Abstract
Glutamate transporters are membrane proteins found in neurons and glial cells, which play a critical role in regulating cell signaling by clearing glutamate released from synapses. Although extensive biochemical and structural studies have shed light onto different aspects of glutamate transport, the time-resolved molecular mechanism of substrate (glutamate or aspartate) translocation, that is, the sequence of events occurring at the atomic level after substrate binding and before its release intracellularly remain to be elucidated. We identify an energetically preferred permeation pathway of ≈23 Å between the helix HP1b on the hairpin HP1 and the transmembrane helices TM7 and TM8, using the high resolution structure of the transporter from Pyrococcus horikoshii (GltPh) in steered molecular dynamics simulations. Detailed potential of mean force calculations along the putative pathway reveal 2 energy barriers encountered by the substrate (aspartate) before it reaches the exit. The first barrier is surmounted with the assistance of 2 conserved residues (S278 and N401) and a sodium ion (Na2); and the second, by the electrostatic interactions with D405 and another sodium ion (Na1). The observed critical interactions and mediating role of conserved residues in the core domain, the accompanying conformational changes (in both substrate and transporter) that relieve local strains, and the unique coupling of aspartate transport to Na+ dislocation provide insights into methods for modulating substrate transport.
Sodium-dependent glutamate transporters, also known as excitatory amino acid transporters (EAATs), are membrane proteins involved in regulating excitatory signal transmission by clearing excess l-glutamate released at synapses (1). The concentration of these excitatory amino acid neurotransmitters at the extracellular (EC) space may increase by several thousandfold during the periods of synaptic activation (2–4). Efficient removal of excess glutamate is critical to protect neurons against excitotoxicity. Glutamate uptake in the brain is coupled to the cotransport of 3 sodium ions (Na+) and 1 proton (H+), and followed by the countertransport of 1 potassium ion (K+) (2–4). In addition, in the transporter, glutamate activates an uncoupled chloride (Cl−) current, which is known to be important for glutamatergic neurotransmission (5–7).
The first crystal structure of an archaeal orthologue (GltPh, from Pyrococcus horikoshii) of the eukaryotic EAATs was resolved by Gouaux and coworkers (8) in 2004. This structure offered the possibility of exploring the molecular interactions that drive substrate transport via structure-based models and simulations. Additional insights were provided in a more recent article by Boudker et al. (9), which reported another conformation of GltPh, resolved in the presence of the bound substrate and thallium ions. Radio labeled flux experiments revealed that GltPh preferentially mediates the uptake of aspartate over glutamate (9).
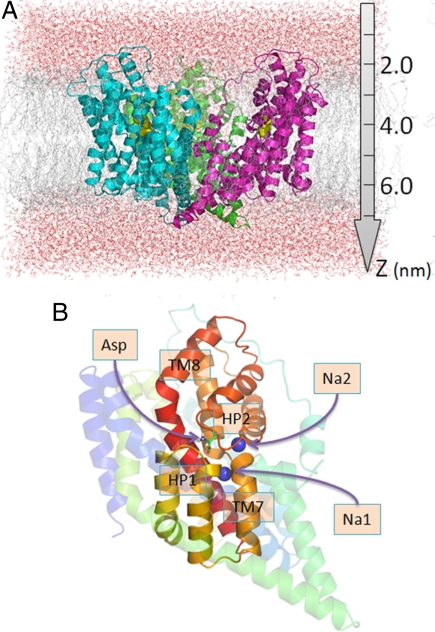
These X-ray crystallographic structures showed that GltPh is a bowl-shaped homotrimer, with a solvent-filled basin exposed to the EC region (Fig. 1A). The basin extends almost halfway through the membrane bilayer. Each monomer consists of 8 transmembrane helices (TM1-TM8) and 2 helical hairpins (HP1 and HP2), organized into 2 domains (Fig. 1B). The C-terminal “core domain” facing the central basin is composed of the structural elements HP1, TM7, HP2, and TM8; and the N-terminal domain is composed of the helices TM1-TM6 on the outer part of the membrane protein. The core domain is essential in mediating Na+-dependent substrate transport. It is surrounded and secured within the TM helices of the N-terminal domain.
Fig. 1.
Schematic view of GltPh and surrounding lipid and water molecules. (A) Side view of the simulation box. GltPh is a bowl-shaped homotrimer. Its 3 monomers are colored green, cyan, and magenta. It is embedded in the lipid bilayer (gray) and water (red) molecules. Aspartates are shown as yellow spheres in (A). (B) Close view of a monomer. Noncore residues are rendered translucent. The core domain (TM7, TM8, HP1, and HP2) is colored in rainbow scheme. The aspartate is displayed in stick representation and the 2 sodium ions as blue spheres. The structural coordinates are taken from the PDB entry 2NWX.
One of the most conserved regions of the sequence is the N310MDGT314 motif at the partially unwound portion of TM7, between the 2 helical segments TM7a and TM7b. This motif has been pointed out to play a role in binding the substrate and sodium ions, as also confirmed by the GltPh structure (4, 8). In addition, residues in the mammalian EAATs equivalent to the GltPh residues D394 and R397 on TM8 and the serine rich motif (S277-S278-S279) at the HP1 tip have been proposed to be involved in glutamate binding, in line with the interactions with the bound aspartate observed in the GltPh structure (8–10).
