Abstract
Aptamers are nucleic acid molecules that have been selected in vitro to bind to their molecular targets with high affinity and specificity. Typically, the systematic evolution of ligands by exponential enrichment (SELEX) process is used for the isolation of specific, high-affinity aptamers. SELEX, however, is an iterative process requiring multiple rounds of selection and amplification that demand significant time and labor. Here, we describe an aptamer discovery system that is rapid, highly efficient, automatable, and applicable to a wide range of targets, based on the integration of magnetic bead-based SELEX process with microfluidics technology. Our microfluidic SELEX (M-SELEX) method exploits a number of unique phenomena that occur at the microscale and implements a design that enables it to manipulate small numbers of beads precisely and isolate high-affinity aptamers rapidly. As a model to demonstrate the efficiency of the M-SELEX process, we describe here the isolation of DNA aptamers that tightly bind to the light chain of recombinant Botulinum neurotoxin type A (with low-nanomolar dissociation constant) after a single round of selection.
Keywords: microchannel, recombinant Botulinum neurotoxin type A, systematic evolution of ligands by exponential enrichment
Aptamers are nucleic acid molecules—either RNA or DNA—that have been selected in vitro to bind to molecular targets with high affinity and specificity (1). Since their initial description (2, 3), aptamers have been discovered for a wide variety of molecular targets including small molecules (2), proteins (3–5), cell surfaces (6), whole organisms (7), and inorganic materials (8), and they have matured as a useful tool for disease diagnostics and therapeutics as well as basic research (9–14). Once the nucleic acid sequence of the aptamer is identified for a particular target, it is produced synthetically—a distinct advantage over traditional affinity reagents such as antibodies, which require biological processes (e.g., hybridoma). In addition, many aptamers can be chemically modified to further enhance their biochemical stability (15), or altered to yield variants that undergo reversible ligand-dependent folding to provide molecular-recognition functionalities (16).
Typically, aptamers are generated through systematic evolution of ligands by exponential enrichment (SELEX), an iterative process of binding, separation, and nucleic acid amplification (1, 2). Multiple rounds of selection (typically 8–15) are generally necessary to isolate aptamers with sufficient specificity and binding affinity (e.g., nanomolar dissociation constant (Kd) for proteins and micromolar for small molecules), which therefore requires significant resources and time (17). To accelerate this lengthy discovery process, a wide variety of molecular separation techniques—beyond traditional filter-binding assays (18) and affinity chromatography (19)—have been explored as means to enhance the efficiency of aptamer selection, including surface plasmon resonance (20), flow cytometry (21), and capillary electrophoresis (CE) (22–25). In particular, CE-based separation methods have shown remarkable selection efficiencies for protein targets that exhibit significant shifts in the electrophoretic mobility (μe) upon binding to aptamers; protein-binding aptamers with Kd in the low-nanomolar range have been isolated after a small number (≈1–4) of selection rounds (22–25). However, the method is less effective for other classes of targets that do not induce sufficient μe shift of aptamers (e.g., small molecules and cell surfaces), or those that cannot be accommodated in the separation apparatus (e.g., whole organisms). On the other hand, magnetic bead-based selection methods have been shown to be effective for virtually any class of molecular targets that can be immobilized on bead surfaces, including small molecules (26), proteins (27, 28), and cell surfaces (21, 29). However, the efficiency of magnetic selection techniques has lagged far behind what can be achieved by CE and other advanced molecular separation methods, and most current methods require multiple time-consuming selection rounds (27–33) or delicate manual manipulation of micron-sized magnetic beads (34).
In pursuit of an aptamer discovery system that is rapid, efficient, universal, and automatable, we have developed a platform that integrates magnetic bead-assisted SELEX with microfluidics technology, and we demonstrate here the capability of this microsystem to isolate high-affinity aptamers after a single round of selection. This chip-based microfluidic SELEX (M-SELEX) system takes advantage of unique phenomena that occur at the microscale to achieve exceptional selection efficiencies. First, the device is capable of reproducibly generating highly localized magnetic field gradients via ferromagnetic patterns imbedded in the microchannel, enabling it to precisely and automatically manipulate small numbers of beads. Second, the negative effects of molecular diffusion on purity are minimized through the use of a multistream, laminar-flow fluidic architecture within the device microchannel. As a model to demonstrate the efficiency of M-SELEX system, we describe here the isolation of DNA aptamers against the light chain of recombinant Botulinum neurotoxin type A (BoNT/A-rLc, 50 kDa, pI 8.7). Because the target is the enzymatic component of one of the most potent toxins known to humankind (35), it has obvious importance for biological research as well as biodefense, cosmetic, and therapeutic applications (36).
