Abstract
We analyzed the distribution of the cauliflower mosaic virus (CaMV) aphid transmission factor (ATF), produced via a baculovirus recombinant, within Sf9 insect cells. Immunogold labeling revealed that the ATF colocalizes with an atypical cytoskeletal network. Detailed observation by electron microscopy demonstrated that this network was composed of microtubules decorated with paracrystalline formations, characteristic of the CaMV ATF. A derivative mutant of the ATF, unable to self-assemble into paracrystals, was also analyzed. This mutant formed a net-like structure, with a mesh of four nanometers, tightly sheathing microtubules. Both the ATF– and the derivative mutant–microtubule complexes were highly stable. They resisted dilution-, cold-, and calcium-induced microtubule disassembly as well as a combination of all three for over 6 hr. CaMV ATF cosedimented with microtubules and, surprisingly, it bound to Taxol-stabilized microtubules at high ionic strength, thus suggesting an atypical interaction when compared with that usually described for microtubule-binding proteins. Using immunofluorescence double labeling we also demonstrated that the CaMV ATF colocalizes with the microtubule network when expressed in plant cells.
Keywords: aphid transmission, microtubule-binding protein, microtubule-associated-protein, cytoskeleton
Cauliflower mosaic virus (CaMV) was the first DNA plant virus to be reported (1); it is the type member of the genus Caulimovirus and is characterized by icosahedral particles 50 nm in diameter. The complete genome is a double-stranded circular DNA of ≈8000 bp encoding at least six genes which are believed to be independently translated from a full genome length polycystronic 35S RNA (for a review, see ref. 2).
CaMV is transmitted between host plants by several aphid species in a noncirculative manner (ref. 3; for a review about plant virus vector transmission, see refs. 4 and 5). After acquisition from an infected plant, the virus does not circulate within the vector’s body and is retained for a relatively short time (few hours), probably on the cuticular lining of the aphid feeding apparatus. During subsequent feeding on a healthy plant, the virus is released to initiate a new infection. Lung and Pirone (6, 7) demonstrated that aphid transmission is regulated by an aphid transmission factor (ATF) that has been identified as the expression product of CaMV gene II (8–11). The ATF is a nonstructural polypeptide of 18 kDa (P18) and appears to have no other function in the CaMV life cycle. Indeed, isolate CM4-184, a naturally occurring mutant which completely lacks gene II, is infectious in host plants but is not aphid transmissible (12).
Gene II of an aphid-transmissible CaMV isolate has been expressed to produce P18 in Sf9 insect cells via a baculovirus recombinant (13), which is biologically active (14). P18 and the virus can be acquired separately; aphids that are first fed through a parafilm membrane with baculovirus expressed P18 become capable of transmitting the naturally non-aphid-transmissible CM4-184 isolate. Native P18 accumulates, both in baculovirus recombinant-infected Sf9 cells and in CaMV-infected plants, as a well-characterized mostly cytoplasmic paracrystalline structure, as seen by negative staining and electron microscopic observations. The paracrystals appear as rod-shaped structures of variable width and length consisting of a repetitive series of parallel layers of closely associated subunits. The interlayer periodicity and the distance between subunits is 13 nm and 4 nm, respectively, and this is true for all P18s originating from a CaMV aphid-transmissible isolate tested so far. We proposed that the paracrystals might be the storage form of the ATF within infected plant cells. Indeed, the paracrystals can be solubilized in small amounts to provide the active form of the CaMV ATF (15).
Members of several other plant virus genera are vector transmitted using a comparable “helper-dependent” molecular strategy (5). Various conflicting hypotheses can be proposed to explain the molecular mechanisms of P18 activity and, by analogy, the activity of the other ATF-like factors. (i) P18 could be a bifunctional molecule with one domain interacting with the virus particle and the other with a receptor in the vector feeding apparatus, thus reversibly bridging between the two. (ii) P18 could alternatively embed the virions, thereby creating a protective shield against proteolytic enzymes, thus maintaining the virus in an infectious form within its vector for a sufficient length of time. (iii) The virus and P18 may be released together by the vector in the new host plant. In this case, P18 might be involved in the very early stages of the new host plant infection.