We recently examined the substrate recognition and binding events of GltPh (11) by molecular dynamic (MD) simulations in a fully solvated lipid-bilayer. The helical hairpin HP2 was observed to open up and close down multiple times during 40 ns runs performed in the absence of substrate. These simulations thus confirmed that the HP2 hairpin acts as an “EC gate” that readily fluctuates between open and closed states. Moreover, they showed that the open state is predisposed for substrate binding within nanoseconds, and the closed state is favored upon Na+ binding. No net correlation was observed between the motions of the 3 EC gates, confirming that the subunits function independently with respect to their own substrate neighborhoods. However, no further permeation or release to the cell interior could be observed. The translocation of substrate has indeed a timescale of milliseconds, beyond the reach of conventional MD.
In the present study, we focus on the translocation of the substrate from its binding site near the EC gate into the transporter interior and its release to the cytoplasm. Our objective is to elucidate the translocation pathway(s) and corresponding free energy profile, and understand the significance of Na+ cotransport. To this aim, we conducted a series of steered MD simulations (12) using GltPh structure and its substrate, aspartate. Steered MD permits us to accelerate the translocation process and thereby identify the pathway(s) preferentially selected by the substrate, and the coupling to Na+ cotransport. A highly preferred pathway, called P1, is revealed between the helices HP1b (of hairpin HP1), TM7, and TM8, along with an alternative, less probable outlet (P2). The Na+-dependent stochastic events that facilitate the channeling of the substrate to the exit point are visualized at the atomic level detail. We calculated the potential of mean force (PMF) profile using umbrella sampling and a weighted histogram analysis method (WHAM) (13, 14) applied to a total of 46 windows along the 23-Å-long permeation pathway. Two local energy barriers were identified. The conformational changes coupled to Na+ dislocation undergone by the substrate and surrounding transporter residues for relieving the local strains near the energy barriers provide us with insights into the molecular mechanism and atomic interactions that control substrate transport by this important family of transporters.
Results and Discussion
Substrate Permeation Occurs via 2 Well-Defined Pathways, One More Probable than the Other.
The high resolution structure of GltPh with a bound substrate (aspartate) on each of the 3 subunits (9) was used as input structure in steered MD simulations (see Methods). Our previous simulations have shown that the substrate is highly stable and resistant to displacement away from its original binding site within the timescale (tens of nanoseconds) of conventional MD. We applied here the constant-velocity AFM pulling method along the z axis (Fig. 1A) to enable the permeation of substrate within the time frame of simulations. A threshold value of 2,500 pN was needed to initiate the translocation. At this point, as shown in Fig. 2, the substrate was able to surmount a first local energy barrier, corresponding to conformation B1 described in atomic detail below. The passage over this barrier was evidenced by the abrupt decrease in the force experienced (Fig. 2A) and the accompanying change in the position of the aspartate (Fig. 2B), which rapidly settled in another relatively stable position ≈1 nm further along the z axis (downward). This new position, shown by PMF calculations below to be a local energy minimum, is designated as M.
Fig. 2.
Force and displacement profiles of substrate observed in simulated pulling experiments. Results for subunits A, B, and C are colored red, green, and blue, respectively. (A and B) Results starting from the initial structure. The force increases linearly consistent with the deformation of the elastic spring appended at one end to the substrate, and exhibits sharp decreases when the substrate surmounts local barriers and dislocates along the z axis. The diagonal lines in B describe the instantaneous positions of the opposite ends of the springs used in the The force experienced by the aspartate increases when the separation between the two ends increases.
Strikingly, all 3 subunits exhibited identical (macroscopic) stress-strain profiles in the first half of the trajectory, despite minor differences in their microscopic features (i.e., the fluctuations in the instantaneous positions and velocities of the individual atoms in the 3 subunits). This portion of the trajectory spanning the range 4 ≤ z ≤ 5 nm, approximately, is referred to as phase 1. Two distinct pathways were selected near 5 nm along the z axis: one, selected by the aspartates in subunits B and C, with minor differences in the instantaneous coordinates of interacting residues, hereafter referred to as pathway 1 (P1); and the other, selected by the aspartate in subunit A (P2). P1 was between HP1b, TM7a, and TM8; and P2, between HP1b, TM7a, and TM2. The motion along P1 required less work than that along P2 [see supporting information (SI) Fig. S1A], and the 2 pathway directions deviated by approximately 20°. We accordingly defined phase 2 as the portion from z = 5 nm to 6.3 nm with 2 alternative pathways. Translocation of aspartate along pathway P1 all the way to the intracellular (IC) region can be seen in Movie S1.
