Abstract
The aging suppressor gene Klotho encodes a single-pass transmembrane protein. Klotho-deficient mice exhibit a variety of aging-like phenotypes, many of which are similar to those observed in fibroblast growth factor-23 (FGF23)-deficient mice. To test the possibility that Klotho and FGF23 may function in a common signal transduction pathway(s), we investigated whether Klotho is involved in FGF signaling. Here we show that Klotho protein directly binds to multiple FGF receptors (FGFRs). The Klotho-FGFR complex binds to FGF23 with higher affinity than FGFR or Klotho alone. In addition, Klotho significantly enhanced the ability of FGF23 to induce phosphorylation of FGF receptor substrate and ERK in various types of cells. Thus, Klotho functions as a cofactor essential for activation of FGF signaling by FGF23.
The Klotho gene encodes a 130-kDa single-pass transmembrane protein with a short cytoplasmic domain (10 amino acids) and is expressed predominantly in the kidney. Mice carrying a loss-of-function mutation in the Klotho gene develop a syndrome resembling human aging, including shortened life span, skin atrophy, muscle atrophy, osteoporosis, arteriosclerosis, and pulmonary emphysema (1). Conversely, overexpression of the Klotho gene extends the life span and increases resistance to oxidative stress in mice (2–4). These observations suggest that the Klotho gene functions as an aging suppressor gene. The extracellular domain of Klotho protein is shed and secreted in the blood (2, 5), potentially functioning as a humoral factor that signals suppression of intracellular insulin/IGF1 signaling, which partly contributes to its anti-aging properties (2). However, a signaling pathway(s) directly activated by Klotho protein, including the identity of the Klotho receptor, has not been determined. The function of the transmembrane form of Klotho protein also remains to be determined.
Fibroblast growth factor-23 (FGF23)2 was originally identified as a gene mutated in patients with autosomal dominant hypophosphatemic rickets (6), where mutations in the FGF23 gene conferred resistance to inactivation by protease cleavage, resulting in elevated serum levels of FGF23 (7–12). FGF23 inhibits phosphate transport in renal proximal tubular cells and in proximal tubules perfused in vitro (13). Consistent with these findings, mice defective in FGF23 expression show increased renal phosphate reabsorption and hyper-phosphatemia (14). Although FGF23 binds to multiple FGF receptors (FGFRs) (15), it has modest receptor affinity (KD = 200–700 nM) and often requires cofactors such as heparin or glycosaminoglycan (15, 16) to activate FGF signaling in cultured cells and to inhibit phosphate transport in proximal tubules perfused in vitro (13).
Klotho-deficient mice (Klotho−/− mice) and FGF23 deficient mice (Fgf23−/− mice) develop many common phenotypes, including shortened life span, growth retardation, infertility, muscle atrophy, hypoglycemia, and vascular calcification in the kidneys. Notably, they both have increased serum levels of phosphate (14, 17). These observations have led us to the hypothesis that Klotho and FGF23 may function via a common signal transduction pathway. In this report we show that Klotho binds to multiple FGFRs and functions as a cofactor necessary for FGF signaling activation by FGF23.
MATERIALS AND METHODS
Expression Vectors
Complementary DNA containing the mouse FGFRs coding region (IMAGE Clone, Invitrogen, supplemental Fig. 1) were cloned into pcDNA3.1(+) expression vector (Invitrogen). Before subcloning, a V5-epitope tag was added to the C terminus and appropriate restriction enzyme sites to the both ends using synthetic oligonucleotides and polymerase chain reaction. Expression vectors for the mouse FGF23 resistant to proteolytic inactivation (R179Q) (18), the transmembrane form of mouse Klotho, and the extracellular domain of mouse Klotho were cloned into pEF1 vector (Invitrogen) in the same way except that a FLAG-epitope tag was added to the C terminus.