Results
Theoretical Framework of High-Stringency Selection.
It has been theoretically shown that selection conditions that promote highly stringent competitive binding yield aptamers with high affinities (37). Such conditions may be created by exposing a very small quantity of target molecules to the oligonucleotide library during incubation. In this case, within the limits of mass-action kinetics, the average value of Kd of the aptamers that bind to the target can be analytically expressed as Eq. 1:
 |
where [S] is the concentration of exposed unbound target; [AT] is the total concentration of aptamers irrespective of binding state; [ATS] is the total concentration of aptamers that are bound to the target; [AT, i] is the total concentration of aptamers of type i, and Kd(i) is the dissociation constant of these aptamers. Within the limit of highly stringent competitive binding, where [S] → 0, Eq. 1 reduces to
 |
This result indeed suggests that at equilibrium, the use of smallest amount of the target during the selection leads to generation of aptamers with highest affinity. By using conventional methods, manual manipulation of a small number of target proteins and magnetic beads requires delicate handling (34). In contrast, microfluidics technology provides the means to transport small numbers of particles reproducibly and automatically for highly efficient molecular selection.
Target Protein Immobilization and Characterization.
To ensure highly competitive aptamer–target binding conditions (Eq. 2), we controlled the number of target proteins bound to the carboxylic acid-functionalized magnetic beads via carbodiimide coupling chemistry (38). In this coupling reaction, carboxylic acids are activated by the formation of a reactive N-hydroxysuccinimide (NHS) ester at low pH, proceeding through the formation of a less stable ester with 1-[3-(dimethylamino) propyl]-3-ethylcarbodiimide (EDC). The activated esters are then exposed to amino groups in the target protein (BoNT/A-rLc) at a higher pH to accelerate deprotonation of the amine, which then reacts with the NHS-ester to yield a stable amide bond. The number of target molecules per bead was ≈2 × 104 as measured by fluorescence assay [supporting information (SI) Fig. S1]. This surface density is similar to that of many membrane proteins on cell surfaces. The target proteins were verified as being immobilized in their native active conformation by measuring their enzymatic activity on the bead surface with a molecular beacon assay (Fig. S2).
Overview of the M-SELEX Process.
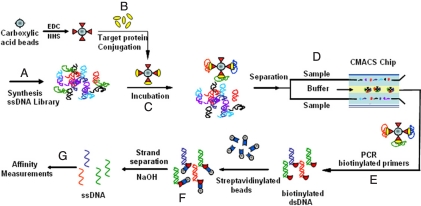
The oligonucleotide library was synthesized with ≈1014 copies of ssDNA, where each molecule contains a 60-base random region flanked by 2 20-base primer regions (Table S1). Five microliters of 100 μM library (Fig. 1A), 0.5 μL of BoNT/A-rLc-coated magnetic beads (≈2 × 1010 BoNT/A-rLc molecules on 106 beads) (Fig. 1B), and 494.5 μL of binding buffer were mixed and incubated at room temperature for 2.5 h (Fig. 1C). Before adding BoNT/A-rLc-coated beads, the DNA library in binding buffer was heated at 95 °C for 10 min, then quickly cooled down to 0 °C on ice, and subsequently incubated for 5 min at room temperature.
Fig. 1.
Overview of the M-SELEX process. (A) The starting ssDNA library consists of ≈1014 unique sequences, each containing a 60-base internal randomized region flanked by 2 20-base PCR primer-specific sequences. (B) The target protein (BoNT/A-rLc) is conjugated to the magnetic beads through carbodiimide coupling. (C) Target-conjugated beads are incubated with the heat-treated ssDNA library. (D) Aptamers that bind to the target protein are separated in the CMACS device. (E and F) The aptamers bound on the target-coated beads are amplified via PCR and single-stranded products are generated. (G) The binding kinetics of the resulting aptamers is measured.
The sample was then separated in the continuous-flow magnetic activated chip-based separation (CMACS) device (Fig. 1D) to purify the DNA bound to the BoNT/A-rLc-coated beads. The volumetric throughput was 1 mL/hr, and thus it took ≈30 min to complete the separation. The aptamers bound to the target-coated beads were collected at the Product outlet of the device (Fig. 1D) and were amplified via PCR with Alexa Fluor-labeled forward primer and a biotin-labeled reverse primer (Fig. 1E). The unbound oligonucleotides were eluted through the Waste outlet of the device. After PCR, fluorescently labeled ssDNAs were isolated by incubating the PCR amplicons with streptavidin-coated magnetic beads and eluting the nonbiotinylated strand with 15 mM NaOH, followed by neutralization with 1 M HCl (Fig. 1F).
Design of the Microfluidic Separation Chip.