The instantaneous pulling force on the substrate was significantly lowered, but not abolished at the “stable” position near the local minimum M (t = 1.6 ns in Fig. 2A). To allow the protein and substrate to completely relax at this position, we removed the pulling force and equilibrated the system for 1 ns, and then repeated the steered MD simulations for the second phase. Fig. S1B displays the results from this additional run. Again, two of the aspartates (in subunits A and B, this time) permeated through P1, and the third (in subunit C) through P2. No coupling between the translocation of the aspartates was observed. The 3 subunits are generally accepted to function independently (10), whereas intersubunit couplings have also been suggested by disulfide cross-linking experiments and analytical models. Present simulations show that the subunits do function independently in so far as substrate translocation is concerned. Additional calculations performed by changing the steering direction (see SI Text and Fig. S2) showed that the pathway P1 is the prominent pathway preferred by substrate in the context of the transporter structure and energetics. In the following, we analyze more closely this “easier” (or more probable) pathway, because the substrate-conducting channel intrinsically favored by the core architecture of the subunits.
Free Energy Profile Along the Most Probable Translocation Path Reveals 2 Local Energy Barriers.
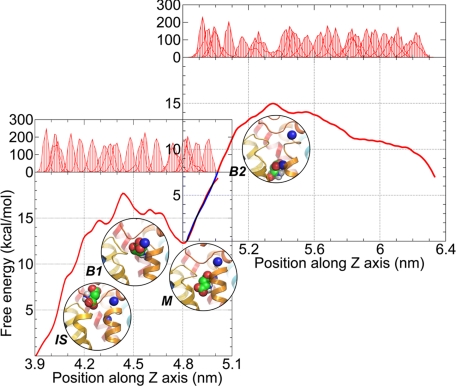
Fig. 3 displays the PMFs resulting from umbrella sampling and the WHAM (see Methods) calculations. Calculations were performed separately for the phases 1 and 2 (pathway P1) of the substrate channeling pathway. Each phase covers a displacement of 10 Å, approximately. We have combined the results into one figure to visualize the complete free energy profile. As suggested by the time dependencies of the pulling force and substrate displacements presented above, we identified 2 energy barriers. The first is located at z = 4.4 nm (B1) and the second at 5.4 nm (B2). These are separated by the intermediate state, or local energy minimum (M), near 4.8 nm. Fig. 3 displays 4 snapshots representing the neighborhood of the substrate at the initial state (IS), and 3 conformations, B1, M, and B2 along the permeation pathway.
Fig. 3.
Free energy profiles along the putative permeation pathway. The bottom left and upper right figures refer to the first and second phases (pathway P1), respectively. The curve in the overlapping region in the upper right figure is colored blue. The position distributions of aspartate obtained via umbrella sampling MD simulations for a series of windows along the z axis are shown in the upper abscissa. Four snapshots corresponding to the initial state (IS), the first energy barrier (B1), the local energy minimum (M), and the second energy barrier (B2) are shown, which highlight the relative positions of aspartate and sodium ions. See Fig. 4 for more details on these snapshots.
The free energy difference between the initial state and the first barrier B1, and that between M and B2, were >15 kcal/mol, summing to a total of >25 kcal/mol. Interestingly, these numbers closely agree with the activation enthalpies of 95 kJ/mol and 120 kJ/mol observed for EAAC1 by Grewer and coworkers (15) for 2 processes involved in the translocation branch of the glutamate transport cycle. Our results and these experiments suggest that the translocation cannot be accomplished at room temperature without a sufficiently large driving potential. In the present study, we introduced an artificial pulling effect to mimic the driving potential, with the objective of examining the interaction between the substrate, Na+, and protein during substrate translocation within accessible timescales. There might be other processes, e.g., cotransport of a proton and a third Na+ not included here, and potassium ion countertransport, which could facilitate the permeation of aspartate and lower the free energy barriers. We also noted that the backbone conformation lining the translocation pathway showed minimal, if any, changes in conformation, in our PMF calculations, although ANM calculations performed for GltPh indicated that the core domain tends to “relax” via collective modes that are intrinsically accessible to the transporter. Such effects on the effective energy barrier may be further explored by the recently introduced ANM-steered MD simulations (16). Finally, the free energy profile suggests that the reverse transfer of aspartate from the IC to the EC side is not less favorable than the forward passage. The substrate translocation cycle has indeed been observed to be reversible (17).
Key Interactions with Residues Lining the Permeation Pathway.
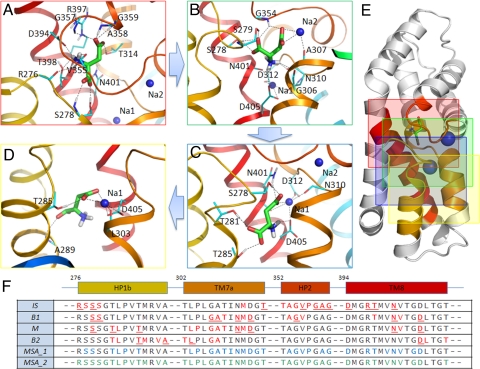
Fig. 4 A–D illustrates the near neighborhood of the substrate in the conformations IS (A), B1 (B), M (C), and B2 (D) distinguished along the free energy profile. Fig. 4E illustrates the positions of these 4 frames on the core domain of the subunits (the frame colors match those of the borders in Fig. 4 A–D). Comparison of the panels shows that the packing density near the substrate is low in B2. This type of weaker interaction with the surrounding amino acids may partly explain why the IS and M states, and even B1, have lower free energies than B2. We furthermore note that in the B2 conformation, there are very few polar or charged residues in the neighborhood of aspartate. The sequence fragments shown in Fig. 4F highlight (in red) the residues closely interacting with the aspartate in each conformation.