Cell Culture and Transfection
All cells except PC12 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. PC12 cells were maintained in the same medium with additional 10% horse serum. Stable transformants of 293 cells expressing the full-length transmembrane form of Klotho (293KL) or the extracellular domain of Klotho (293KLΔTM) were isolated after selection with G418 (Invitrogen) for 14 days. Subconfluent 293, 293KL, and 293KLΔTMcells were transfected with the FGFR expression vector plasmids using the Lipofectamine transfection reagent (Invitrogen) according to the manufacturer’s protocol. A CHO cell line that stably expresses the full-length Klotho (CHOKL) is a gift from Kyowa Hakko Kogyo Co. Ltd. (5).
Adenovirus Construction
A cDNA encoding the full-length transmembrane form of mouse Klotho with a C-terminal myc-eptope tag or green fluorescent protein (GFP) was inserted into the multiple cloning site of pShuttle- CMV (Qbiogene). After linearization, the shuttle vector was introduced into an electroporation-competent Escherichia coli BJ5183-AD-1 harboring the adenoviral backbone pAdEasy-1 (Stratagene). The recombinant vector was introduced into the adenovirus packaging cell line QBI-HEK293A (Qbiogene) using FuGENE 6 (Roche Applied Science). The viruses were amplified by several rounds of infection in QBI-HEK293A cells. Subconfluent HeLa cells or PC12 cells were infected with the adenovirus expressing Klotho or GFP (m.o.i. = 3 for HeLa and m.o.i. = 10 for PC12) 36 h before stimulation with FGF23 and then subjected to immunoblot analysis of FGF signaling pathway as described below.
Immunoprecipitation and Immunoblotting
To prepare cell lysate, cells were snap-frozen in liquid nitrogen and lysed in the lysis buffer containing inhibitors for phosphatase and proteinase as described previously (2). The lysate of 293KL or 293KLΔTM cells transfected with expression vectors for FGFRs was incubated with agarose beads conjugated with anti-V5 antibody (Sigma) or anti-FLAG antibody (Sigma) at 4 °C for 3 h. The beads were washed three times with Tris-buffered saline (TBS) containing 1% Triton X-100 (TBST) and three times with TBS. The washed beads were suspended in SDS-sample loading buffer and subjected to SDS-PAGE. The protein transferred to Hybond C Extra membrane (Amersham Biosciences) was incubated with anti- Klotho rat monoclonal antibody KM2119 (19) or anti-V5 antibody (Invitrogen) and then with horseradish peroxidase-linked secondary antibodies (Amersham Biosciences). The signals were detected with SuperSignal West Dura system (Pierce). For detecting Klotho binding to endogenous FGFRs in 293KL cells, cell lysate was immunoprecipitated with anti-FLAG-agarose in the same way as described above and then immunoblotted with antibodies against FGFR1 (Santa Cruz Biotechnology), FGFR2 (Santa Cruz Biotechnology), FGFR3 (Sigma), or KM2119.
Preparation of Conditioned Medium Containing FGF23 (R179Q)
Serum-free conditioned medium was prepared by transfecting 293 cells with the mouse FGF23 (R179Q) expression vector. 293KL cells were stimulated with various doses of the conditioned medium and subjected to immunoblot analysis using anti-phospho-ERK antibody. The FGF23 activity in the conditioned medium was determined by comparing ERK phosphorylation with that induced by recombinant human FGF23 of known concentrations. The conditioned medium with the FGF23 activity equivalent to that of 10 ng/ml recombinant human FGF23 was used for the experiments. The same amount of serum-free conditioned medium from mock-transfected 293 cells was used as a negative control.
