Abstract
We report the first chemoenzymatic synthesis of the stable isotope-enriched heparin from a uniformly labeled [13C,15N]N-acetylheparosan (-GlcA(1,4)GlcNAc-) prepared from E. coli K5. Glycosaminoglycan (GAG) precursors and heparin were formed from N-acetylheparosan by the following steps: chemical N-deacetylation and N-sulfonation leading to N-sulfoheparosan (-GlcA(1,4)GlcNS-); enzyme-catalyzed C5-epimerization and 2-O-sulfonation leading to undersulfated heparin (-IdoA2S(1,4)GlcNS-); enzymatic 6-O-sulfonation leading to the heparin backbone (-IdoA2S(1,4)GlcNS6S-); and selective enzymatic 3-O-sulfonation leading to the anticoagulant heparin, containing the GlcNS6S3S residue. Heteronuclear, multidimensional nuclear magnetic resonance spectroscopy was employed to analyze the chemical composition and solution structure of [13C,15N]N-acetylheparosan, precursors, and heparin. Isotopic enrichment was found to provide well-resolved 13C spectra with the high sensitivity required for conformational studies of these biomolecules. Stable isotope-labeled heparin was indistinguishable from heparin derived from animal tissues and is a novel reagent for studying the interaction of heparin with proteins.
Introduction
Heparin and heparan sulfate (HS) participate in many important biological processes, including blood anticoagulation, viral and bacterial infection and entry, angiogenesis, inflammation, cancer, and development.1–3 Heparin/HS carry out their biological functions primarily by their interaction with proteins, in which sulfo and carboxyl groups electrostatically interact or hydrogen-bond with basic amino acids of the target protein.1 In a number of cases, heparin/HS has been demonstrated to bind specifically and with high affinity to proteins, regulating their biological functions.2,3 Structural analysis, relying on either NMR or X-ray crystallography, has been reported for several heparin/HS–protein complexes, revealing the relative importance of certain negatively charged groups in specific positions of the uronic acid (UA) and/or glucosamine residues.4–6 Unfortunately, in most cases there is a lack of general approaches for the identification and positioning of the key groups within heparin/HS required for protein-binding specificities.
Heparin and HS are acidic linear polysaccharides having closely related structures. Both are isolated by extraction from animal tissues and are members of the glycosaminoglycan (GAG) family. These GAGs have an average molecular weight of 10–15 kDa and a polydispersity of 1.05–1.6.7 Heparin and HS are comprised of a repeating disaccharide structure of 1,4-linked hexuronic acid and glucosamine residues. The most common disaccharide unit of heparin is composed of a 2-O-sulfo-α-L-iduronic acid 1,4-linked to 6-O-sulfo-N-sulfo-α-D-glucosamine, -IdoA2S(1,4)GlcNS6S-. HS has a similar but less sulfated structure containing primarily β-D-glucuronic acid (GlcA) and N-acetyl-α-D-glucosamine (GlcNAc).8 Heparin, a clinical anticoagulant, binds to the serine protease inhibitor antithrombin III (ATIII), causing it to undergo a conformational change resulting in ATIII inhibition of thrombin and other coagulation cascade proteases.2,8 ATIII binds to a specific pentasaccharide sequence, contained within the heparin/HS, having the structure GlcNAc/NS6S(1,4)GlcA(1,4)GlcNS3S-6S(1,4)IdoA2S(1,4)GlcNS6S.9
Methods for analyzing heparin/HS sequence and conformation are important for understanding biological activity and structure–activity relationships.8 Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) have become the primary tools for establishing the chemical structure of heparin/HS. NMR has also provided insight into the conformational and dynamic solution properties of short-chain heparin/HS oligosaccharides, prepared from animal sources or by chemical synthesis.9–11 Mulloy and co-workers12 utilized a joint NMR and molecular-modeling approach, in which models at atomic resolution were generated for heparin and de-N-sulfonated, re-N-acetylated heparin. While a number of high-quality crystal structures have been reported for heparin/HS oligosaccharides in complex with various proteins, no high-resolution crystal structures have been reported for unbound heparin/HS polysaccharides or oligosaccharides, due to difficulties in both purification and crystallization. As a result, much of our understanding of heparin/HS-derived oligosaccharide structure is derived from crystallographic models of these carbohydrates in complex with heparin-binding proteins.6 These structures often reveal carbohydrates in distorted conformations and offer little information on the flexibility of the carbohydrate and the number of conformational forms. NMR is ideally suited to determine the conformational and dynamic features of heparin/HS in solution. Unfortunately, the extensive proton spectral overlap, the extended and flexible conformation, and the inherent microheterogeneity (sequence variability and dispersity) of heparin/HS, isolated from animal tissues, limit conformational analysis by NMR to short oligosaccharides and make analysis in complex with proteins nearly intractable.9–11
A principal way for reducing spectral overlap and resolving proton resonances is the correlation of chemical shifts of heteronuclei in additional spectral dimensions.13 Heteronuclear NMR is highly effective in resolving heparin/HS spectra due to the surprisingly high level of 13C dispersion. However, nearly all heteronuclear NMR experiments that probe higher ordered structures require isotopic enrichment for acceptable sensitivity. While isotopic enrichment of animal-derived polysaccharides is theoretically possible by feeding stable isotopic precursors, the cost of such an approach is prohibitive. Chemoenzymatic synthesis has recently been introduced as an alternative method to prepare various heparin/HS derivatives for biological and clinical evaluation.14,15 The biosynthesis of heparin/HS GAGs has been extensively studied.16,17 A repeating 1,4-glycosidically linked copolymer of β-D-GlcA and α-D-GlcNAc, called N-acetylheparosan, is synthesized through the stepwise addition of uridine diphosphate-activated sugars. Bacteria, including Escherichia coli and Pasteurella multocida, also biosynthesize N-acetylheparosan.18,19 During heparin/HS biosynthesis in animal cells, the N-acetylheparosan linear homocopolymer is modified sequentially through the action of N-deacetylase/N-sulfotransferase, C5-epimerase, and 2-, 6-, and 3-O-sulfotransferases (OSTs). Complete or nearly complete modification of a chain results in heparin, while partial modification results in HS.16,17 These biosynthetic enzymes have been cloned, expressed, and used for controlled synthesis of heparin/HS.15,20
N-Acetylheparosan and its derivatives have been the subject of recent interest in an effort to prepare heparin-like therapeutics from non-animal sources.14,21 Enzymatic sulfonation of these derivatives offers products that closely resemble the heparins isolated from animal tissues, suggesting a utility in the development of new pharmaceuticals.15,20,22 Unfortunately, despite the decreased heterogeneity of these heparin derivatives, structural studies using NMR are still plagued by the low natural abundance of heteronuclei. Heteronuclear NMR has been extremely beneficial in solving the solution structures of proteins and nucleic acids.23,24 Studies on 13C-labeled chondroitin sulfate oligosaccharides25 and stable isotope-labeled hyaluronan and hyaluronan oligosaccharides26 suggested to us that a similar approach might be possible for heparin. As a demonstration, the solution structures of N-acetylheparosan, heparin, and intermediate GAGs have been analyzed here using heteronuclear NMR and molecular dynamics (MD) with time-averaged restraints (Tar-MD).
Experimental Section
Preparation of [13C,15N]N-Acetylheparosan
13C,15N-isotopically labeled N-acetylated heparosan (-GlcA(1,4)GlcNAc-) was prepared by fermentation of E. coli K5 strain in minimal media containing 13C-D-glucose and 15NH4Cl.