Fig. 4.
Close neighborhood of aspartate in the conformations IS (A), B1 (B), M (C), and B2 (D) depicted in Fig. 3. The protein backbone is shown in diagram representation, using the same color scheme as in Fig. 1B. The aspartate (green) and residues (cyan) interacting with aspartate are shown as sticks, with their O, N, and H atoms colored blue, red, and white, respectively. Sodium ions are shown as blue spheres. Hydrogen bonds are shown as dashed lines. (E) The locations of the 4 snapshots colored by the respective panel frames. The sequence alignment in (F) shows the fragments containing the residues (colored red) that interact with aspartate in the 4 conformations (rows 1–4), and those conserved across 13 sequences (row 5, highlighted in blue) and across 7 archaeal sequences (row 6, highlighted in green). Underlined residues in rows 1–4 refer to those explicitly shown in A–D. Bars above the table indicate the secondary structures and corresponding starting residue numbers in GltPh. Note that only the relevant portions of the sequence are shown.
Several groups have drawn attention to conserved residues that interact with substrate and sodium ions, even before the crystal structure with bound aspartate and sodium ions was resolved (8). However, there remain several highly conserved residues, among different species, whose role and functional interactions are unclear. To our surprise, most of the residues distinguished here as key players in mediating the protein-substrate interactions at the 4 conformations (colored red in the rows 1–4 of the fragments aligned in Fig. 4F) are conserved in the glutamate transporter family. Row 5 highlights (in blue) the conserved residues among 13 sequences (seven archaeal plus 6 mammalian sequences), and row 6 highlights (in green) those fully conserved across 7 archaeal sequences in the same family (see also Table S1). The close correspondence between the residues observed here to be involved in mediating aspartate transport and those highly conserved across family members reinforces the functional significance of the residues identified here. The present analysis permits us to propose hypotheses on the specific role of these residues at various stages of the translocation process, including substitutions that may potentially enhance or abolish these specific interactions yet to be tested and established by experiments (see below).
Conformations and Global Reorientation Preferred by Aspartate During Permeation.
Aspartate has an amino group and 2 carboxylate (COO−) groups: an α-carboxylate near the amino group, and a β-carboxylate corresponding to the side chain COO−. In the bound state resolved by X-ray crystallography (conformation IS), the aspartate is oriented such that its amino group occupies a position above the main chain COO− (α-carboxylate), whereas, in conformation B1, the relative positions of these two groups are inverted (Fig. 4A and B). It is interesting to note that during this significant change in conformation, the α-carboxylate maintains its hydrogen bonds with the 3-Ser motif residues S278 and S279 on the HP1 loop, whereas the remaining portions of the molecule exhibit significant tumbling. The β-carboxylate, originally interacting with the HP2 loop glycines (G357 and G359) and TM8 arginine R397 (A), is “detached” from the HP2 loop, presumably with the assistance of the sodium ion Na2 (B). The conformational change of the aspartate involves not only a rigid body rotation and translation, but also a rotation in the Cα-Cβ dihedral angle χ1 from the gauche+ to the trans state (the χ1 values in the 4 respective states are −67.4°, −178.0°, −61.2°, and −72.7°). As a result of the dihedral angle change, the two COO− groups are momentarily located at a close proximity Fig. 4B). This may be a factor contributing to the instability of the B1 conformation, which rapidly relaxes to the M state (C) by a dihedral angle jump back to the gauche+ state. In the M state, the α-COO− group readjusts its orientation to optimize its interaction with the surrounding polar residues T281 and T285 on HP1, whereas the β-COO− engages in electrostatic interactions with the sodium ion Na1 and with D312 (via Na1). D312 exhibits a high avidity for interacting with the substrate, as one of the “frustrated” residues located at the “broken” portion of the helix TM7, between TM7a and TM7b.
After surmounting the barrier B1, the aspartate was observed to generally maintain its orientation with slight rearrangements, with the amino group being the leading group (pointing toward the IC side) and the β-carboxylate as the tailing group. This particular reorientation thus emerges as an intrinsic property imparted by the geometry at the core region of the protein. Because there were no membrane potentials or Na+ concentration gradient in our simulations, we suggest that the protein itself tends to stabilize a conformation with such a polarity during the permeation process.
Two Residues Distinguished by Their Persistent Mediating Role: S278 and N401.