All three hypotheses are consistent with a specific P18-virus particle interaction, as recently demonstrated (16). However, Schmidt et al. (16) concluded that although the P18–virion interaction was a specific requirement, alone it was not sufficient for aphid transmission to occur. They suggested that to understand the biological activity of the ATF, other P18 properties need to be elucidated. These putative properties are most likely to relate to P18–vector interaction(s), to P18–host cell interaction(s), or to both.
Here we report an additional characteristic of P18, that it exhibits the properties of a microtubule (MT)-binding protein. We demonstrate that when produced in Sf9 cells, P18 paracrystals specifically decorate MTs both in vivo and in vitro. We further characterize the interaction by showing that the MT–P18 complex is resistant to extensive dilution-, cold-, and calcium-induced MT disassembly as well as to a combination of all three. Surprisingly, the P18–MT interaction is not disrupted and occurs at high ionic strength. Finally, we show that P18 also specifically associates with plant MTs in vivo, thus demonstrating that this property is not an artifact linked to the baculovirus expression system.
MATERIALS AND METHODS
Sf9 Insect Cell Cultures and Production of P18.
We used the AcSLP10 baculovirus expression system (17) to express P18 from gene II of the aphid-transmissible CaMV isolate Cabb B-JI (18). The construction of baculovirus recombinants, as well as the conditions for insect cell cultures and insect cell infections, were previously described (19). A baculovirus recombinant producing a P18 mutant (16) was also used in this study. This P18 mutant, referred to as P18 157m, has an amino acid substitution at residue 157 from Ile to Asn. As a result of this amino acid change, P18 157m was biologically inactive; it could not form paracrystals and was no longer able to bind to virus particles.
MT Isolation.
MTs were Taxol-purified from both healthy Sf9 cells and Sf9 cells infected with a baculovirus recombinant, using a modification of the technique previously described (20), according to Cheley et al. (21).
Alternatively, MT-associated protein (MAP)-free neurotubulin was purified from fresh pig brain as described (22). Purified tubulin was cycled once more, resuspended in AB buffer (100 mM Pipes, pH 6.6/1 mM EGTA/1 mM MgCl2) at 4 mg/ml and stored as aliquots in liquid nitrogen.
Preparation of P18 Crude Extracts.
Unfortunately, we have been unable to develop a satisfactory P18 purification protocol, either from CaMV infected plants or from baculovirus recombinant-infected Sf9 cells. Due to a strong self-assembly tendency, P18 is very poorly soluble in buffers compatible with MT polymerization. The addition of 500 mM NaCl to various buffers, including AB, increased P18 solubility up to 100 μg/ml, and therefore most experiments were carried out using a crude salt-soluble extract from infected Sf9 cells as the P18 source. Sf9 cells (2 × 107) infected for 2 days were submitted to a freeze/thaw cycle at −20°C in 10 ml solubilizing buffer (S buffer, 500 mM NaCl in AB buffer) and centrifuged at 100,000 × g for 1 hr. The resulting supernatant will hereafter be referred to as the salt-soluble P18 extract. All attempts at concentrating or desalting P18 resulted in aggregation and precipitation of the protein, and all attempts at increasing P18 solubility using ionic detergents or chaotropic agents resulted in the complete and irreversible loss of P18 biological activity.
Cloning of the CaMV Gene II and Its Derivative Mutant for P18 Expression in Plant Cell Protoplasts.