Co-precipitation of Endogenous Klotho and FGFRs from Mouse Kidney
Kidney from a 129 mouse (200 mg) was homogenized in 2 ml of homogenizing buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 0.5 mM EDTA) containing protease inhibitors. The homogenate was incubated for 30 min at 4 °C after the addition of Triton X-100 (final 1%) and then centrifuged for 12 min at 18,000 × g two times to remove debris. The supernatant was precleared with 40 µl of protein A-Sepharose (Amersham Biosciences) conjugated with 20 µg of normal rabbit IgG for 2.5 h at 4 °C. The precleared lysate was incubated with 20 µl of protein A-Sepharose conjugated with 16 µg of anti-FGFR1 antibody or normal rabbit IgG for 2.5 h at 4 °C. The beads were processed in the same way as described above for immunoblot analysis using KM2119 and anti-FGFR1 antibody. The lysate of the kidney immunoprecipitated with anti-Klotho antibody KL11-A (Alpha Diagnostic International) was used as a positive control for Klotho. The mouse FGFR1c with a V5-epitope tag expressed in 293 cells was used as a positive control for FGFR1.
FGF23 Pull-down Experiments
Lysate of 293 cells or 293KL cells transfected with the FGFR expression vectors was applied to anti-V5-agarose at 4 °C for 3 h. Serum-free conditioned medium of 293 cells or 293KLΔTM cells was applied to anti-FLAG-agarose at 4 °C for 3 h. The beads were washed four times with TBST and then incubated with conditioned medium of 293 cells transfected with the mouse FGF23 (R179Q) expression vector supplemented with 0.5% Triton X-100 at 4 °C for 3 h. The beads were washed three times with Krebs-Ringer-HEPES buffer containing 1% Triton X-100 and then three times with the same buffer without Triton X-100. The washed beads were suspended in SDS-sample loading buffer and subjected to immunoblot analysis using anti-V5 antibody, KM2119, or anti-FGF23 antibody (R&D Systems).
Immunoblot Analysis of the FGF Signaling Pathway
Subconfluent 293, 293KL, and 293KLΔTM cells grown on 6-well plates were serum-starved over-night and then treated with various concentrations of recombinant human FGF23 (0, 0.1, 1, 10, or 100 ng/ml, Genzyme), acidic or basic FGF (Upstate) for 15 min. Conditioned medium of 293 cells transfected with the expression vector for mouse FGF23 (R179Q) was also used as a source of FGF23 where indicated. The cell lysates were subjected to immunoblot analysis using antiphospho-FRS2-α antibody (Cell Signaling), anti-phospho-p44/42 MAP kinase (ERK1/2) antibody (Cell Signaling), or anti-ERK antibody (Cell Signaling).
RESULTS
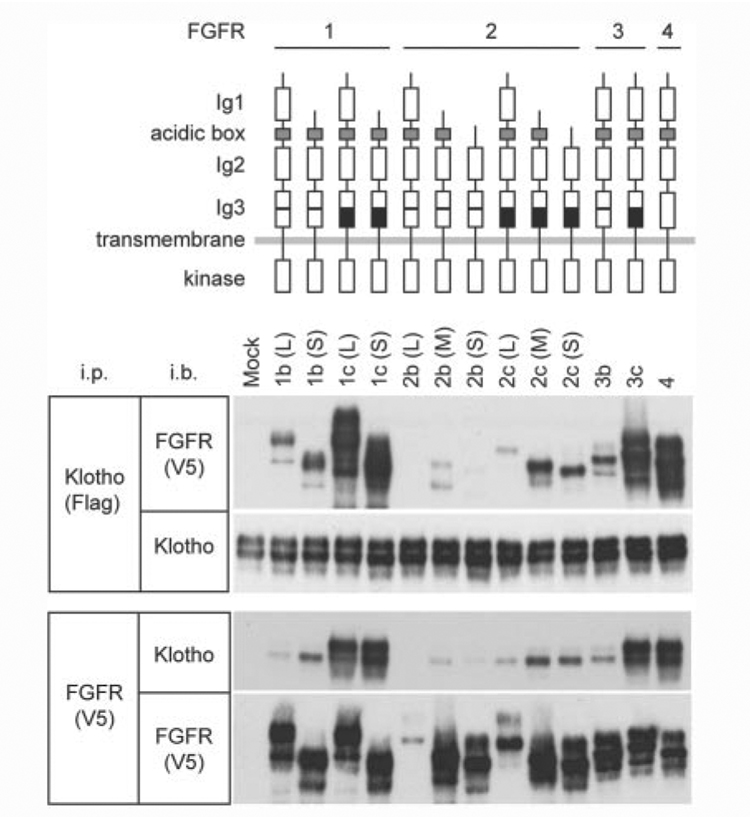