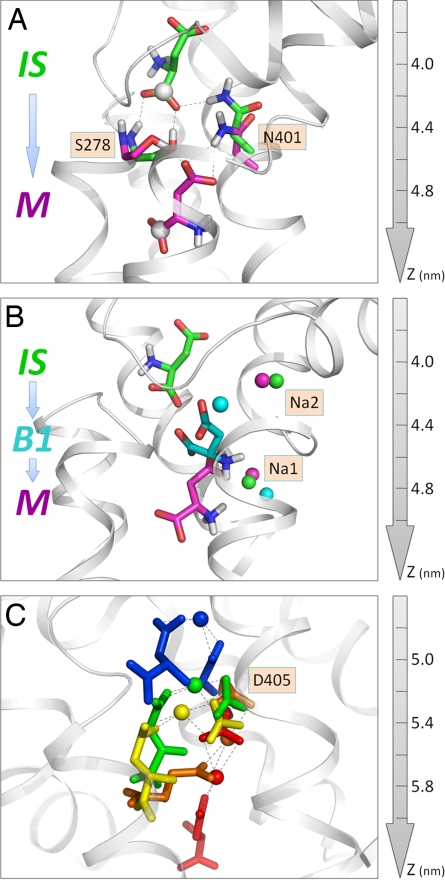
Two polar residues, S278 and N401, were distinguished by their continual coordination of the substrate in the first phase of translocation, and a third residue, M311 (data not shown), on the NMDGT motif played a mediating role via hydrophobic contacts. Fig. 5A illustrates the interactions of S278 and N401 with the aspartate in the IS (green) and M (magenta) states, suggesting that these 2 residues play an important role, not only in coordinating the substrate through interactions with its COO− groups, but also in guiding the substrate along the permeation pathway toward the IC side.
Fig. 5.
Roles of (A) S278 and N401, (B) sodium ions, and (C) D405 in assisting the translocation of the substrate. (A) Two snapshots: the initial state (IS), and intermediate state (M). The backbone atoms of the substrate, S278 and N401 are colored green and magenta in the respective conformations. The α-COO− carbon atom of the substrate is displayed as a white sphere. (B) Three successive conformations (IS, B1, and M) of the substrate and accompanying displacements of the 2 sodium ions (colored green, cyan, and magenta in the respective conformations). Na2 temporarily approaches the substrate to assist in the passage over B1, and returns to its original position (binding site in the X-ray structure). Na1 starts to interact with aspartate in the M state (magenta) only, and remains coupled throughout the remainder of the trajectory, as also shown in C. (C) The ternary interactions between aspartate, D405, and Na1 in 5 successive snapshots, succeeding state M. Each snapshot is shown by a different color: blue, green, yellow, orange, and red, from top to bottom. The snapshots elucidate the conformational changes in D405 accompanying the translocation of the substrate and the coupling to Na1 as the aspartate moves toward the intracellular exit. Hydrogen bonds are shown by dotted lines.
Notably, S278 and N401 interact successively with both COO− groups of aspartate in multiple conformations. In the initial state, S278 forms 2 hydrogen bonds with the α-carboxylate group, via its main chain NH and side chain OH groups; N401 also forms a hydrogen bond with the α-COO−. In the M state, both S278 and N401 switch to hydrogen bonds with the β-COO− group, i.e., the original hydrogen bonds are broken during the passage over the barrier B1, but new ones are quickly formed in the state M. Our simulations thus provide compelling evidence supporting the view that these two highly conserved residues facilitate the translocation of substrate in the permeation pathway over an ≈10-Å-long portion of the translocation pathway.
Successive Coupling to the Two Sodium Ions Is Essential for Surmounting the Local Energy Barriers.
There are 2 sodium ions in the high-resolution crystal structure (Protein Data Bank ID: 2NWX) of GltPh (9). In both steered MD simulations and free energy calculations with umbrella sampling, one of them [designated as Na2 (9)] exhibited a strong tendency to remain at its binding site; whereas the other (Na1) first encountered the substrate around z = 4.8 nm to stabilize the M state, and remained closely coupled to the aspartate all the way through to the IC side.
The role of the Na2 is presumably 2-fold. On the one hand, it stabilizes the core conformation with the HP2 gate in the closed form, thus ensuring that the substrate does not escape to the EC aqueous cavity; however, it is extremely interesting to see that it moves out of its binding site for a short period to engage in a charge-charge interaction with the β-COO− group of aspartate, assisting it in overcoming the energy barrier B1. As soon as the barrier is crossed, Na2 moves back to its original position to restore its “gatekeeper” function (Fig. 5B). The role of Na2 in sealing the binding site has also been reported in recent simulations of the glutamate transporter (18). Interestingly, a similar role has been proposed for a Na+ in a leucine transporter (19).
While Na2 moves back to its original binding site, the second Na+, Na1, comes into play: it is, in a sense, “captured” by the β-COO− group that apparently seeks a positively charged partner after its dissociation from Na2. Once the β-COO− group locates the Na1 in the vicinity, it engages in a coulombic interaction that persists throughout the remaining portion of the translocation (Fig. 5C). Binding of Na1 to aspartate not only neutralizes its negative charge, but also alters its interaction with the surrounding residues. We note in particular a preferential interaction with D405 (see below) upon the mediating effect of Na1 that serves as a cation bridge. The movement of Na1 here does not imply that Na1 is the driving force on aspartate: Na+ essentially accompanies the aspartate. The simulations thus shed light on the importance of the coupling to Na+ during substrate translocation. We propose that the aspartate-Na+ association observed in simulations provides a plausible description of the Na+-dependent transport under physiological conditions.