Expression of Recombinant HS Biosynthetic Enzymes and Heparin Lyases
The catalytic domains of human C5-epimerase, hamster 2-OST, hamster 6-OST-1, mouse 6-SOT-3, and mouse 3-OST-1 were recombinately expressed in E. coli and purified.20 Heparin lyases I, II, and III were cloned from the genomic DNA of Flavobacterium heparinum. The expression of the recombinant heparin lyases was also carried out for E. coli.28
Preparation of Heparin and Its Precursors
13C,15N-Labeled N-acetylheparosan (5 mg) was N-deacetylated using NaOH, neutralized with HCl, and N-sulfonated with (CH3)3N · SO3.29 Three modification steps were used to convert 13C,15N-labeled N-sulfoheparosan to anticoagulant heparin: C5-epimerization/2-O-sulfonation, 6-O-sulfonation, and 3-O-sulfonation. Each step was performed in buffer in the presence of a PAPs regeneration system.15
NMR
1H and 13C NMR, 2D 1H–13C HMQC, and constant-time HSQC and 3D HCCH-COSY, HCCH-TOCSY, and (H)CCH-TOCSY spectra were recorded on 13C,15N-labeled N-acetylheparosan, N-sulfoheparosan, undersulfated heparin, and heparin samples in D2O. The proton and carbon chemical shifts were calibrated against 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS). A series of 3D 13C-separated NOE spectra were also acquired for each sample at mixing times of 20, 40, and 100 ms.
Disaccharide Composition Analysis of Anticoagulant Heparin
Heparin and anticoagulant heparin (20 μg, respectively) were incubated in 50 mM sodium phosphate buffer, pH 7.0, with heparin lyase I, II, and III mixture (100 m-units) at 37 °C for 10 h. Completely digested products were heated in a boiling water bath for 10 min to halt the reaction. The denatured protein was removed by centrifugation at 12000g for 10 min, and disaccharides were analyzed by HPLC-ESI-MS.30
APTT Activity and ATIII-Binding
13C,15N-Labeled heparin and anticoagulant heparin (100–1500 ng) were subjected to APTT assay, and anticoagulant activity was calculated from a standard curve prepared using the same concentrations of pharmaceutical heparin.31 The 3-O-[35S] sulfated, anticoagulant heparin was incubated with ATIII and captured using ConA-Sepharose. Binding of 35S-labeled anticoagulant heparin to ATIII was determined by scintillation counting.
Molecular Dynamics Simulations
For all the MD simulations, with or without time-averaged restraints (tar), a heptasaccharide (residues labeled 1–7 with 1 being a nonreducing end residue), prepared from the repeating disaccharide of each GAG structure, was employed, starting with a GlcNAc residue at the nonreducing end. Two independent simulations were performed for undersulfated heparin and heparin, one with the IdoA in the 1C4 conformation and one with the IdoA in the 2SO skew boat geometry.
Tar-MD Simulations in Explicit Water with NOE-Based Restraints
Tar-MD simulations were performed with AMBER32 to 6.0 (parm94),33 implemented with GLYCAM 06 parameters34 and with parameters computed for the sulfo and sulfamo groups,35 to accurately simulate the conformational behavior of the sugar moiety and the sulfo and sulfamo groups, respectively. The tar-MD used 1805, 1977, 1993, and 2135 water molecules for N-acetylheparosan, N-sulfoheparosan, undersulfated heparin, and heparin, respectively. In all cases, the starting geometries were generated from the available data12 deposited in the Protein Data Bank (pdb code 1hpn) and modified accordingly. Final trajectories were run using an exponential decay constant of 400 ps and a simulation length of 4 ns. Experimental cross-relaxation rates were derived from the NOESY-HSQC experiments acquired with 20, 40, and 100 ms mixing times. The experimental distances and experimental NOEs were compared to those computed for the tar-derived ensembles and for single conformers of the model heptasaccharides using the isolated spin-pair approximation and a full-matrix relaxation approach with software developed in-house. An isotropic reorientation model was inadequate and unable to simultaneously accommodate the experimental intra- and inter-residue NOEs for the different residues. Thus, a symmetric top model with correlation time values τ⊥= 8 ns and τ|| = 0.16 ns was employed for heparin and the related molecules, as described by Mulloy and co-workers.12
Molecular Modeling without Restraints in Explicit Water
The solute molecule was first immersed in a TIP3P36 bath of water molecules with the LEAP module. The simulation was performed using periodic boundary conditions and the particle-mesh Ewald approach37 to introduce long-range electrostatic effects. The SHAKE algorithm for hydrogen atoms, which allows using a 2 fs time step, was also employed. Finally, a 9 Å cutoff was applied to Lennard-Jones interactions (see Supporting Information for details).
Results and Discussion
Preparation of Uniformly Labeled [13C,15N]N-Acetylheparosan
[13C,15N]N-Acetylheparosan was afforded by fermentation of E. coli K5 strain on medium containing uniformly labeled 13C-D-glucose and 15NH4Cl (Figure 1A). The average molecular weight of the [13C,15N]N-acetylheparosan was ~75 kDa,15 and its isotopic purity was 94% based on disaccharide analysis using high-performance liquid chromatography (HPLC) and detection with electrospray ionization mass spectrometry (ESI-MS) (see Supporting Information).
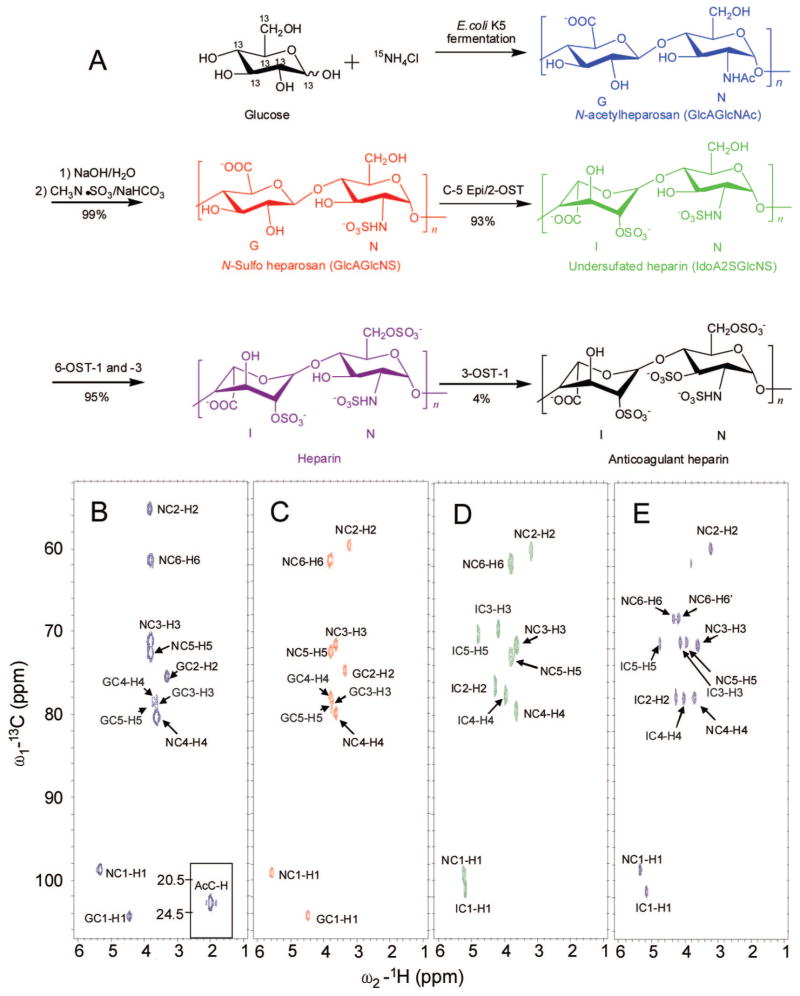
Figure 1.