To test the hypothesis that Klotho may be involved in FGF signaling, we investigated whether Klotho could directly bind to FGFRs. Four receptor tyrosine kinases are designated as the high affinity FGFRs (FGFR1–4). Alternative RNA splicing generates multiple FGFR isoforms. The extracellular domain of FGFR1 and FGFR2 is composed of either two or three immunoglobulin-like (Ig-like) ligand-binding domains. Alternative splicing in the third Ig-like domain exists in FGFR1–3, generating “b” and “c” isoforms (20). We trans-fected 293KL cells that stably expressed the full-length transmembrane form of Klotho with various FGFR expression vectors and asked if any of the FGFRs would be immunoprecipitated with Klotho. Klotho bound to almost all FGFR isoforms tested (Fig. 1). However, significant difference in the ability of Klotho to co-precipitate FGFRs was observed between the isoforms: 1) c isoforms were more efficiently co-precipitated with Klotho than b isoforms. The difference between b and c isoforms is located in the C-terminal half of the third Ig-like ligand-binding domain and known to affect binding affinity to FGFs as well (21). 2) FGFR2 was less efficiently co-precipitated with Klotho than FGFR1, FGFR3, and FGFR4. Thus, Klotho may bind to multiple FGFRs with different affinity.
Figure 1. Klotho binds to multiple FGF receptors.
Lysates of 293KL cells transfected with expression vectors for different FGFR isoforms were immunoprecipitated with Klotho using anti-FLAG antibody and probed either with FGFRs using anti-V5 antibody or with Klotho using KM2119 (upper two panels). The lysates were also immunoprecipitated with FGFRs using anti-V5 antibody and probed either with Klotho using KM2119 or with FGFRs using anti-V5 antibody (lower two panels). Antibodies used for immunoprecipitation (i.p.) and immunoblotting (i.b.) were indicated. The possibility that Klotho could bind to FGFR2b(L) was not excluded because of its low expression. Schemes for FGFR isoforms used in this study are shown above the data. The difference between b and c isoforms in FGFR1-3 resides in the C-terminal half of the third immunoglobulin-like (Ig3) domain as indicated in the scheme by white and black boxes, respectively. The difference in genomic structure among FGFR isoforms was summarized in supplemental Fig. 1.
To test whether binding of Klotho to FGFRs may affect interaction between FGFRs and FGF23, the ability of FGFRs to pull down FGF23 was examined in the presence and absence of Klotho. FGF23 was pulled down with FGFR1c, -3c, and -4 only in the presence of Klotho (Fig. 2). We tested the possibility that FGF23 might directly bind to Klotho. However, the extracellular domain of Klotho alone failed to pull down FGF23 under these experimental conditions (Fig. 2). These observations indicate that FGF23 show stronger interaction with the Klotho-FGFR complex than with Klotho or FGFR alone.
Figure 2. FGF23 preferentially binds to the Klotho-FGFR complex.
Agarose beads bound to FGFR, Klotho, or the Klotho-FGFR complex were incubated with conditioned medium containing mouse FGF23(R179Q) and probed with FGF23, FGFR (anti-V5 anti-body), or Klotho by immunoblotting.