We note that only 1 Na+ is cotransported in our simulations with aspartate, whereas the other remains bound at its X-ray crystallographic binding site, apart from momentarily assisting the substrate surmount a local energy barrier (B1). The transport of one Na+ departs from the experimental data for EAATs, which indicate the cotransport of 3 sodium ions. One possible explanation is the lack of a Na+ concentration gradient, or driving force for Na+ flux, in simulations. Yet, irrespective of such an external field, which would favor the Na+ transport in the required direction, even the internal dynamics of the transporter and its interactions with the substrate seem to favor the conduction of a Na+. Alternatively, given that the crystal structure of GltPh also reveals 2 sodium ions, GltPh may differ from its EAAT orthologues in that 2 sodium ions, rather than 3, are cotransported, similar to the members of the neurotransmitter:sodium symporter family (e.g., the leucine transporter and GABA transporter) (20).
Role of D405 in Facilitating Na+-Assisted Translocation.
The aspartic acid D405 on TM8 plays an important role in binding Na1. The mutation D405N has been reported to diminish aspartate binding by ≈100-fold (9). Our simulations also support the functional importance of D405 during the permeation process (Fig. 5C). Not only does D405 harbor 1 Na+, it also acts like a “hand” holding the Na+ together with aspartate throughout the permeation process, until its release to the cytoplasm. This has the net effect of facilitating the translocation of the aspartate along the pathway P1. In our simulation setup, D405 was deprotonated. This is its preferred state at pH 7. The uptake of glutamate in EAATs is known to occur with the cotransport of 1 proton (2–4). Our simulation system does not include protons. However, it was important to have D405 in the deprotonated (charged) state to ensure its mediating role in substrate translocation in GltPh. We propose that the protonation state of D405 may affect the energy barrier and the permeation rate. This hypothesis is currently under investigation by site-directed mutagenesis experiments in our laboratory. Furthermore, water molecules have been known to mediate proton transfer in membrane proteins (21). Our previous simulations (11) also drew attention to the continual hydration of the substrate binding site, and the possibility of a water molecule serving as a potential proton donor in EAATs, which is being further investigated by quantum chemical calculations.
Conclusion
The present study elucidates for the first time a permeation pathway for the translocation of substrate (aspartate) by a structurally resolved member (GltPh) of the glutamate transporter family. The pathway is ≈23 Å long. It consists of 2 phases. The first phase, invariably reproduced in all independent runs by all subunits, involves the passage over a first energy barrier, B1, assisted with coupling the aspartate β-carboxylate group to one of the sodium ions, whereas 2 of the 3-Ser motif residues, S278 and S279, coordinate the α-carboxylate group; the NMDGT motif N310 and D312, on TM7a, and N401, on TM8, coordinate the amino group of the substrate. In particular S278 and N401 are distinguished by their close interaction with various functional groups of the substrate at all stages (IS, B1 and M) of phase 1. The second phase, however, exhibits a highly preferred path (P1), and an alternative (P2) path, accessible with a lower probability. We focused on the prominent path P1. This path is lined by HP1b, TM7a, and TM8, in accord with the original proposition of Gouaux and coworkers (9). Notably, P1 also involves a passage over a locally strained region, manifested by an energy barrier (B2) in the free energy profile. This “bottleneck” was surmounted by the assistance of the sodium ion, Na1. D405 (on TM8) is distinguished in this phase as a key residue that significantly facilitates the translocation of the substrate until its release to the cytoplasm. The interaction of the substrate with D405 is mediated by Na1 over a trajectory of >10 Å.
Of particular interest are the multiple roles of the Na+ ions observed in simulations. Na2 was definitely found to favor the closed state of the EC gate (hairpin HP2) in our previous simulations (11). This role is confirmed in the present simulations as well. However, this does not prevent this ion from momentarily leaving its binding site to assist the substrate in surmounting the barrier B1, and then going back to its preferred binding site to continue its gatekeeper role. Na1, however, comes into play at the second phase, and remains “coupled” to the substrate until its release to the cytoplasm. Both sodium ions thus play a key role in enabling substrate transport. Overall, our simulations end with the cotransport of 1 Na+ (Na1). It is clear that the dislocation of the second Na+ that favors the closed state of the EC gate is required for opening the gate to the next substrate molecule. However, the role of a possible third Na+ could not be assessed, and it remains to be established whether GltPh has 3 Na+ (rather than two, observed in the X-ray structure) cotransported.