(A) Scheme of stepwise chemoenzymatic synthesis of 13C,15N-labeled anticoagulant heparin. The major saccharide units are shown (with the exception of anticoagulant heparin which shows a minor fully modified sequence). The GlcN residues are labeled N and the GlcA/IdoA residues are labeled G/I. HMQC spectra of (B) 13C,15N-labeled N-aceylheparosan (blue), (C) 13C,15N-labeled N-sulfoheparosan (red), (D) 13C,15N-labeled undersulfated heparin (green), and (E) 13C,15N-labeled heparin (purple).
Preparation of Uniformly Labeled Heparin and Its Precursors
13C,15N-Labeled heparin and its precursors were prepared by a stepwise chemoenzymatic synthesis in milligram amounts from [13C,15N]N-acetylheparosan. The initial chemical N-deacetylation/N-sulfonation steps result in a partial depolymerization N-acetylheparosan, reducing the average chain length of all of the precursors to ~20 disaccharide units,15 and ultimately giving a heparin molecular weight of ~12 kDa. The percent conversion of each step (Figure 1A) was calculated on the basis of disaccharide analysis (see Supporting Information).15,20
Treatment with 2-OST and C5-Epi afforded an undersulfated heparin that was treated with 6-OST to afford heparin. Finally, treatment with 3-OST-1 results in an anticoagulant heparin. The presence of 13C,15N-label permitted the unambiguous assignment of all proton and carbon resonances. It is notable that, while excess enzyme and PAPS cofactor were used in each step, none of the enzymatic modifications were complete. The final products, both heparin and anticoagulant heparin, however, resemble the pharmaceutical heparin products extracted from animal tissues38 in disaccharide composition (see Supporting Information), with 86–89% of their structure consisting of the trisulfated, IdoA2S(1,4)GlcNS6S, disaccharide repeating unit.
Chemical Structure Analysis of Uniformly Labeled Heparin and Its Precursors
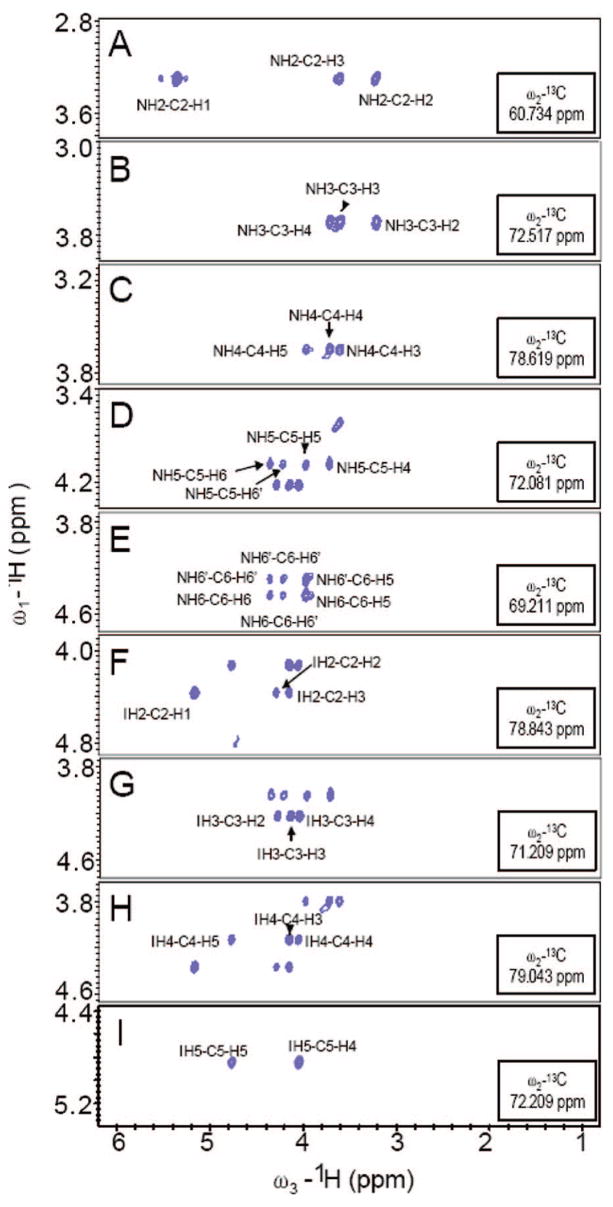
1D, 2D, and 3D NMR were applied to confirm the structures of N-acetylheparosan, N-sulfoheparosan, undersulfated heparin, and heparin with significant improvement in resolution. 1H–13C HMQC, 1H–13C constant-time (CT)-HSQC, 3D HCCH-COSY, HCCH-TOCSY, (H)CCH-TOCSY, and 13C-separated NOESY spectra were applied to unambiguously assign the spectra (Table 1, the GlcN signals are designated N1–N6 and the GlcA/IdoA signals are designated G/I1–G/I6). NMR assignments for N-acetylheparosan and heparin, found in the literature,12,39 were comparable to those presented in Table 1. Examples of 1H–13C HMQC spectra of N-acetylheparosan, N-sulfoheparosan, undersulfated heparin, and heparin are shown in Figure 1B–E (see Supporting Information). The HMQC spectra show both highly degenerate proton and well-dispersed carbon resonances. A high degree of chemical homogeneity is evident and the differences in the chemical structures of these GAGs are clearly reflected in these spectra. Chemical differences at the chain termini were not observable since they comprise low relative concentrations (<5%) as a result of the high chain length. The 3D-HCCH-COSY (Figure 2) spectra were instrumental in the assignment of severely overlapped proton signals that could not be resolved in homonuclear spectra. Even in cases where overlap in both the 1H and 13C dimensions was observed, such as for N3, N4, and N5 of N-acetylheparosan and N-sulfoheparosan, constant-time encoding of 13C chemical shifts was effective at resolving and aiding in the assignment of cross-peaks. The chemical shifts of 15N and corresponding 1H were assigned on the basis of 1H–15N HSQC (Supporting Information). The 1H and the corresponding 15N signals were observed at 8.26 and 123.25 ppm, and they shifted upfield to 5.92 and 92.5 ppm, respectively, after N-deacetylation and N-sulfonation.
Table 1.