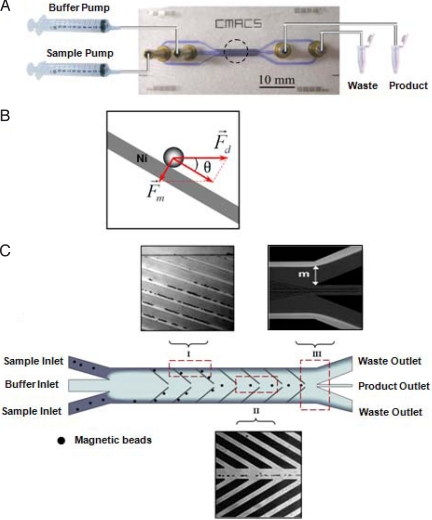
The CMACS device was designed and fabricated to perform the molecular partitioning step of the M-SELEX process (Fig. S3). This device utilizes multistream laminar fluidic architecture (39) to achieve extremely high molecular partition efficiencies (23) and uses microfabricated nickel ferromagnetic structures (40) to reproducibly create highly localized magnetic field gradients within the microchannel that allow efficient manipulation of a small number of magnetic particles. The device has 2 inlets (Sample and Buffer) that are driven by 2 independently controlled syringe pumps, and 2 outlets (Product and Waste) (Fig. 2A). When magnetic beads with immobilized target proteins and bound aptamers travel through the CMACS device, a combination of hydrodynamic (Fd) and magnetophoretic forces (Fm) is imposed on them (Fig. 2B). Assuming spherical geometry of the bead, Fd can be approximated by Stokes' law as Fd = 6πη·Rp·v (where η is the viscosity of the medium, Rp is the bead radius, and v is the velocity). On the other hand, because the beads are superparamagnetic, Fm can be approximated as Fm = msat∇B (where msat is the saturated magnetization of the bead, and B is the magnetic flux density). When an external magnetic field is applied to the CMACS device, the differences in magnetic permeability between the nickel structures (μNickel = 600μ0) and the buffer (μbuffer = ≈μ0) automatically creates a large magnetic field gradient at their interface, generating Fm that attracts the magnetic beads to the edges of the nickel structures [Fig. 2C (I)]. The CMACS device is designed to operate such that Fm exceeds the component of Fd perpendicular to the nickel strips (i.e., Fm > Fd·sinθ). Therefore, magnetic beads bound to the desired aptamers are selectively transported from the sample stream into the buffer stream by traveling along the nickel structures [Fig. 2c (I)]. After the magnetic particles reach the buffer stream at the center of the device [Fig. 2C (II)], they are eluted through the Product outlet. In contrast, unbound oligonucleotides follow the sample stream and are eluted through the Waste outlet [Fig. 2C (III)]. Because the nickel structures are passivated with a thin silicon dioxide layer, there is no direct contact between the nickel structures and the fluids.
Fig. 2.
Design and physics of the CMACS device. (A) Photograph of the CMACS device with the various inlets and outlets labeled. The inputs are driven by 2 independently controlled syringe pumps. An external magnet is placed in the dotted area to magnetize the Ni strips embedded within the microchannel. A blue food dye was used to enhance the visibility of the microchannel. (B) Schematic of the hydrodynamic (Fd) and magnetophoretic (Fm) forces on the magnetic particle in the device, relative to the nickel patterns in the CMACS microchannel. (C) Schematic of the flow pattern and deflection of magnetic beads within the microchannel. The laminar flow architecture prevents the sample and the buffer streams from mixing. Target protein-coated magnetic beads are selectively guided by the combined mechanical forces along the Ni strips and eluted through the product outlet, whereas unbound aptamers are directed into the waste outlets. Optical micrographs of the device in operation near the inlet ends (I) and near the outlet ends (II) demonstrate the partition process of the magnetic beads. The nickel lines show bright color in (I) because of the reflection of the light by the magnets. The flow pattern of the sample stream at the outlet (III) is demonstrated by adding fluorescein into the sample. The distance m is controlled to prevent diffusion of unbound aptamers into the product channel.
DNA Separation Performance of the CMACS Device.
Partition efficiency (PE) is widely used as a benchmark for molecular separation methods (9). In the CMACS device, we define PE as the ratio between the amount of DNA collected through the Waste outlet compared with those collected through Product outlet, when a standard number of beads are processed without any immobilized target proteins (to account for any nonspecially bound DNA to the beads). Thus, larger PE values indicate higher purity separation. We measured the PE of CMACS by quantifying the amount of DNA in the Waste and Product outlets via quantitative real-time PCR (Fig. S4). The CMACS device was operated under typical conditions, in which buffer was pumped at 5 mL/hr and samples containing 1 μM ssDNA and magnetic beads (2 × 106 beads per milliliter) were pumped at 1 mL/hr. The measured PE of CMACS was (1.4 ± 0.6) × 106, which significantly exceeded most conventional separation methods (e.g., filters and columns) and is comparable with that of capillary electrophoresis (23).