The expression system used is based on the tomato bushy stunt virus and was recently described elsewhere (23). Plasmid pHST2-14 is an expression cassette allowing the cloning of any foreign gene in place of the tomato bushy stunt virus coat protein which is dispensable for its replication. Gene II of the CaMV B-JI isolate and its derivative mutant were PCR amplified using AGAGAGCGGCCGCATGAGCATTACGGGTCAACCG and AGAGAGAGCTCTTAAGATTAGCCATTTTAGCC as forward and reverse primers, respectively. Such PCR products contained the complete CaMV gene II sequence flanked by NotI and SacI restriction sites that were used for insertion of the genes at the corresponding sites of the pHST2-14 expression cassette. Recombinant plasmids were designated pHST218 when bearing the CaMV gene II and pHST218m when bearing its derivative mutant. Both plasmids were linearized at the SmaI site and in vitro transcription was performed using T7 RNA polymerase (Stratagene) in the presence of m7GpppG (New England Biolabs) as a cap analogue.
Isolation and Inoculation of Protoplasts and Membrane Ghosts Preparation.
Protoplasts were isolated from Nicotiana tabacum cv. Xanthi and inoculated by electroporation as described (24) using a Bio-Rad gene pulser with 100 μl of in vitro transcription reaction mixture of either pHST2-14, pHST218, or pHST218m. After incubation of the protoplasts for 20 to 48 hr, membrane ghosts were prepared according to Marchant (25) to overcome problems linked to autofluorescence of chloroplasts.
Immunofluorescence.
The MTs and P18 associated with membrane ghosts were visualized by indirect immunofluorescence staining. Specimens were fixed for 30 min at room temperature with 3% paraformaldehyde in PBS-EGTA buffer (PBS, pH 6.9, containing 3 mM EGTA), rinsed in the same buffer, and submitted to 10-min extraction in pure methanol followed by three additional buffer rinses. Antibodies were applied in PBS-EGTA containing 0.05% Tween 20 and rinses were performed in PBS-EGTA. For double labeling of tubulin and P18 (or P18 157m), a mixture of an anti-P18 made in rabbit (14) and an anti-α tubulin made in mouse (N356, Amersham) was applied as the primary antibody. The secondary antibody solution consisted in a mixture of an anti-rabbit conjugated to fluorescein (Sigma) and an anti-mouse conjugated to rhodamine (Sigma). Samples were mounted in PBS-EGTA containing 50% glycerol (vol/vol) and observed using a Nikon fluorescence microscope.
RESULTS
P18 Decorates Sf9 MTs in Vivo.
While observing P18 distribution in baculovirus recombinant-infected Sf9 cells, we noted some abnormal changes in the cytoskeleton. Very high amounts of linear MT-like structures were scattered throughout the cytoplasm of P18 producing cells (Fig. 1a) and, to an even greater extent, of P18 157m producing cells (Fig. 2a). Though they resemble cellular MTs, the structures observed in those cells had a larger diameter (35 to 40 nm instead of 25 nm). Such MT-like elements were never observed in wild-type baculovirus-infected Sf9 cells (Fig. 1d), thus indicating that P18 (and P18 157m) might be either directly or indirectly involved in their formation. Consistent to this was the P18-specific gold labeling of infected cell thin sections demonstrating that both P18 and P18 157m colocalize with the MT-like elements (Figs. 1a and 2 a1 and d).
Figure 1.
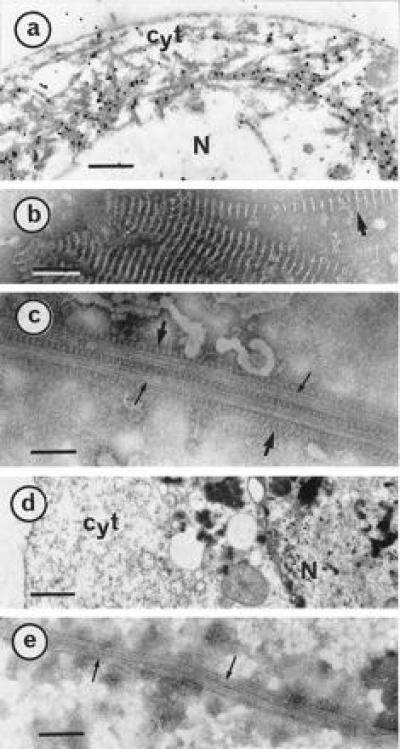
P18 paracrystals decorate MTs within Sf9 cells. Cells infected for 3 days were harvested and processed for electron microscopic observations and gold-labeling as described (15). (a) A thin section of an infected Sf9 cell was gold-labeled by a P18 antibody. As reference, negatively stained characteristic P18 paracrystals (15) are shown in b. (c) Infected cells were treated with Taxol and disrupted in AB buffer before negative staining. P18 paracrystals and well-shaped MTs are indicated with large and small arrows, respectively. (d) Thin section of a cell infected with a wild type baculovirus. (e) Negative staining of a similar cell disrupted after a Taxol treatment. Cyt, cytoplasm; N, nucleus. [Bars = 1 μm (a and d), 100 nm (b), and 75 nm (c and e).]