Chemical shifts (δ, ppm) for the Major N and G/I Residues Present in N-Acetylheparosan, N-Sulfoheparosan, Undersulfated Heparin, and Heparin (see Figure 1A for Structures)
|
δ (ppm) |
|||||
|---|---|---|---|---|---|
| N-acetylheparosan | N-sulfoheparosan | undersulfated heparin | heparin | ||
| 1H NMR | |||||
| UA (G/I) | H1 | 4.43 | 4.47 | 5.19 | 5.16 |
| H2 | 3.31 | 3.35 | 4.29 | 4.28 | |
| H3 | 3.63 | 3.73 | 4.19 | 4.14 | |
| H4 | 3.68 | 3.80 | 3.97 | 4.05 | |
| H5 | 3.74 | 3.77 | 4.80 | 4.76 | |
| GlcN (N) | H1 | 5.31 | 5.56 | 5.25 | 5.35 |
| H2 | 3.78 | 3.22 | 3.19 | 3.22 | |
| H3 | 3.78 | 3.63 | 3.63 | 3.60 | |
| H4 | 3.62 | 3.63 | 3.63 | 3.72 | |
| H5 | 3.78 | 3.78 | 3.80 | 3.96 | |
| H6 | 3.81 | 3.81 | 3.81 | 4.35 | |
| H6′ | 3.78 | 3.76 | 3.80 | 4.20 | |
| CH3 | 1.97 | - | - | - | |
| 13C NMR | |||||
| UA (G/I) | C1 | 105.5 | 105.3 | 102.1 | 102.3 |
| C2 | 76.6 | 75.7 | 77.4 | 78.8 | |
| C3 | 79.2 | 79.5 | 70.6 | 71.2 | |
| C4 | 79.1 | 78.9 | 78.7 | 79.0 | |
| C5 | 76.5 | 79.3 | 71.2 | 72.2 | |
| C6 | 178.2 | 178.3 | 179.2 | 179.1 | |
| GlcN (N) | C1 | 99.8 | 100.1 | 100.3 | 99.6 |
| C2 | 56.3 | 60.6 | 61.2 | 60.7 | |
| C3 | 72.2 | 72.6 | 71.7 | 72.5 | |
| C4 | 81.4 | 80.8 | 80.4 | 78.6 | |
| C5 | 73.7 | 73.5 | 73.9 | 72.1 | |
| C6 | 61.4 | 62.4 | 62.7 | 69.2 | |
| CH3 | 23.9 | - | - | - | |
| CO | 176.6 | - | - | - | |
Figure 2.
Strip plots from the 3D-HCCH-COSY spectrum of 13C,15N-labeled heparin. Panels A–E illustrate a sequential walk from C2 of GlcN to C6 of GlcN; panels F–I under C2 of IdoA to C5 of IdoA.
The percent conversions of 97% (99%) for N-deacetylation and N-sulfonation, 87% (93%) for C5-epimerization and 2-O-sulfonation, and 90% (95%) for 6-O-sulfonation, obtained from NMR and (disaccharide analysis, Figure 1A), were comparable to those calculated from these spectra. The chemoenzymatic process used in GAG synthesis results in a polydisperse product containing minor structural heterogeneity as a result of incomplete modification. Thus, the assignments obtained (Table 1) correspond to the major disaccharide-repeating unit present in each GAG. While saccharide residues at the reducing and nonreducing ends of the GAG chains can also contribute to heterogeneity, NMR did not readily observe these end groups, as they corresponded to <5% of the saccharidic units. The percent conversion of heparin to anticoagulant heparin, resulting from 3-O-sulfonation, was too low to be unambiguously confirmed and/or quantified by NMR or by disaccharide analysis.
Heparin structure varies between species and organs; for example, porcine intestinal and bovine lung heparins (both pharmaceutical heparins) have distinctly different structures.1,38 On treatment with heparin lyases, individual chains can afford as many as many as eight different disaccharides and several resistant oligosaccharides to as few as one repeating disaccharide, -IdoA2S(1,4)GlcNS6S-. This microheterogeneity of animal-derived heparin results from variability of the activity, specificity, and accessibility of enzymes in the Golgi at the time of biosynthesis. Heparan sulfate, a closely related polysaccharide, has an even more complex structure than heparin, and it is often an impurity in pharmaceutical heparin isolated from animal tissues. Chemoenzymatically synthesized heparin contains no heparan sulfate impurity and no residual linkage region impurity, and since it is lacking N-acetyl-substituted glucosamine residues it shows a simpler disaccharide composition than pharmaceutical heparin.
Uniform 13C/15N isotopic labeling facilitated the NMR-based structural analysis of the GAGs. The well-dispersed 13C-spectra enabled sensitive application of a wide array of multidimensional heteronuclear experiments by overcoming limitations resulting from severe overlap of proton resonances that have historically hindered the interpretation of GAG NMR data. One-bond 1H–15N couplings in N-acetylheparosan were sensitively detected, but rapid solvent exchange in N-sulfoheparosan decreased their value in the structural assignment of N-sulfonated GAGS. In the future we anticipate that 15N labeling will be important in the NMR studies of these GAGs due to the sensitivity of 15N chemical shift to the local environment. 15N dispersion can be instrumental in overcoming spectral degeneracies by combining 15N- and 13C-separated dimensions within an NMR experiment. This dual heteronuclear labeling strategy should facilitate sequence-specific assignment and provide structural data at atomic resolution for heparin oligosaccharides in the future, particularly when used for heparin–protein complexes.
Characterization of Anticoagulant Heparin
In the transformation of heparin to anticoagulant heparin, the small number of sites undergoing 3-O-sulfonation afforded no detectable change by NMR. To determine percent conversion,13C,15N heparin was treated with 3-OST-1 and [35S]PAPS, affording 3-O-[35S]sulfo-13C,15N anticoagulant heparin. Measurement of [35S] showed that 1 μg (83 pmol) of heparin incorporated 115 pmol of [35S]sulfo group, representing ~1.5 3-O-[35S] sulfo groups/chain. This corresponds to the sulfonation of ~7.5% of heparin’s GlcN-3-hydroxyl groups.
The ATIII binding activity of this anticoagulant heparin was also determined.20 After 3-OST-1 modification, scintillation counting demonstrated that only 35.2% of the 35S-labeled anticoagulant heparin chains bound to ATIII, confirming that the 3-OST-1 modified heparin binds to ATIII.
Anticoagulant activity was next assessed using activated thromboplastin time (APTT).40 The activity of chemoenzymatically synthesized heparin was 20 ± 6 U/mg, while chemoenzymatically anticoagulant heparin was 180 ± 15 U/mg, comparable to the 170 U/mg displayed by a standard pharmaceutical heparin. Previously, we had reported anti-Xa and anti-IIa activities comparable to those of pharmaceutical heparin for heparin chemoenzymatically synthesized without isotopic labeling.15
While chemoenzymatically synthesized heparin showed only four disaccharide peaks in HPLC-ESI-MS (see Supporting Information), anticoagulant heparin showed additional (4%) heparin lyase-resistant peaks as a result of the incorporation of 3-O-sulfo groups. 35S incorporation studies confirmed that ~7.5% of the GlcN residues were 3-O-sulfonated and that approximately one-third of these chains bind with high affinity to ATIII, closely resembling the percentage of chains in pharmaceutical heparin with high affinity for ATIII.38 APTT assay showed the activity of anticoagulant heparin to be comparable to that of standard pharmaceutical heparin.
Conformational Analysis of Heparin and Its Precursors
From the fully assigned spectra (Table 1) and quantitative NOEs (Table 2), the conformation of the starting N-acetylheparosan, N-sulfoheparosan, undersulfated heparin, and heparin were examined with experimentally restrained MD simulations using a protocol41 similar to that recently applied by our group on a dermatan sulfate oligosaccharide.42 Existing models of GAGs and related glycans were used to evaluate NOE cross-peak assignments and degeneracy.43 Once the assignment process was completed, the intensities of the NOE cross-peaks at the various mixing times were normalized to the corresponding diagonal peaks. The interproton distances were deduced from a full-matrix relaxation approach on model chair and skew-boat geometries.44 Spin-diffusion was essentially absent at 20 ms NOE mixing time and gave a very minor contribution at 40 ms NOE mixing time. A maximum error of 10%, estimated from spectral signal-to-noise, resulted from noncorrected spin-diffusion contributions and resonance overlap, problems largely eliminated by our 3D 13C-separated approach. The observed data correspond to an average for all the glycosidic linkages of the GAG and should show identical ensemble-averaged behavior on the long time scale required for measuring NOE values.
Table 2.
