β signaling, this provides a second link between these proteins, and raises the possibility of a reinforcing feedback loop. HEF1 mRNA and protein are strongly induced by serum treatment, but as yet most of the serum factors directing transcription of the HEF1 are poorly understood.
It is extremely likely that post-transcriptional events will be found to modulate the expression and action of HEF1 in aggressive cancers. Phosphorylation not only regulates HEF1 scaffolding capacity but can also regulate HEF1 protein turnover. HEF1 phosphorylation is induced by a number of key factors linked with cancer progression, including FAK (commonly activated in advanced melanomas), TGF-β (24), PDGF (4), Abl (2), and BCR-ABL (25). This last observation is of particular interest as the literature describing a role for HEF1 in hematopoietic malignancies becomes increasingly compelling ((15), and discussed comprehensively in (7)). Conversely, for solid tumors, a particularly appealing idea is that changes in the cellular microenvironment, such as the increasing rigidity of stroma organized around an invasive tumor (26), may directly condition HEF1 activity. The HEF1 paralog p130Cas has recently been recognized as a mechanosensor hyperphosphorylated in response to stretch (27): HEF1 conserves all the structural elements necessary for a similar response.
Prospect for therapeutic exploitation of HEF1
At present, HEF1 is an attractive biomarker of metastatic melanomas. Whether this will be true for other cancers remains to be established; for instance, while HEF1 promotes metastatic behavior in glioblastomas (4), reduced levels of HEF1 transcript characterize an MDA-MB-231 breast cancer cell line selected by serial in vivo passages for efficient metastasis to the lung in mice (9). This may represent tumor type-specific HEF1 action. Alternatively, given that the stochiometry of HEF1 and its binding partners is likely to be critical to the signaling function of HEF1, it is possible that these data simply reflect an inhibition of HEF1 function – achieved either by loss of gene product or by altered protein stochiometry resulting from overexpression. Future delineation of these different possibilities will be important for determining HEF1 function in metastatic cancer. No serious attempt has as yet been made to target HEF1 for drug development; as a scaffolding protein with no assigned catalytic function, drug selection strategies are clearly more challenging than for enzymes. However, several points suggest a HEF1-directed targeting strategy is feasible. A peptide aptamer screen has identified discrete peptides that bind and stabilize the HEF1 protein from degradation, implying it may be possible to identify agents that destabilize the protein (28). Significantly, although HEF1−/− mice manifest some developmental and migration defects, these animals are viable and fertile (15), indicating that loss of HEF1 function can probably be well tolerated in adults. Drugs targeting FAK, Src, BCR-ABL, and TGF-β already in the clinic may be particularly effective in tumors overexpressing HEF1, through limiting HEF1 phosphorylation. Finally, early evidence suggests a potential role for HEF1 in other pathological conditions, including stroke, rheumatoid arthritis, and HTLV-1 infection, as well as leukemias and lymphomas (reviewed in (6, 7)). A decade after its first description, HEF1 is poised to yield exciting insights into the process of metastasis, and may provide an important new target in chemotherapy.
Acknowledgments
We apologize to our colleagues whose outstanding publications we have not been able to directly cite due to space constraints. GMO'N is supported by a Career Development Fellowship from the NSW Cancer Council. EAG and IGS are supported by NIH RO1 CA63366, a Department of Defense OCRF IDEA Award, and Pennsylvania Tobacco Settlement Funds (to EAG), and by an Appropriation from the Commonwealth of Pennsylvania, and by NIH core grant CA06927 (to Fox Chase Cancer Center). SS is supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Ministry of Health, Labor, and Welfare of Japan.