This high-purity molecular separation is enabled by the multiple-stream laminar flow architecture of the CMACS device. More specifically, by independently controlling the flow rate of the sample and buffer streams, we precisely regulate the distance between the Product outlet and the interface of the 2 streams [denoted by m in Fig. 2C (III)] such that m is significantly larger than the diffusion length of unbound DNA (Ldiff). Here, Ldiff is approximately equal to , where DDNA is the diffusivity of the unbound DNA, and tresidence is the time for an average molecule to travel through the device. We typically operate the system such that m (> 300 μm) is much larger than Ldiff (≈4.2 μm). In this way, we effectively prevent unbound DNA from diffusing into the Product outlet and thereby achieve high PE reproducibly without any washing steps.
Affinities of Aptamers Generated in a Single Round of Selection.
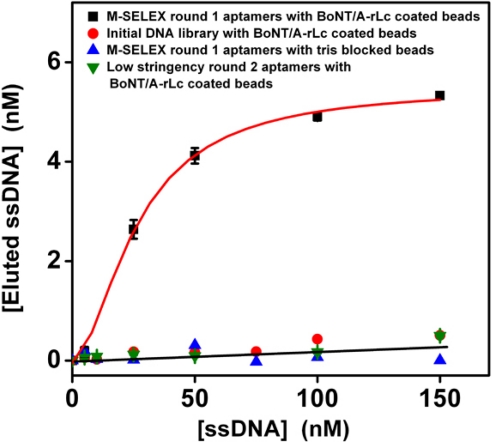
After a single round of microfluidic separation, PCR amplification and ssDNA generation, the bulk binding affinity of the enriched aptamer pool was subsequently measured via a fluorescence binding assay (33). As expected, the initial library showed no significant binding affinity to the target protein (Fig. 3, red circles. In contrast, assuming a 1:1 Langmuir binding model, the Kd of the enriched aptamer pool after a single round of M-SELEX was 33 ± 8 nM (Fig. 3, black squares), and it showed negligible affinity to magnetic beads without the target (Fig. 3, blue triangles). Furthermore, we note that the total amount of aptamers eluted from the beads (≈6 nM) was close to the estimated amount of bead-conjugated target used in this assay (7 ± 2 nM), suggesting that the binding is indeed specific to BoNT/A-rLc.
Fig. 3.
Affinity measurements of selected aptamer pools. Fluorescence measurements show that the average Kd of the aptamer pool selected via single-round M-SELEX is 33 ± 8 nM, assuming a Langmuir 1:1 binding model (black squares). Three different negative controls were measured: Negligible binding affinity was observed among (i) the initial library and the BoNT/A-rLc coated beads (red circles), (ii) M-SELEX-enriched aptamers and Tris-blocked beads without the target protein (blue triangles), and (iii) enriched aptamer pool after 2 rounds of low-stringency selection (green triangles).
Subsequently, individual aptamer sequences were cloned from the selected aptamer pool via insertion of aptamer DNA molecules into the pCR4-TOPO vector and transformation into competent bacterial cells. Fifteen colonies were randomly picked and sequenced, and 4 of the isolated sequences were synthesized and measured for their binding affinity to BoNT/A-rLc via surface plasmon resonance (SPR). The dissociation constants of the 4 aptamers ranged from 34 to 86 nM, which is consistent with the average Kd obtained from the selected aptamer pool. Sequences of the core regions of these 4 aptamers are listed in Table 1. We modeled the secondary structures of these 4 aptamers using mfold software (41) and found that the consensus regions for the 4 sequences form small protruding loops and stems, which presumably make them more accessible to target binding.
Table 1.