Experimental Key Proton-Pair Distances (Å) As Deduced from the NOE-Based 3D NOESY-HSQC Experiments Employing the Isolated Spin-Pair Approximation (As Described in the Experimental Section) from Three Different NOESY Experiments, at 20, 40 and 100 ms Mixing Times (Estimated Experimental Error is 10%)
| H1 GlcA–H4 GlcNAc | H1 GlcA–H3 GlcNAc | H1 GlcA–H6 GlcNAc | H1 GlcA–H6 GlcNAc | H1 GlcNAc–H3 GlcA | H1 GlcNAc–H4 GlcA | |
|---|---|---|---|---|---|---|
| N-acetylheparosan | 2.5 | 4.5 | 2.7a | 2.7a | 2.7 | 3.6 |
| N-sulfoheparosan | 2.7 | 4.5 | 2.5a | 2.5a | 2.7 | 3.8 |
| H1 IdoA–H4 GlcNS | H1 IdoA–H3 GlcNS | H1 IdoA–H6 GlcNS | H1 IdoA–H6 GlcNS | H1 GlcNS–H3 IdoA | H1 GlcNS–H4 IdoA | |
|
| ||||||
| undersulfated heparin | 2.5 | 4.5 | 2.7a | 2.7a | 2.5 | 2.7 |
| heparin | 2.4 | 4.5 | 2.6a | 3.1a | 2.8 | 2.8 |
No stereospecific assignment was performed, and the restraint was included in the tar-MD simulations to the corresponding C-6 carbon atom (adding 0.8 Å).
Tar-MD simulations were employed to generate 3D models of the different GAGs. For comparison purposes, the same simulations were also performed without time-averaged restraints (7–9 ns of simulation time), providing similar results. Tar-MD45–48 simulations were performed for heparin and its precursors to obtain a conformer ensemble having distances comparable to those estimated by NMR. The NOE-derived distances were included as time averaged distance constraints in two different trajectories using either 〈r−6〉−1/6 or 〈r−3〉−1/3 restraints to cope with different internal motion timescales. In all cases, tar-MD trajectories were collected in the presence of explicit water, and unrestrained MD simulations were also computed for comparison purposes.
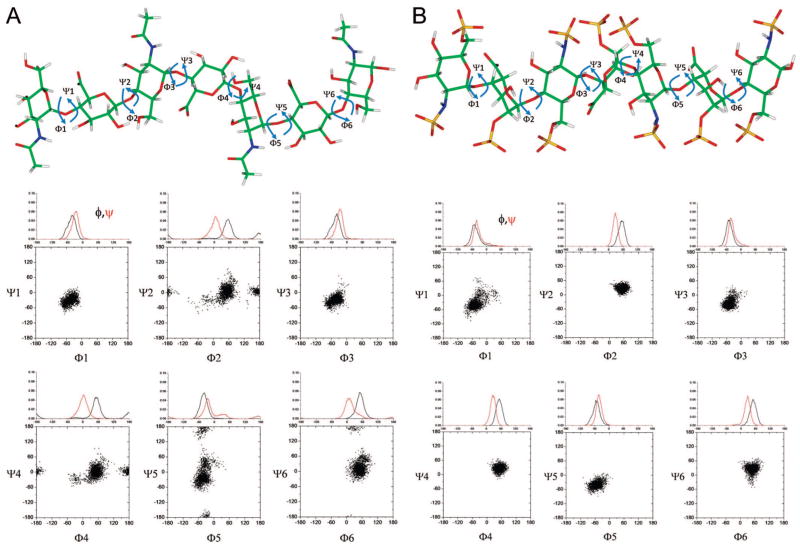
Results from the tar-MD simulations for N-acetylheparosan (Table 3 and Figure 3A) show close agreement, in numerical terms, between the distances found in the refined models and experimental data using a 4 ns tar-MD simulation with explicit water. The φ/ψ distribution maps indicate that the glycosidic linkages cover a similar area of the allowable conformational space, independent of their position in the sequence (Figure 3A). In terms of the glycosidic angle φ, only exoanomeric orientations were adopted, demonstrating the goodness of the employed force field. Only a very minor proportion (<2%) of non-exoanomeric conformers were predicted for the linkage between residues 2 and 3 of the heptasaccharide sequence undergoing MD simulation. Both GlcNAc(1,4)GlcA and GlcA(1,4)GlcNAc linkages show conformers with a negative-ψ angle highly populated with only minor incursions for the anti-ψ and positive-ψ values. Analogous φ/ψ plots are also provided heparin (Table 4 and Figure 3B) and for N-sulfoheparosan, and undersulfated heparin (see Supporting Information). The ensemble shows that the conformational space accessible for each glycosidic linkage covers a well-defined area of the total available φ/Ψ energy surface. Flexibility is apparent in N-acetylheparosan, which cannot be defined by a single, rigid chain model, with a behavior indeed similar to that reported for hyaluronic acid (HA), a similar structure having a -4)GlcA(1,3)GlcNAc(1- repeating disaccharide unit, with almost four sugar rings per helix turn.49 The reported standard deviations of the glycosidic linkages in this polymer are ~13°, while in our tar-MD approach for N-acetylheparosan, the estimated maximum deviations for φ and ψ angles from the major value are ~35°.
Table 3.
Results of the Tar-MD Simulations (〈r−6〉−1/6/〈r−3〉−1/3 Averaging) for the Different Glycosidic Linkages of N-Acetylheparosana
| H1 GlcA–H4 GlcNAc | H1 GlcA–H3 GlcNAc | H1 GlcA–H6a GlcNAc | H1 GlcA–H6b GlcNAc | H1 GlcNAc–H4 GlcA | H1 GlcNAc–H3 GlcA | |
|---|---|---|---|---|---|---|
| 1→2 | - | - | - | - | 2.5/2.5 | 3.3/3.4 |
| 2→3 | 2.5/2.6 | 3.6/4.4 | 2.6/2.9 | 2.6/2.9 | - | - |
| 3→4 | - | - | - | - | 2.5/2.5 | 3.4/3.4 |
| 4→5 | 2.4/2.6 | 4.4/4.4 | 2.7/3.0 | 2.6/2.9 | - | - |
| 5→6 | - | - | - | - | 2.5/2.5 | 3.3/3.3 |
| 6→7 | 2.6/2.5 | 3.0/4.2 | 2.7/2.7 | 2.7/2.6 | - | - |
| MD-average | 2.5/2.6 | 3.5/4.3 | 2.6/2.9 | 2.6/2.8 | 2.5/2.5 | 3.3/3.4 |
| experimental | 2.5 | 4.5 | 2.7b | 2.7b | 2.7 | 3.6 |
The experimental key proton-pair distances (Å) as deduced from the NOE-based 3D NOESY-HSQC experiments were implemented as structural restraints with a 10% margin, using a flat well potential.
No stereospecific assignment was performed and the restraint was set to the corresponding C-6 carbon atom (adding 0.8 Å).
Figure 3.
φ/ψ distribution for (A) N-acetylheparosan and (B) heparin. In the tar-MD simulation a heptasaccharide (saccharide residues 1–7) is used. Each panel shows the φ/ψ plot between adjacent saccharide residues.
Table 4.