References
- 1.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys.Res.Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 2.Law SF, Estojak J, Wang B, Mysliwiec T, Kruh GD, Golemis EA. Human Enhancer of Filamentation 1 (HEF1), a novel p130Cas-like docking protein, associates with FAK, and induces pseudohyphal growth in yeast. Mol. Cell. Biol. 1996;16:3327–3337. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta-1 integrin-mediated signaling in lymphocytes. J. Exp. Med. 1996;184:1365–1375. doi: 10.1084/jem.184.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan M, Stewart JE, Golemis EA, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–1732. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- 5.Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Singh MK, Cowell L, Seo S, O'Neill GM, Golemis EA. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis, and cell cycle. Cell Biochem Biophys. 2007 doi: 10.1007/s12013-007-0036-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo S, Ichikawa M, Kurokawa M. Structure and Function of Cas-L and Integrin-Mediated Signaling. Critical Reviews In Immunology. 2006;26:391–406. doi: 10.1615/critrevimmunol.v26.i5.20. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill GM, Fashena SJ, Golemis EA. Integrin signaling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 2000;10:111–119. doi: 10.1016/s0962-8924(99)01714-6. [DOI] [PubMed] [Google Scholar]
- 9.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–946. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dadke D, Jarnik M, Pugacheva EN, Singh MK, Golemis EA. Deregulation of HEF1 impairs M-phase progression by disrupting the RhoA activation cycle. Mol Biol Cell. 2006;17:1204–1217. doi: 10.1091/mbc.E05-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fashena SJ, Einarson MB, O'Neill GM, Patriotis CP, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J. Cell. Sci. 2002;115:99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- 13.Cai D, Felekkis KN, Near RI, et al. The GDP exchange factor AND-34 is expressed in B cells, associates with HEF1, and activates Cdc42. J Immunol. 2003;170:969–978. doi: 10.4049/jimmunol.170.2.969. [DOI] [PubMed] [Google Scholar]
- 14.Regelmann AG, Danzl NM, Wanjalla C, Alexandropoulos K. The hematopoietic isoform of cas-hef1-associated signal transducer regulates chemokine-induced inside-out signaling and T cell trafficking. Immunity. 2006;25:907–918. doi: 10.1016/j.immuni.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Seo S, Asai T, Saito T, et al. Crk-associated substrate lymphocyte type is required for lymphocyte trafficking and marginal zone B cell maintenance. J Immunol. 2005;175:3492–3501. doi: 10.4049/jimmunol.175.6.3492. [DOI] [PubMed] [Google Scholar]
- 16.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 17.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Inamoto S, Iwata S, Inamoto T, et al. Crk-associated substrate lymphocyte type regulates transforming growth factor-beta signaling by inhibiting Smad6 and Smad7. Oncogene. 2006;26:893–904. doi: 10.1038/sj.onc.1209848. [DOI] [PubMed] [Google Scholar]
- 19.Javelaud D, Mohammad KS, McKenna CR, et al. Stable overexpression of smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 20.Kuehn EW, Walz G, Benzing T. Von hippel-lindau: a tumor suppressor links microtubules to ciliogenesis and cancer development. Cancer Res. 2007;67:4537–4540. doi: 10.1158/0008-5472.CAN-07-0391. [DOI] [PubMed] [Google Scholar]
- 21.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007 doi: 10.1016/j.cell.2007.04.035. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Rendon E, Hale SJ, Ryan D, et al. Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia. Stem Cells. 2006 doi: 10.1634/stemcells.2006-0398. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki T, Iwata S, Okano HJ, et al. Nedd9 Protein, a Cas-L Homologue, Is Upregulated After Transient Global Ischemia in Rats. Possible Involvement of Nedd9 in the Differentiation of Neurons After Ischemia. Stroke. 2005;36:2457–2462. doi: 10.1161/01.STR.0000185672.10390.30. [DOI] [PubMed] [Google Scholar]
- 24.Zheng M, McKeown-Longo PJ. Regulation of HEF1 expression and phosphorylation by TGF-beta 1 and cell adhesion. J Biol Chem. 2002;277:39599–39608. doi: 10.1074/jbc.M202263200. [DOI] [PubMed] [Google Scholar]
- 25.de Jong R, van Wijk A, Haataja L, Heisterkamp N, Groffen J. BCR/ABL-induced leukemogenesis causes phosphorylation of Hef1 and its association with Crkl. J. Biol. Chem. 1997;272:32649–32655. doi: 10.1074/jbc.272.51.32649. [DOI] [PubMed] [Google Scholar]
- 26.Paszek MJ, Zahir N, Johnson KR, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Sawada Y, Tamada M, Dubin-Thaler BJ, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serebriiskii IG, Mitina O, Pugacheva EN, et al. Detection of peptides, proteins, and drugs that selectively interact with protein targets. Genome Res. 2002;12:1785–1791. doi: 10.1101/gr.450702. [DOI] [PMC free article] [PubMed] [Google Scholar]