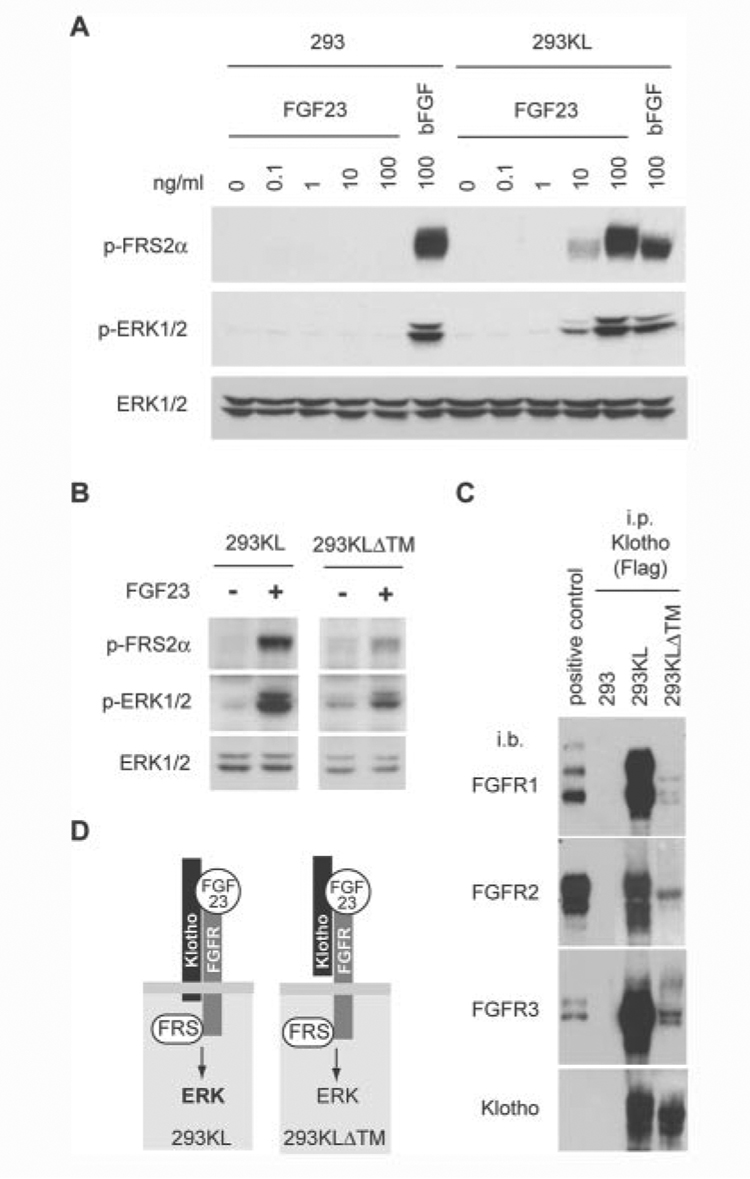
Since Klotho increased binding of FGF23 to FGFRs, it might enhance the ability of FGF23 to activate FGF signaling. To test this possibility, we stimulated 293KL cells or 293 cells with various concentration of recombinant human FGF23 and compared phosphorylation of FGF receptor substrate-2α (FRS2α) and p44/42 MAP kinase (ERK1/2). 3 nM (100 ng/ml) FGF23 failed to induce phosphorylation of FRS2α and ERK1/2 in 293 cells. In contrast, 0.3 nM (10 ng/ml) FGF23 activated FRS2αand ERK1/2 in 293KL cells (Fig. 3A), indicating that Klotho enhanced the cellular sensitivity to FGF23 >10 times without the help of heparin or glycosaminoglycan. The similar results were obtained using two independent 293KL clones (data not shown). In addition, we observed FGF23-induced FRS2α and ERK phosphorylation in 293KLΔTM cells (Fig. 3B), although the levels were reduced relative to those in 293KL cells. Consistent with these findings, endogenous FGFRs in 293KL cells were co-precipitated with the full-length transmembrane Klotho and, to a smaller extent, with the extracellular domain of Klotho (Fig. 3C). We also confirmed that the extracellular domain of Klotho bound to exogenously expressed FGFRs in the same pattern as the full-length Klotho did (supplemental Fig. 2). Based on these observations, we conclude that both the full-length Klotho and the extracellular domain of Klotho function as cofactors necessary for efficient activation of FGF signaling by FGF23 (Fig. 3D).