Results of the Tar-MD Simulations (〈r−6〉−1/6/〈r−3〉−1/3 Averaging) for the Different Glycosidic Linkages of Heparina
| H1 IdoA– H4 GlcNS | H1 IdoA– H3 GlcNS | H1 IdoA– H6 GlcNS | H1 IdoA– H6 GlcNS | H1 GlcNS– H4 IdoA | H1 GlcNS– H3 IdoA | |
|---|---|---|---|---|---|---|
| 1→2 | - | - | - | - | 2.6/2.6 | 2.6/2.6 |
| 2→3 | 2.6/2.6 | 4.5/4.5 | 2.3/2.2 | 3.7/3.7 | - | - |
| 3→4 | - | - | - | - | 2.6/2.6 | 2.5/2.6 |
| 4→5 | 2.6/2.6 | 4.5/4.5 | 2.3/2.2 | 2.9/3.7 | - | - |
| 5→6 | - | - | - | - | 2.6/2.7 | 2.5/2.5 |
| 6→7 | 2.4/2.6 | 4.4/4.4 | 2.3/2.3 | 3.7/3.8 | - | - |
| MD-average | 2.5/2.6 | 4.4/4.5 | 2.3/2.2 | 3.2/3.7 | 2.6/2.6 | 2.5/2.6 |
| experimental | 2.4 | 4.5 | 2.6b | 3.2b | 2.8 | 2.8 |
The experimental key proton-pair distances (Å) as deduced from the NOE-based 3D NOESY-HSQC experiments were implemented as tar-constraints with a 10% margin.
No stereospecific assignment was performed and the restraint was set to the corresponding C-6 carbon atom (adding 0.8 Å).
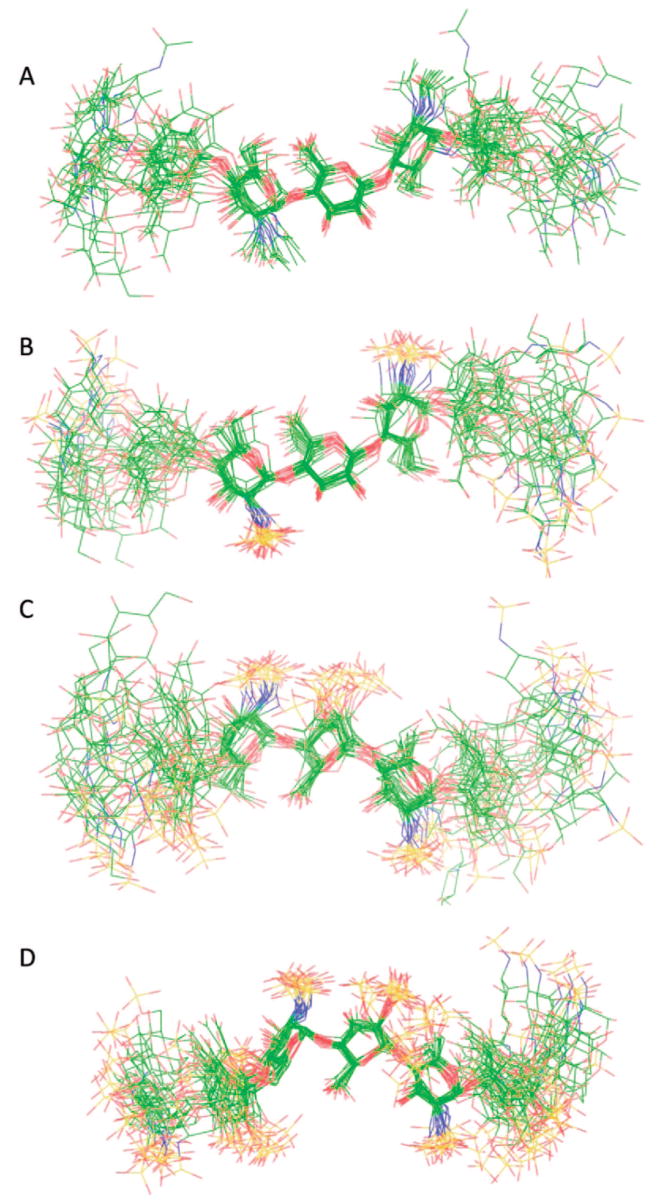
MD simulations were similarly performed on all GAG precursors and heparin. A superimposition of 20 conformers, randomly collected from the tar-MD simulations, is shown in Figure 4. After discarding data from the first nanosecond, the root-mean-square deviation (rmsd) of the skeleton atoms of all the sampled structures was 2.1 Å, compared to the averaged structure. The unrestrained simulations showed a very similar behavior. The simulation began with all seven saccharide residues 1–7 of the heptasaccharide in the 4C1 form. While the NMR data were acquired on a polysaccharide of approximately 20 disaccharide units, our simulations were performed on heptasaccharide models of the polysaccharides. Therefore, special attention was paid to the motion of the central residues, as a paradigm for the motion of the disaccharide entities within the intact polysaccharide. At the end of the simulation no interconversion into other ring forms was observed for either GlcNAc or GlcA residues. In contrast, in the case of the IdoA residues, interconversion between 1C4 and 2SO forms was observed in both the restrained and unrestrained simulations. These results are consistent with observations that GlcNAc and GlcNS residues were solely in the 4C1 conformation based on 3JH–H values.5 Additionally, the pyranose form of GlcA also has been shown to prefer the more stable 4C1 form.50 Tar-MD simulations were also used here to examine the conformation of N-sulfoheparosan. The ring geometries and the φ/ψ values of N-sulfoheparosan in the generated structural models were nearly identical to those of N-acetylheparosan resulting, in a similar local and global conformation (Figure 4B, also see Supporting Information). The length (N–N distance between the GlcN residues at positions 1 and 13) and shape of the two chains were similar when oligosaccharides having 13 residues were modeled from the more populated values of the tar-MD Φ/Ψ distributions of the central residues for each structure (Figure 5). The lengths of heparosan and N-sulfoheparosan were 57.4 and 58.4 Å, differing by only 1.0 Å. The pairwise rmsd between the 13-mer N-acetylheparosan and N-sulfoheparosan chains was 4.9 Å, and the pairwise rmsd for the central trisaccharide was 1.0 Å. Thus, it appears that N-sulfonation does not modify the conformation of this GAG (Figure 5A). Neither anti-conformer nor non-exo conformer was found in the final ensemble of N-sulfoheparosan, and the models showed even less conformational variation (rmsd 1.9 Å) than that of N-acetylheparosan. The observed increase in the ensemble-averaged H1GlcA–H4GlcNS distance results from an increase in positive ψ-conformers for this linkage, as can be observed in the corresponding curves in Figure 3A for heparosan (5 ± 65°) and those in Figure 3B for N-sulfoheparosan (25 ± 60°). Epimerization of the GlcA C5 position and 2-O-sulfonation affords undersulfated heparin. The structural impact of these modifications was explored through the tar-MD simulations on undersulfated heparin (Figure 4C, also see Supporting Information). In solution, IdoA2S pyranose exhibits both the 2SO and 1C4 forms in equilibrium.50,51 As a means of evaluating bias in the resulting structures, dual sets of simulations were initiated with IdoA2S in the 2SO and half in the 1C4 conformer. Both sets of simulations converged within 4 ns, with an rmsd value of 2.4 Å. The estimated NOE-based H2IdoA2S–H5IdoA2S distance is 3.8Å(± 10%). Since the idealize dH2IdoA2S–H5IdoA2S distances for the 1C4 and 2SO conformers are 4.0 and 2.4 Å, respectively, this value is consistent with a large, >85% population of the 1C4 conformer of IdoA2S. There was a decrease in the length of the corresponding 13-mer chains, from 58.4 Å for N-sulfoheparosan to 51 Å for undersulfated heparin with all of the IdoA2S residues in the 1C4 conformation (Figure 5). Nevertheless, deviations from an idealized helical geometry are found throughout the resulting ensemble members.