Sequences of the core regions of selected aptamers
| Aptamer ID | Core region of the selected aptamers | Kd, nM |
|---|---|---|
| a | 5′-TCTTTCATACCGAGGTTGATCCAGTAGTATTCTTATCTAATTTGTTTTGTTCGTATGTGC-3′ | 65 |
| b | 5′-TGGTCTTGTCTTTCATTCCAGGTGGCGAGTCAAATCGATACTGATCCGCTGACACAGGAT-3′ | 86 |
| c | 5′-CTTGAGTGTCATGGACGTTCCGGTCTTGGGCGGGATATTTGTTTGTTTTCTGCCTATGTT-3′ | 34 |
| d | 5′-TCAGATGGCCGGACTCAGGCACTAGACCACTATGCTTCCGTTGCCATTCATCGGCACGT-3′ | 53 |
Discussion
In this work, we report a microfluidics-based SELEX strategy that we believe represents a promising step toward the development of an efficient and automatable system for the rapid generation of affinity reagents. As a test of our M-SELEX method, we have demonstrated the isolation of DNA aptamers against BoNT/A-rLc protein with low-nanomolar Kd after a single round of selection. This notable efficiency of M-SELEX is achieved through the convergence of several enabling factors. First, because oligonucleotides are negatively charged, we have greatly suppressed nonspecific binding to magnetic bead surfaces by using carboxylic acid-coated beads. During the carbodiimide immobilization of the target protein, we purposely left ≈50% of the carboxylic acid groups on the bead surface unreacted, and the resulting negative charge dramatically inhibited nonspecific binding of DNA via electrostatic interactions during the SELEX process (Fig. 3).
Second, the molecular separation performance of the CMACS device benefited significantly from a number of physical phenomena at the microscale that would be difficult to achieve with conventional macroscale technologies. For example, the laminar flow characteristics of the CMACS device (Reynolds number ≈0.1) enabled the establishment of a multistream fluidic architecture that gave rise to exceptionally high-purity [PE ≈ (1.4 ± 0.6) × 106]. In addition, the microfabrication techniques allowed the construction microchannel with imbedded ferromagnetic features that enabled precise control over magnetophoretic and fluidic forces that govern the separation process; the micropatterned ferromagnetic structures generated high B field gradients (≈104 T/m) and magnetophoretic forces (≈nN) that were appropriately balanced against the hydrodynamic forces (≈100 pN) to precisely and reproducibly manipulate very small quantities of magnetic particles and target proteins (≈33 fmol) with high purity, recovery, and throughput during the separation step.
This capability is critical, because it significantly influences the stringency of selection. Conventional SELEX methods typically require multiple rounds of selection (usually 8–15 rounds) to generate high-affinity binders because of the nonstringency of target concentration used in the first binding step and the low partition efficiencies of subsequent separation steps. In the M-SELEX process, we make use of higher molar ratios between the DNA library and protein targets (Rlibrary/target) to yield higher-efficiency selection. For example, in conventional SELEX, Rlibrary/target ranges between 10 and 1,000 (2, 3, 33, 42). In contrast, methods that used higher Rlibrary/target, such as CE-based SELEX (22, 23) or Monolex (19), require significantly fewer rounds of selection, and their Rlibrary/target values range from 5 × 104 to 2.3 × 109. In this M-SELEX experiment, the Rlibrary/target value was 1 × 106, which yielded aptamers with low-nanomolar affinity after a single round of selection. It is interesting that when we performed 2 rounds of selection against the same target under otherwise similar conditions at a ≈6,500-fold lower stringency (Rlibrary/target ≈ 150), the resulting aptamers showed negligible binding affinities for the protein target (Fig. 3, green triangles).
Although the selection efficiency of M-SELEX is extraordinary, the approach has the drawback of requiring target molecule immobilization on a supporting bead. In contrast, alternative separation methods such as CE have been demonstrated to support highly efficient, label-free SELEX (12–14). It is important to note, however, that electrophoretic separation methods such as CE are limited to targets that can cause significant mobility shift of aptamers upon their binding. CE typically requires careful tuning depending on the specific μe of each target–apamer complex and is thus not suitable for targets that do not cause significant changes in μe, such as small molecules. In comparison, M-SELEX is advantageous in that it works equally well for any bead-bound targets; once a target is immobilized on the magnetic beads, the separation process and parameters (e.g., magnetic actuation and flow rates) remain the same and do not require any tuning. Because of this important property, we believe that our selection approach may be readily extended for its use with cells toward rapid generation of aptamers for cell surface markers.
In conclusion, we have demonstrated a promising path toward rapid, universal, and automatable generation of affinity reagents based on microfluidics technology. The efficiency of the M-SELEX is fueled by the convergence of multiple factors including the use of extremely small amount of target (high Rlibrary/target), utilization of electrostatic interactions to reduce nonspecific binding, and development of high-purity microfluidic separation systems. Although it is not demonstrated here, we believe that the specificity of the selected aptamers can be significantly increased via negative selection by using the same devices and methods that were used for positive selection. Moreover, it may prove interesting to consider further miniaturization and integration of other functions of molecular selection processes into a microfluidic chip, toward the development of an automated microsystem capable of generating affinity reagents “on-demand.”
Materials and Methods
Library Preparation and Target Molecule Immobilization.