Figure 3. FGF23 requires Klotho to activate FGF signaling.
A, 293 cells or Klotho-expessing 293 cells (293KL) were stimulated with various concentration of recombinant human FGF23 or basic FGF (100 ng/ml) for 15 min. Activation of FGF signaling was determined by immunoblot analysis using anti-phospho-FRS2a antibody (p-FRS2a), anti-phospho-ERK1/2 antibody (p-ERK1/2), or anti-ERK1/2 antibody (ERK1/2). B, 293KL cells or 293KLDTM cells were stimulated with the conditioned medium containing mouse FGF23 (R179Q) (~10 ng/ml) for 15 min. The same volume of conditionedmedium from mock-transfected 293 cells was used as a negative control (FGF23−). C, detection of endogenous FGFRs bound to Klotho in 293KL cells. Lysates of 293, 293KL, and 293KLDTM cells were immunoprecipitated with Klotho using anti-FLAG antibody and then immunoblotted with antibodies against FGFR1, FGFR2, FGFR3, and Klotho. Lysates of 293 cells transfected with expression vectors for mouse FGFR1c, FGFR2c, and FGFR3c were immunoprecipitated with anti-V5 antibody and used as positive controls for immunoblotting of FGFR1, FGFR2, and FGFR3, respectively. D, a model for interaction between Klotho, FGFR, FGF23, and FGF signaling.
The primary function of FGF23 is to suppress phosphate reabsorption in the renal tubular cells. Indeed, interaction between endogenous Klotho and FGFR was observed in the mouse kidney (supplemental Fig. 3). To test whether the activity of Klotho to enhance FGF23 action might be observed in non-kidney cells as well, we infected PC12 and HeLa cells, which originated from pheo-chromocytoma and cervical carcinoma, respectively, with adenovirus expressing the full-length Klotho and then stimulated with FGF23. In addition, we stimulated stable transformants of ovary-derived CHO cells expressing the full-length Klotho with FGF23. These non-kidney cells acquired the ability to respond to FGF23 when Klotho was expressed (Fig. 4).
Figure 4. Klotho functions as a regulator of FGF23 signaling in various types of cells.
PC12 and HeLa cells were infected with adenovirous expressing the full-length Klotho or GFP as a negative control and then stimulated with mouse FGF23 (R179Q) for 15 min. CHO cells and Klotho-expressing CHO cells (CHOKL) were also stimulated in the same way. Cell lysates were immunoblotted with anti-Klotho antibody (KM2119) to confirm Klotho expression. Activity of FGF signaling was analyzed in the same way as Fig. 3.
Since Klotho binds to multiple FGFRs, it may affect the activity of FGFs other than FGF23. We stimulated 293 or 293KL cells with various doses of acidic and basic FGF and found that Klotho did not enhance their ability to activate FGF signaling but slightly suppressed basic FGF action (Fig. 3A and supplemental Fig. 4).
DISCUSSION
The fact that FGF23 requires Klotho to activate FGF signaling may explain why Klotho−/− mice develop all the phenotypes observed in Fgf23−/− mice. However, Klotho−/− mice show many phenotypes not described in Fgf23−/− mice, including arteriosclerosis, ectopic calcification in extra-renal tissues, skin atrophy, neuronal degeneration, and pulmonary emphysema (1). This may imply that Klotho affects the activity of multiple FGFs through binding to multiple FGFRs, although the effect of Klotho on acidic and basic FGF was small when compared with its robust effect on FGF23 (supplemental Fig. 4). Precise comparison between Klotho−/− and Fgf23−/− phenotypes may provide a clue to understanding of potential effects of Klotho on the other FGFs besides FGF23.