Figure 4.
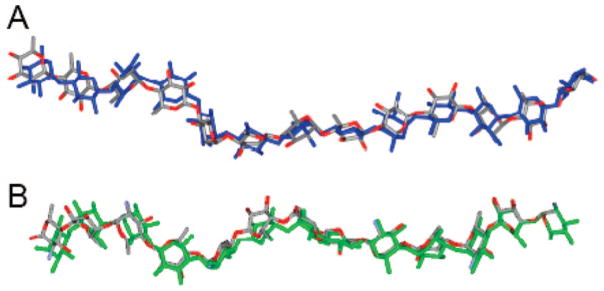
Superimposition of 20 conformers randomly taken from the tar-MD simulations for each GAG. The three central residues have been chosen for the superimposition to show the propagation of the flexibility around the GAG chain: (A) N-acetylheparosan, (B) N-sulfoheparosan, (C) undersulfated heparin, and (D) heparin. Carbon is shown in green, oxygen in red, nitrogen in blue, and sulfur in yellow.
Figure 5.
Superimposition of simulated 13-mers. (A) N-Acetylheparosan (gray and red) is shown superimposed with N-sulfoheparosan (blue). (B) Undersulfated heparin (gray and red) is shown superimposed with heparin (green).
Heparin’s repeating unit was next afforded by 6-O-sulfonation of the undersulfated heparin. In contrast to its undersulfated precursor, when the force field matched the H1GlcNS6S–H4IdoA2S and H1GlcNS6S–H3IdoA2S distances, the NOE-based H2IdoA2S–H5IdoA2S distance was calculated at 2.8 Å (±10%), a value that, according to the idealized values given above, is consistent with >80% of the IdoA2S being in the 2SO form, with an rmsd value of 2.2 Å. These results clearly demonstrate that the addition of a 6-O-sulfo group onto GlcNS residues markedly shifts the equilibrium of adjacent IdoA2S residues from the 1C4 to the 2SO conformer. The balance of the 1C4 to 2SO equilibrium in an IdoA residue within a GAG chain had been previously shown to depend on both its own substitution with a 2-O-sulfo group and the substitution of the adjacent GlcN residues.12,52 Mulloy and co-workers50 showed that the overall conformation of the chain (i.e., the optimal angles around the inter-residue glycosidic linkages) is not greatly affected by the position of the 1C4 to 2SO equilibrium. Our observations are similar with only a very minor 0.7 Å difference between the lengths of the 13-mer chains built using 1C4 and 2SO geometries for the IdoA2S. The helical structure of the chain only becomes apparent in the fully modified heparin (Figures 4D and 5B and see Supporting Information). While the global structures of the members of the tar-MD heparin ensemble deviate from the idealized geometry in models proposed by Mulloy and co-workers,12 there is good agreement at a local structural level. The ring geometries and glycosidic linkages in the tar-refined heparin ensemble are consistent with previous structural studies performed on short heparin oligosaccaharides.10,11 Medium-range to long-range structural ordering is also consistent with the previous studies and, in the current study, heparin forms a left-handed helix with rotational and translational parameters similar to those of short heparin oligosaccharides. Therefore, the key helical parameters in heparin, including helical width, groove depth, and axial rise, are not significantly influenced by overall chain length. Local conformational flexibility is evident in the tar-refined ensemble, consistent with previous reported observations. However, this flexibility does not appear to result in accordion-like stretching along the helical axis (Figure 5). Some rotational freedom about the glycosidic linkages was observed in the heparin ensemble, and this fact results in small inter-residue deviations that would be expected to propagate along the extended heparin structure, resulting in long-range disorder or bending of the helical axis. These global distortions are representative of the conformational space explored in solution by the dynamic heparin polysaccharide.
The structure of heparin has been extensively studied since its discovery.7,8 The advent of NMR was instrumental in defining heparin’s major uronic acid residue as IdoA and has greatly improved our understanding of heparin’s primary structure.53 NMR-based conformational analyses of heparin and heparin oligosaccharides have resulted in an improved understanding of its ring conformations and their flexibility.9–11,54 Molecular modeling coupled with NMR12 has led to structures widely used for analyzing heparin binding to proteins.4–6 However, NMR has had limited success in high-resolution structure determination of the heparin molecule portions within these complexes. X-ray crystallography has offered an independent method to look at heparin’s structure. Despite the publication of a number of crystal structures of heparin oligosaccharides complexed with heparin-binding proteins,6 unbound heparin has only been studied at low resolution in fiber structures.55
Future studies will be required to map the fine structural features of heparin and compare these to various pharmaceutical heparins prepared from animal tissues. Additional studies will focus on the use of uniformly stable isotope-labeled GAGs as binding partners for various heparin-binding proteins. In such studies, it should be possible to study conformational changes in the bound and unbound GAG using NMR. Moreover, it should be possible to prepare structurally homogeneous oligosaccharide binding partners56 uniformly labeled with NMR-sensitive isotopes that enable high-resolution structural determination of these complexes. This should afford the improved understanding of heparin structure–activity relationships required for the design of a novel and specific class of heparin-based therapeutic agents.
Supplemental Materials
Acknowledgments
National Institutes of Health Grants GM38060 and HL62244 (to R.J.L.) and AI50050 (to J.L.), and the Ministry of Education and Science of Spain for Grant CTQ-2006-10874-C02-01 (to J.J.-B.) and for Ramón-y-Cajal contract (to F.C.) are gratefully acknowledged for supporting this work.
Footnotes
Supporting Information Available: HPLC-ESI-MS disaccharide analysis of N-acetylheparosan; CT-HSQC NMR of N-acetylheparosan and N-sulfoheparosan; 3D-HCCH-COSY of N-acetylheparosan, N-sulfoheparosan, and undersulafted heparin; 3D HCCH-NOESY of N-acetylheparosan, N-sulfoheparosan, undersulfated heparin, and heparin; φ/ψ plots of N-sulfoheparosan and undersulfated heparin; stereoview of heparin helix; disaccharide composition of heparin and anticoagulant heparin; key proton-pair distances by tar-MD for N-sulfoheparosan and undersulfated heparin. This information is available free of charge via the Internet at http://pubs.acs.org.