The DNA library was synthesized and purified by Integrated DNA Technologies. Each member of the library contains a central random region of 60 bases flanked by 2 specific 20-base sequences (Table S1) that function as primer-binding sites for PCR. One hundred microliters of Dynabeads M-270 carboxylic acid-terminated magnetic beads (Invitrogen Dynal) were washed twice with 100 μL of 25 mM 2-(N-morpholino)ethane sulfonic acid (Mes) (pH 5.0) for 10 min. 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) (50 mg/mL) and 50 mg/mL N-hydroxysuccinimide (NHS) were freshly prepared in cold 25 mM Mes (pH 5.0). Fifty microliters of EDC solution and 50 μL of NHS solution were added to the washed beads, which were mixed well and then incubated with slow rotation at room temperature for 30 min. After incubation, the activated beads were washed 4 times in a magnetic particle concentrator (MPC) separator (Invitrogen Dynal) for 2 min, and the supernatant was removed. Solution containing 1 μg of target protein BoNT/A-rLc in reconstitution buffer (List Biological Laboratories), and 25 mM Mes (pH 5.0) was added to the beads to a final volume of 100 μL. BoNT/A-rLc is known to be quite stable in aqueous solutions and retains its activity for at least 6 months at −20 °C. This mixture was then incubated at room temperature for 2 h with slow rotation. After incubation, the unbound BoNT/A-rLc was removed by MPC separator. The protein-coated beads were then incubated with 50 mM Tris (pH 7.4) for 15 min to quench the unreacted carboxylic acid groups, then washed 4 times with 100 μL of PBS buffer containing 0.1% Tween-20 and finally stored at 4 °C in the same buffer.
Selection of Aptamers.
Five-hundred picomoles of ssDNA library (≈1014 copies) in 494.5 μL of 1× binding buffer [20 mM Hepes, 150 mM NaCl, 2 mM KCl, 2 mM MgCl2, 2 mM CaCl2 (pH 7.4)] without glycerol were heated at 95 °C for 10 min, rapidly cooled for 5 min in an ice bath, and then incubated for another 5 min at room temperature. BoNT/A-rLc-coated beads (1 × 106) were then added and incubated with the library for 2.5 h at room temperature. After incubation, 50 μL of glycerol was added into the mixture to prevent bead precipitation during the separation step and was then separated in the CMACS device. The aptamers bound to target-coated beads were collected at the Product outlet in a final volume of ≈680 μL.
Pilot PCR.
We prepared a PCR mixture containing 25 μL of HotStarTaq Master Mix (Qiagen), 0.25 μL of 100 μM forward primer (FP), 0.25 μL of 100 μM biotinylated reverse primer (RP-Biotin), and 24.5 μL of DNA sample collected from CMACS. After preheating at 95 °C for 15 min, samples were subjected to 35 thermal cycles of heating at 95 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s. Five microliters of PCR mixture were removed during the elongation step at specific cycles and stored on ice until all samples were collected. The optimal PCR amplification cycle number was determined by resolving PCR products on a 10% PAGE-TBE gel.
ssDNA Generation.
The collected aptamer pool was amplified via PCR at the optimized cycle number determined by the pilot PCR described above by using the forward primer (either FP, FP-Alexa or FP-A24) and biotinylated reverse primer (RP-Biotin). The biotin-labeled double-stranded (ds) PCR product was incubated with Dynabeads MyOne Streptavidin C1 (Invitrogen Dynal) for 2 h at room temperature to separate the dsDNA from the PCR mixture, according to the manufacturer's protocol. ssDNA was then generated in solution by incubating the beads with 15 mM NaOH for 4 min at room temperature. The supernatant was collected and neutralized with 1 M HCl. The amount of ssDNA in solution was determined at 260 nm via UV-vis absorption. The purity of the ssDNA was examined on a 10% PAGE-TBE Urea gel.
Cloning, Sequencing, and Structure Analysis of Selected Aptamers.
One percent of the collected aptamer pool was PCR amplified with unlabeled forward and reverse primers at the optimized cycle number determined by the pilot PCR. The PCR products were purified by using the MiniElute PCR Purification kit (Qiagen) and cloned with the TOPO TA Cloning kit (Invitrogen). Fifteen colonies were randomly picked, purified, and sequenced. Plasmids were purified by using the QIAprep Spin Miniprep kit from Qiagen and sequenced at the University of California, Berkeley, DNA Sequencing Facility. Secondary structure analysis was performed with mfold software (www.idtdna.com/Scitools/Applications/mFold/).
Determination of Kds.