In the absence of Klotho, FGF23 requires exogenous heparin or glycosaminoglycan as a cofactor to stimulate FGF signaling (13, 15, 16), indicating that the help of sugar chains are critical for the biological activity of FGF23. Klotho is a glycoprotein and detected as two bands by immunoblot analysis (Fig. 1), which represent different glycosylation (5). We noticed that the upper band of Klotho was enriched when Klotho was co-precipitated with the high affinity binding partners such as FGFR1c, -3c, and -4 (Fig. 1), suggesting that a sugar chain(s) on the upper band of Klotho may facilitate interaction with these FGFRs. Furthermore, binding of FGFRs to the upper band of Klotho was associated with the ability of the Klotho-FGFR complex to pull down FGF23 (Fig. 2). It is possible that a particular sugar chain(s) on Klotho protein may be involved in the high affinity interaction between FGFRs and FGF23. We also noticed that 293KLΔTM cells predominantly expressed the lower band (supplemental Fig. 2), which might partly explain why 293KLΔTM cells showed weaker response to FGF23 than 293KL cells (Fig. 3B).
The fact that the extracellular domain of Klotho can increase cellular sensitivity to FGF23 (Fig. 3B) has raised the possibility that it may function as a paracrine factor in the kidney. Since Klotho is expressed in the distal convoluted tubules (1) and the extracellular domain of Klotho is shed and secreted (2, 5), it may act on adjacent proximal tubules and work cooperatively with FGF23 to inhibit phosphate reabsorption. It remains to be determined whether extracellular Klotho peptide could function as a paracrine and/or an endocrine factor in the regulation of FGF23 signaling.
It was recently reported that mice defective in βKlotho, a protein that structurally resembles Klotho, showed increased synthesis and excretion of bile acids (22). Interestingly, these phenotypes are identical with those observed in FGFR4 knock-out mice (23, 24). The Klotho gene family may have evolved in the regulation of FGF signaling. Further studies on Klotho are expected to promote better understanding of the complex FGF signaling system and its relation to aging.
Supplementary Material
Footnotes
This work was supported in part by grants from Endowed Scholar Program at the University of Texas Southwestern (to M. K.), Pew Scholars Program in Biomedical Science (to M. K.), Eisai Research Fund (to M. K.), High Impact/High Risk Research Program at The University of Texas Southwestern (to M. K.), The Ellison Medical Foundation (to M. K.), and by National Institutes of Health Grants R01AG19712 (to M. K.), R01AG25326 (to M. K. and K. P. R.), and R01DK065842 (to M. B.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1–Fig. 4.
The abbreviations used are: FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; Ig-like, immunoglobulin-like; TBS, Tris-buffered saline; CHO, Chinese hamster ovary; GFP, green fluorescent protein; m.o.i., multiplicity of infection; ERK, extracellular signal-regulated kinase; MAP, mitogen-activated protein.