Perhaps the important implications of the present study are those that can be made for human EAATs residues, especially those conserved between the archaeal and mammalian transporters. Many residues presently identified are indeed conserved between archaeal and mammalian transporters, and the observations made here for the GltPh help us build testable hypotheses for the EAATs. For example, the counterpart of the highly conserved GltPh D405 in EAAT1 is D487, or counterparts of the respective residues S278 and N401 are S365 and N483. Some residues distinguished here by their critical role have already been observed in previous experimental studies to be important for particular functions. For example, the 3-Ser motif residues S364-S366 in EAAT1 have been shown by mutation and/or sulfydryl modification experiments to be critically important for substrate uptake (11, 22), or the significance of the NMDGT motif (N398MDGT402 in EAAT1) for Na+ binding is well established in previous studies (23, 24) or our recent review (1). However, the significance of other residues such as EAAT1 D487 or N483 on TM8, inferred from their GltPh counterparts, has yet to be established by further experimental studies.
EAATs, the human orthologues of GltPh, are essential for termination of signal transmission mediated by glutamate, and for prevention of neurotoxicity. Dysfunction of these transporters leads to neurodegenerative and neuropsychiatric diseases. These transporters thus serve as therapeutic targets for these potentially fatal diseases. The atomic level details of protein-substrate-ion interactions along the translocation pathway, provided by the present study, can potentially be used as a basis to design experiments aimed at demystifying the transport mechanism in human EAATs.
Methods
Simulation System Setup.
We adopted as starting structure the recently determined crystal structure of GltPh (PDB ID: 2NWX). The missing residues in the loop between TM3 and TM4 were built by homology modeling using SWISS-MODEL (25). Side chains were completed using SCWRL (26). The transporter was embedded in a lipid bilayer of 393 POPC molecules, with the 3-fold noncrystallographic symmetry (NCS) axis (z axis) perpendicular to the membrane. The system was solvated using the simple point charge model for water molecules, which led to an initial simulation box of 123 × 123 × 88 Å3 (Fig. 1A). Twelve chloride ions were added to make the system electroneutral. Periodic boundary conditions were applied. All simulations were carried out using the GROMACS package version 3.3.1 (27) with the GROMOS 43a1 force field (28). Electrostatic interactions were calculated using the particle mesh Ewald method (29). The LINCS algorithm (30) was used to constrain the bond lengths, thus permitting us to use integration time steps of 2 fs. Isothermal conditions (T = 310 K) were implemented by Berendsen temperature coupling to the solute, lipid, and water molecules. Semiisotropic pressure coupling was used to maintain a constant pressure of 1 bar.
Steered MD Simulations.
We used the GROMACS built-in AFM pulling method. A soft elastic spring of force constant 1,000 kJ·mol−1·nm−2 was attached to the mass center of the aspartate and pulled from the opposite end at constant velocity along the NCS axis (see Fig. S3). Because the 3 subunits transport the substrate independently (10, 31, 32) and the core domains undergo uncorrelated dynamics at the timescale of MD simulations (11), we applied pulling forces on the aspartates in each monomer. Simultaneous examination of the 3 aspartates increased the events observed by a factor of 3, thus enhancing the statistical accuracy of our results and permitting us to verify the reproducibility of the results. Simulations were repeated with different pulling velocities. Larger velocities drive a faster permeation process, but potentially induce some unphysical deformation on the structure because of the lack of sufficient time for the molecule to relax in response to the applied stress. We chose a pulling rate of 0.001 nm·ps−1 as an optimal velocity, which, on the one hand, gives the protein enough time to relax, and in contrast, drives the aspartate onto the IC side within time frames accessible to MD simulations.
Umbrella Sampling.
The PMF along the putative permeation pathway was calculated using umbrella sampling technique, together with the WHAM. The movements of the aspartate were confined to successive regions of interest by applying a harmonic potential of force constant 3,000 kJ·mol−1·nm−2 to its mass center. This approach allowed sampling for the entire permeation pathway and ensuring sufficient overlap in adjacent windows. The probability distributions extracted from the trajectories at each position along the permeation pathway were combined to map the free energy profile along the full range of the reaction coordinate. Two series of umbrella sampling calculations were performed for 2 successive phases of the transport process: a first phase, consisting of 21 windows between the positions z = 4.0 and 5.0 nm along the z axis (Fig. 3), and a second, of 25 windows, spanning the axial position between z = 5.0 and 6.2 nm. Trajectories of 1 ns were generated for each window, and the last 900 ps of each run was used for calculating the PMFs.
Supplementary Material
Acknowledgments.
This work was supported, in whole or in part, by National Institutes of Health grants 5R01GM086238-02 (to I.B.) and MH080726 (to S.G.A.). We also gratefully acknowledge useful discussions with J. Jiang, S.D. Watts and J.K. Vries.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812299106/DCSupplemental.