JA8026345
References
- 1.Capila I, Linhardt RJ. Angew Chem, Int Ed. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 2.Munoz EM, Linhardt RJ. Arterioscler Thromb Vasc Biol. 2004;24:1549–1557. doi: 10.1161/01.ATV.0000137189.22999.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raman R, Sasisekharan V, Sasisekharan R. Chem Biol. 2005;12:267–277. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. Proc Natl Acad Sci USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrini M, Guglieri S, Beccati D, Torri G, Viskov C, Mourier P. Biochem J. 2006;399:191–198. doi: 10.1042/BJ20060656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulloy B, Linhardt RJ. Curr Opin Struct Biol. 2001;11:623–628. doi: 10.1016/s0959-440x(00)00257-8. [DOI] [PubMed] [Google Scholar]
- 7.Casu B. Adv Carbohydr Chem Biochem. 1985;43:51–134. doi: 10.1016/s0065-2318(08)60067-0. [DOI] [PubMed] [Google Scholar]
- 8.Linhardt RJ. J Med Chem. 2003;46:2551–2554. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 9.Ragazzi M, Ferro DR, Perly B, Sinaÿ P, Petitou M, Choay J. Carbohydr Res. 1990;195:169–185. doi: 10.1016/0008-6215(90)84165-q. [DOI] [PubMed] [Google Scholar]
- 10.Mikhailov D, Linhardt RJ, Mayo KH. Biochem J. 1997;328:51–61. doi: 10.1042/bj3280051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angulo J, Hricovíni M, Gairi M, Guerrini M, de Paz JL, Ojeda R, Martín-Lomas M, Nieto PM. Glycobiology. 2005;15:1008–1015. doi: 10.1093/glycob/cwi091. [DOI] [PubMed] [Google Scholar]
- 12.Mulloy B, Forster MJ, Jones C, Davies DB. Biochem J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homans SW. Biochem Soc Trans. 1998;26:551–560. doi: 10.1042/bst0260551. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl U, Li JP, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emeis J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. J Med Chem. 2005;48:349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Avci FY, Munoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindahl U, Feingold DS, Roden L. Trends Biochem Sci. 1986;11:221–225. [Google Scholar]
- 17.Esko JD, Selleck SB. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 18.Navia JL, Riesenfeld J, Vann WF, Lindahl U, Rodén L. Anal Biochem. 1983;135:134–140. doi: 10.1016/0003-2697(83)90741-8. [DOI] [PubMed] [Google Scholar]
- 19.DeAngelis PL, Gunay NS, Toida T, Mao WJ, Linhardt RJ. Carbohydr Res. 2002;337:1547–1552. doi: 10.1016/s0008-6215(02)00219-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Jones CL, Liu J. Chem Biol. 2007;14:986–993. doi: 10.1016/j.chembiol.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casu B, Grazioli G, Razi N, Guerrini M, Naggi A, Torri G, Oreste P, Tursi F, Zoppetti G, Lindahl U. Carbohydr Res. 1994;263:271–284. doi: 10.1016/0008-6215(94)00172-3. [DOI] [PubMed] [Google Scholar]
- 22.Kuberan B, Beeler DL, Lech M, Wu ZL, Rosenberg RD. J Biol Chem. 2003;278:52613–52621. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]
- 23.Westler WM, Stockman BJ, Hosoya Y, Miyake Y, Kainosho M, Markley JL. J Am Chem Soc. 1988;110:6255–6258. doi: 10.1021/ja00226a057. [DOI] [PubMed] [Google Scholar]
- 24.Nikonowicz EP, Pardi A. Nature. 1992;355:184–186. doi: 10.1038/355184a0. [DOI] [PubMed] [Google Scholar]
- 25.Yu F, Wolff JJ, Amster IJ, Prestegard JH. J Am Chem Soc. 2007;129:13288–13297. doi: 10.1021/ja075272h. [DOI] [PubMed] [Google Scholar]
- 26.Almond A, DeAngelis PL, Blundell CD. J Mol Biol. 2006;358:1256–1269. doi: 10.1016/j.jmb.2006.02.077. [DOI] [PubMed] [Google Scholar]
- 27.Vann WF, Schmidt MA, Jann B, Jann K. Eur J Biochem. 1981;116:359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 28.Duncan MB, Liu M, Fox C, Liu J. Biochem Biophys Res Commun. 2006;339:1232–1237. doi: 10.1016/j.bbrc.2005.11.142. [DOI] [PubMed] [Google Scholar]
- 29.Nadkarni VD, Toida T, Van Gorp CL, Schubert RL, Weiler JM, Hansen KP, Caldwell EEO, Linhardt RJ. Carbohydr Res. 1996;290:87–96. doi: 10.1016/0008-6215(96)00129-2. [DOI] [PubMed] [Google Scholar]
- 30.Warda M, Zhang F, Radwan M, Zhang Z, Kim N, Kim YN, Linhardt RJ, Han J. Glycoconjugate J. 2008;25:441–450. doi: 10.1007/s10719-007-9090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearlman DA, Case DA, Caldwell JW, Ross WR, Cheatham TE, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 32.Kollman PA. University of California; San Francisco: 1999. [Google Scholar]
- 33.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 34.Woods RJ, Dwek RA, Edge CJ, Fraser-Reid B. J Phys Chem B. 1995;99:3832–3846. [Google Scholar]
- 35.Lan J. PhD Thesis. University of Edinburgh; 2007. [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 37.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Linhardt RJ, Gunay NS. Semin Thrombos Hemostas. 1999;25:5–16. [PubMed] [Google Scholar]
- 39.Guerrini M, Naggi A, Guglieri S, Santarsiero R, Torri G. Anal Biochem. 2005;337:35–47. doi: 10.1016/j.ab.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 40.Murugesan S, Park TJ, Yang H, Mousa S, Linhardt RJ. Langmuir. 2006;22:3461–3463. doi: 10.1021/la0534468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corzana F, Busto JH, Jiménez-Osés G, García de Luis M, Asensio JL, Jiménez-Barbero J, Peregrina JM, Avenoza A. J Am Chem Soc. 2007;129:9458–9467. doi: 10.1021/ja072181b. [DOI] [PubMed] [Google Scholar]
- 42.Silipo A, Zhang Z, Cañada FJ, Molinaro A, Linhardt RJ, Jiménez-Barbero J. ChemBioChem. 2007;9:240–252. doi: 10.1002/cbic.200700400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hricovíni M, Nieto PM, Torri G. In: NMR spectroscopy of glycoconjugates. Jimenez-Barbero J, Peters T, editors. Wiley-VCH; Weinheim: 2002. pp. 189–230. [Google Scholar]
- 44.Neuhaus D, Williamson MP, editors. The nuclear Overhauser effect in structural and conformational analysis. Wiley; New York: 1989. [Google Scholar]
- 45.Pearlman DA. J Biomol NMR. 1994;4:1–16. doi: 10.1007/BF00178332. [DOI] [PubMed] [Google Scholar]
- 46.Torda AE, Scheek RM, van Gunsteren WF. J Mol Biol. 1990;214:223–235. doi: 10.1016/0022-2836(90)90157-H. [DOI] [PubMed] [Google Scholar]
- 47.Torda AE, Scheek RM, van Gunsteren WF. Chem Phys Lett. 1989;157:289–294. [Google Scholar]
- 48.Forster M, Jones C, Mulloy B. J Mol Graphics. 1989;7:196–201. doi: 10.1016/0263-7855(89)80002-5. [DOI] [PubMed] [Google Scholar]
- 49.Almond A, Deangelis PL, Blundell CD. J Mol Biol. 2006;358:1256–1269. doi: 10.1016/j.jmb.2006.02.077. [DOI] [PubMed] [Google Scholar]
- 50.Mulloy B, Forster MJ. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 51.Sanderson PN, Huckerby TN, Nieduszynski IA. Biochem J. 1987;243:175–181. doi: 10.1042/bj2430175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Boeckel CA, van Aelst SF, Wagenaars GN, Mellema JR, Paulsen H, Peters T, Pollex A, Sinnwell V. Recl Trav Chim Pays-Bas. 1987;106:19–29. [Google Scholar]
- 53.Perlin AS, Mackie DM, Dietrich CP. Carbohydr Res. 1971;18:185–194. doi: 10.1016/s0008-6215(00)80341-9. [DOI] [PubMed] [Google Scholar]
- 54.Casu B, Petitou M, Provasoli M, Sinaÿ P. Trends Biochem Sci. 1988;13:221–225. doi: 10.1016/0968-0004(88)90088-6. [DOI] [PubMed] [Google Scholar]
- 55.Atkins ED, Nieduszynski IA. Adv Exp Med Biol. 1975;52:19–37. doi: 10.1007/978-1-4684-0946-8_2. [DOI] [PubMed] [Google Scholar]
- 56.Kuberan B, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Nat Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.