REFERENCES
- 1.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima Y. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuin-ness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto M, Clark JD, Pastor JV, Gurnani P, Nandi A, Kurosu H, Miyoshi M, Ogawa Y, Castrillon DH, Rosenblatt KP, Kuro-o M. J. Biol. Chem. 2005;280:38029–38034. doi: 10.1074/jbc.M509039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikushima M, Rakugi H, Ishikawa K, Maekawa Y, Yamamoto K, Ohta J, Chihara Y, Kida I, Ogihara T. Biochem. Biophys. Res. Commun. 2006;339:827–832. doi: 10.1016/j.bbrc.2005.11.094. [DOI] [PubMed] [Google Scholar]
- 5.Imura A, Iwano A, Tohyama O, Tsuji Y, Nozaki K, Hashimoto N, Fujimori T, Nabeshima Y. EBS Lett. 2004;565:143–147. doi: 10.1016/j.febslet.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 6.The ADHR Consortium. Nat. Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- 7.White KE, Carn G, Lorenz-Depiereux B, Benet-Pages A, Strom TM, Econs MJ. Kidney Int. 2001;60:2079–2086. doi: 10.1046/j.1523-1755.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Guo R, Simpson LG, Xiao ZS, Burnham CE, Quarles LD. J. Biol. Chem. 2003;278:37419–37426. doi: 10.1074/jbc.M304544200. [DOI] [PubMed] [Google Scholar]
- 9.Jonsson KB, Zahradnik R, Larsson T, White KE, Sugimoto T, Imanishi Y, Yamamoto T, Hampson G, Koshiyama H, Ljunggren O, Oba K, Yang IM, Miyauchi A, Econs MJ, Lavigne J, Juppner H. N. Engl. J. Med. 2003;348:1656–1663. doi: 10.1056/NEJMoa020881. [DOI] [PubMed] [Google Scholar]
- 10.Yamazaki Y, Okazaki R, Shibata M, Hasegawa Y, Satoh K, Tajima T, Takeuchi Y, Fujita T, Nakahara K, Yamashita T, Fukumoto S. J. Clin. Endocrinol. Metab. 2002;87:4957–4960. doi: 10.1210/jc.2002-021105. [DOI] [PubMed] [Google Scholar]
- 11.Riminucci M, Collins MT, Fedarko NS, Cherman N, Corsi A, White KE, Waguespack S, Gupta A, Hannon T, Econs MJ, Bianco P, Gehron Robey P. J. Clin. Invest. 2003;112:683–692. doi: 10.1172/JCI18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quarles LD. Am. J. Physiol. 2003;285:E1–E9. doi: 10.1152/ajpendo.00016.2003. [DOI] [PubMed] [Google Scholar]
- 13.Baum M, Schiavi S, Dwarakanath V, Quigley R. Kidney Int. 2005;68:1148–1153. doi: 10.1111/j.1523-1755.2005.00506.x. [DOI] [PubMed] [Google Scholar]
- 14.Shimada T, Kakitani M, Yamazaki Y, Hasegawa H, Takeuchi Y, Fujita T, Fukumoto S, Tomizuka K, Yamashita T. J. Clin. Invest. 2004;113:561–568. doi: 10.1172/JCI19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Ibrahimi OA, Goetz R, Zhang F, Davis SI, Garringer HJ, Linhardt RJ, Ornitz DM, Mohammadi M, White KE. Endocrinology. 2005;146:4647–4656. doi: 10.1210/en.2005-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita T, Konishi M, Miyake A, Inui K, Itoh N. J. Biol. Chem. 2002;277:28265–28270. doi: 10.1074/jbc.M202527200. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T, Fujimori T, Nabeshima Y. Endocrinology. 2002;143:683–689. doi: 10.1210/endo.143.2.8657. [DOI] [PubMed] [Google Scholar]
- 18.Shimada T, Muto T, Urakawa I, Yoneya T, Yamazaki Y, Okawa K, Takeuchi Y, Fujita T, Fukumoto S, Yamashita T. Endocrinology. 2002;143:3179–3182. doi: 10.1210/endo.143.8.8795. [DOI] [PubMed] [Google Scholar]
- 19.Kato Y, Arakawa E, Kinoshita S, Shirai A, Furuya A, Yamano K, Nakamura K, Iida A, Anazawa H, Koh N, Iwano A, Imura A, Fujimori T, Kuro-o M, Hanai N, Takeshige K, Nabeshima Y. Biochem. Biophys. Res. Commun. 2000;267:597–602. doi: 10.1006/bbrc.1999.2009. [DOI] [PubMed] [Google Scholar]
- 20.Eswarakumar VP, Lax I, Schlessinger J. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 22.Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y, Nabeshima Y. J. Clin. Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. J. Biol. Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.