The Kd of the selected aptamer pool was determined optically. The Alexa Fluor 488N-labeled aptamer pool was diluted to several different concentrations (from 0 to 200 nM) in 100 μL of 1× binding buffer; these dilutions were heated at 90 °C for 10 min, rapidly cooled down to 0 °C in an ice bath, and then incubated for another 10 min at room temperature. BoNT/A-rLc-coated beads (1 × 108) were washed in 1× binding buffer and resuspended in the heat-treated aptamer solution and then incubated at room temperature for 1 h. After incubation, unbound aptamers were removed by washing 4 times with 1× PBS buffer containing 0.05% Tween-20. The bound aptamers were then eluted into 100 μL of 1× PBS buffer by heating the aptamer-bound protein-coated beads at 95 °C for 8 min with shaking. The quantity of aptamers eluted from the beads was subsequently determined by fluorescence measurements (TECAN microplate reader) and calculated from a calibration curve. The amount of DNA eluted from the beads was plotted as a function of aptamer concentration. The Kd was calculated by fitting to the kinetic model, assuming Langmuir binding (1:1).
Kd was measured for the cloned aptamers by using surface plasmon resonance (SPR) (BIAcore 3000; GE Healthcare). First, ≈1,000 RU of biotinylated oligo(dT)24 were attached to the reference and sample channels of the streptavidin (SA)-coated SPR chip surface by injecting 10 μL of 1 μM oligo(dT)24 solution in the streptavidin–biotin binding buffer [1 M NaCl, 5 mM Tris·HCl, 0.5 mM EDTA (pH 7.5)] at a flow rate of 5 μL/min. The free binding sites were subsequently blocked with biotinylated polyethylene glycol (PEG). Synthesized aptamers, modified with 24-mer poly(A) oligonucleotides at the 5′ end, were injected into the sample channel at a flow rate of 20 μL/min for 2.5 min to hybridize with the complementary biotinylated oligo(dT)24 on the SA chip, resulting in an ≈700-RU increase. A spacer was included between the poly(A) and the aptamer sequence to avoid overlapping the hybridizing region with the aptamer-binding site. Similarly, the initial library was immobilized on the reference channel as a negative control, with a similar RU increase. To improve the DNA hybridization efficiency, samples were pretreated under basic conditions to destroy their secondary structures. More specifically, DNAs were mixed with 20% formamide, 0.3 M NaOH in 1× PBS buffer to a final concentration of 50 nM and incubated at 42 °C for 30 min. After incubation, the solution was neutralized with 1 M HCl. Finally, BoNT/A-rLc in 1× binding buffer from low to high concentrations (5–200 nM) was sequentially injected into both the reference and sample flow cells at a rate of 20 μL/min for 3 min. The Kd was calculated by fitting to the Langmuir kinetic model.
Supplementary Material
Acknowledgments.
We thank Monte J. Radeke for help with real-time PCR experiments, Prof. Kevin Plaxco for his careful reading of the manuscript, and Prof. Patrick Daugherty for the use of the TECAN microplate reader and SPR BIAcore 3000. We gratefully acknowledge support from the University of California Directed Research and Development Program from Lawrence Livermore National Laboratories (Grant 8-594100-69758), U.S. Army Research Office Institute for Collaborative Biotechnologies Grant DAAD1903D004, and Defense Advanced Research Projects Agency/Defense MicroElectronics Activity–Center for Nanoscience Innovation for Defense Grant H94003-05-2-0503.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813135106/DCSupplemental.