References
- 1.Torres GE, Amara SG. Glutamate and monoamine transporters: New visions of form and function. Curr Opin Neurobiol. 2007;17:304–312. doi: 10.1016/j.conb.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Kanner BI, Bendahan A. Binding order of substrates to the sodium and potassium ion coupled l-glutamic acid transporter from rat brain. Biochemistry. 1982;21:6327–6330. doi: 10.1021/bi00267a044. [DOI] [PubMed] [Google Scholar]
- 3.Levy LM, Warr O, Attwell D. Stoichiometry of the glial glutamate transporter GLT-1 expressed inducibly in a Chinese hamster ovary cell line selected for low endogenous Na+-dependent glutamate uptake. J Neurosci. 1998;18:9620–9628. doi: 10.1523/JNEUROSCI.18-23-09620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerangue N, Kavanaugh MP. Flux coupling in a neuronal glutamate transporter. Nature. 1996;383:634–637. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 5.Eliasof S, Jahr CE. Retinal glial cell glutamate transporter is coupled to an anionic conductance. Proc Natl Acad Sci USA. 1996;93:4153–4158. doi: 10.1073/pnas.93.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otis TS, Kavanaugh MP, Jahr CE. Postsynaptic glutamate transport at the climbing fiber Purkinje cell synapse. Science. 1997;277:1515–1518. doi: 10.1126/science.277.5331.1515. [DOI] [PubMed] [Google Scholar]
- 7.Veruki ML, Morkve SH, Hartveit E. Activation of a presynaptic glutamate transporter regulates synaptic transmission through electrical signaling. Nat Neurosci. 2006;9:1388–1396. doi: 10.1038/nn1793. [DOI] [PubMed] [Google Scholar]
- 8.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 9.Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 10.Grewer C, Balani P, Weidenfeller C, Bartusel T, Tao Z, Rauen T. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry. 2005;44:11913–11923. doi: 10.1021/bi050987n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrivastava IH, Jiang J, Amara SG, Bahar I. Time-resolved mechanism of extracellular gate opening and substrate binding in a glutamate transporter. J Biol Chem. 2008;283:28680–28690. doi: 10.1074/jbc.M800889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struct Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules: 1. the method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 14.Roux B. The calculation of the potential of mean force using computer-simulations. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 15.Mim C, Tao Z, Grewer C. Two conformational changes are associated with glutamate translocation by the glutamate transporter EAAC1. Biochemistry. 2007;46:9007–9018. doi: 10.1021/bi7005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isin B, Schulten K, Tajkhorshid E, Bahar I. Mechanism of signal propagation upon retinal isomerization: Insights from molecular dynamics simulations of rhodopsin restrained by normal modes. Biophys J. 2008;95:789–803. doi: 10.1529/biophysj.107.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grewer C, Gameiro A, Zhang Z, Tao Z, Braams S, Rauen T. Glutamate forward and reverse transport: From molecular mechanism to transporter-mediated release after ischemia. IUBMB Life. 2008;60:609–619. doi: 10.1002/iub.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang ZJ, Tajkhorshid E. Dynamics of the extracellular gate and ion-substrate coupling in the glutamate transporter. Biophys J. 2008;95:2292–2300. doi: 10.1529/biophysj.108.133421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi L, Quick M, Zhao YF, Weinstein H, Javitch JA. The mechanism of a neurotransmitter: Sodium symporter - inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanner BI, Zomot E. Sodium-coupled neurotransmitter transporters. Chem Rev 108:1654–1668. Physiol Rev. 2008;83:475–579. doi: 10.1021/cr078246a. [DOI] [PubMed] [Google Scholar]
- 21.Luecke H, Richter HT, Lanyi JK. Proton transfer pathways in bacteriorhodopsin at 2.3 Angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 22.Seal RP, Leighton BH, Amara SG. A model for the topology of excitatory amino acid transporters determined by the extracellular accessibility of substituted cysteines. Neuron. 2000;25:695–706. doi: 10.1016/s0896-6273(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 23.Leighton BH, Seal RP, Watts SD, Skyba MO, Amara SG. Structural rearrangements at the translocation pore of the human glutamate transporter, EAAT1. J Biol Chem. 2006;281:29788–29796. doi: 10.1074/jbc.M604991200. [DOI] [PubMed] [Google Scholar]
- 24.Tao Z, Zhang Z, Grewer C. Neutralization of the aspartic acid residue Asp-367, but not Asp-454, inhibits binding of Na+ to the glutamate-free form and cycling of the glutamate transporter EAAC1. J Biol Chem. 2006;281:10263–10272. doi: 10.1074/jbc.M510739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 26.Canutescu AA, Shelenkov AA, Dunbrack RL. 20030 A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 28.Scott WRP, et al. The GROMOS biomolecular simulation program package. J Phys Chem A. 1999;103:3596–3607. [Google Scholar]
- 29.Darden T, York D, Pedersen L. Particle mesh Eward: An N. log(N) method for Eward sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 30.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 31.Koch HP, Brown RL, Larsson HP. The glutamate-activated anion conductance in excitatory amino acid transporters is gated independently by the individual subunits. J Neurosci. 2007;27:2943–2947. doi: 10.1523/JNEUROSCI.0118-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leary GP, Stone EF, Holley DC, Kavanaugh MP. The glutamate and chloride permeation pathways are colocalized in individual neuronal glutamate transporter subunits. J Neurosci. 2007;27:2938–2942. doi: 10.1523/JNEUROSCI.4851-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.