References
- 1.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment—RNA ligands to bacteriophage-T4 DNA-polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 5.Agresti JJ, Kelly BT, Jäschke A, Griffiths AD. Selection of ribozymes that catalyse multiple-turnover Diels–Alder cycloadditions by using in vitro compartmentalization. Proc Natl Acad Sci USA. 2005;102:16170–16175. doi: 10.1073/pnas.0503733102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris K, Jensen K, Julin C, Weil M, Gold L. High affinity ligands from in vitro selection: Complex targets. Proc Natl Acad Sci USA. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldheim DL, Eaton BE. Selection of biomolecules capable of mediating the formation of nanocrystals. Am Chem Soc Nano. 2007;1:154–159. doi: 10.1021/nn7002019. [DOI] [PubMed] [Google Scholar]
- 9.Ravelet C, Grosset C, Peyrin E. Liquid chromatography, electrochromatography and capillary electrophoresis applications of DNA and RNA aptamers. J Chromatogr A. 2006;1117:1–10. doi: 10.1016/j.chroma.2006.03.101. [DOI] [PubMed] [Google Scholar]
- 10.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: An emerging class of therapeutics. Annu Rev Med. 2005;56:555–558. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 11.Tombelli SMM, Mascini M. Aptamers-based assays for diagnostics, environmental and food analysis. Biomol Eng. 2007;24:191–200. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Willner I, Zayats M. Electronic aptamer-based sensors. Angew Chem Int Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 13.Shamah SM, Healy JM, Cload ST. Complex target SELEX. Acc Chem Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 14.Ulrich H, et al. DNA and RNA aptamers: From tools for basic research towards therapeutic applications. Comb Chem High Throughput Screen. 2006;9:619–632. doi: 10.2174/138620706778249695. [DOI] [PubMed] [Google Scholar]
- 15.Kirby R, et al. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal Chem. 2004;76:4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Ed. 2006;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 17.Bowser MT. SELEX: Just another separation? Analyst. 2005;130:128–130. doi: 10.1039/b412492h. [DOI] [PubMed] [Google Scholar]
- 18.Gopinath SCB. Methods developed for SELEX. Anal Bioanal Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- 19.Nitsche A, et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotech. 2007;7:1–12. doi: 10.1186/1472-6750-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misono TS, Kumar PKR. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal Biochem. 2005;342:312–317. doi: 10.1016/j.ab.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels—Selective targeting of endothelial regulatory protein pigpen. J Biol Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 22.Mendonsa SD, Bowser MT. In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 23.Berezovski M, et al. Nonequilibrium capillary electrophoresis of equilibrium mixtures: A universal tool for development of aptamers. J Am Chem Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 24.Drabovich A, Berezovski M, Krylov SN. Selection of smart aptamers by equilibrium capillary electrophoresis of equilibrium mixtures (ECEEM) J Am Chem Soc. 2005;127:11224–11225. doi: 10.1021/ja0530016. [DOI] [PubMed] [Google Scholar]
- 25.Mann D, Reinemann C, Stoltenburg R, Strehlitz B. In vitro selection of DNA aptamers binding ethanolamine. Biochem Biophys Res Commun. 2005;338:1928–1934. doi: 10.1016/j.bbrc.2005.10.172. [DOI] [PubMed] [Google Scholar]
- 26.Berezovski M, Musheev M, Drabovich A, Krylov SN. Non-SELEX selection of aptamers. J Am Chem Soc. 2006;128:1410–1411. doi: 10.1021/ja056943j. [DOI] [PubMed] [Google Scholar]
- 27.Bruno JG, Kiel JL. Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. BioTechniques. 2002;32:178–180. doi: 10.2144/02321dd04. [DOI] [PubMed] [Google Scholar]
- 28.Murphy MB, Fuller ST, Richardson PM, Doyle SA. An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res. 2003;31:3185–3193. doi: 10.1093/nar/gng110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shangguan D, et al. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. J Proteome Res. 2008;7:2133–2139. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 30.Cox JC, Rudolph P, Ellington AD. Automated RNA selection. Biotechnol Prog. 1998;14:845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 31.Cox JC, Ellington AD. Automated selection of anti-protein aptamers. Bioorg Med Chem. 2001;9:2525–2531. doi: 10.1016/s0968-0896(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 32.Cox JC, et al. Automated selection of aptamers against protein targets translated in vitro: From gene to aptamer. Nucleic Acids Res. 2002;30:e108. doi: 10.1093/nar/gnf107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoltenburg R, Reinemann C, Strehlitz B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal Bioanal Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- 34.Tok JB-H, Fischer NO. Single microbead SELEX for efficient ssDNA aptamer generation against botulinum neurotoxin. Chem Commun. 2008;16:1883–1885. doi: 10.1039/b717936g. [DOI] [PubMed] [Google Scholar]
- 35.Attygalle D, Karalliedde L. Unforgettable tetanus: A review. Eur J Anaesthesiol. 1997;14:122–133. doi: 10.1046/j.1365-2346.1997.00105.x. [DOI] [PubMed] [Google Scholar]
- 36.Bigalke H, Shoer LF, Aktories K, Just I. Handbook of Experimental Pharmacology; Bacterial Protein Toxins. Berlin: Springer; 2000. pp. 406–407. [Google Scholar]
- 37.Levine HA, Nilsen-Hamilton M. A mathematical analysis of SELEX. J Comput Biol Chem. 2007;31:11–35. doi: 10.1016/j.compbiolchem.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermanson GT. Bioconjugate Techniques. San Diego: Academic Press; 1996. pp. 146–147. [Google Scholar]
- 39.Hu XY, et al. Marker-specific sorting of rare cells using dielectrophoresis. Proc Natl Acad Sci USA. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inglis DW, Riehn R, Austin RH, Sturm JC. Continuous microfluidic immunomagnetic cell separation. Appl Phys Lett. 2004;85:5093–5095. [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golden MC, Collins BD, Willis MC, Koch TH. Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. J Biotechnol. 2000;81:167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.