Figure 2.
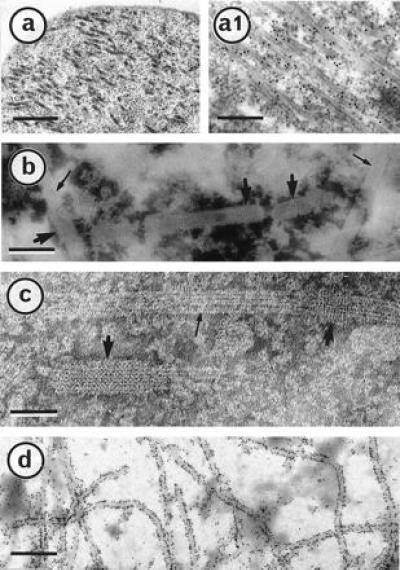
A P18 derivative mutant, P18 157m, is creating a net-like sheath on Sf9 cell MTs. Cells infected for 3 days with a baculovirus recombinant producing P18 157m were prepared for electron microscopic observations as in ref. 15. A thin section of an infected cell (a) was gold-labeled with a P18 antibody (a1). After being treated with Taxol, infected cells were disrupted in AB buffer, negatively stained, and observed (b and c). P18 157m net-like structure and well-shaped MTs are indicated by large and small arrows, respectively. (d) An infected cell was trapped on a microscope grid and gold labeled with a P18 antibody. (Bars = 500, 250, 75, 50, and 400 nm in a, a1, b, c, and d, respectively.)
To further observe these MT-like elements forming an atypical cytoskeletal network, Sf9 cells infected for 2 days with either wild-type baculovirus, P18, or P18 157m recombinant baculoviruses were incubated for an additional 24 hr in the culture medium containing 20 μM Taxol. The cells were then disrupted in AB buffer and observed after negative staining. In infected cells producing P18, the MTs were frequently decorated with characteristic P18 paracrystals that were observed surrounding MTs either completely or partially or in between two different MTs (Fig. 1c).
Consistent with previous findings (16), we did not observe P18 paracrystals in cells infected with a baculovirus recombinant encoding P18 157m. Despite this, the MTs were densely covered with a net-like sheath (Fig. 2b) with a mesh of about 4 nm (Fig. 2c). The presence of the sheath extended the MT’s diameter up to 40 nm and some discontinuities, presumably due to alteration of the sheath, were easily observable. We hypothesized that the main constituent of the sheath might be P18 157m and this was confirmed by immunogold-labeling infected Sf9 cells using an antibody to P18 (Fig. 2d).
Grids on which wild type baculovirus-infected and Taxol-treated Sf9 cells were trapped showed large amounts of MTs, none of which were sheathed nor associated with paracrystalline structures of any sort (Fig. 1e).
All together these results demonstrate that both P18 and P18 157m colocalize, in vivo, with Sf9 cell MTs.
Both P18 and P18 157m Strongly Stabilize MTs.
When insect cell baculovirus infections were extended over 5 days, examination of thin sections revealed the coexistence of intact infected cells and a graded series of more or less dramatically disrupted cells. When wild-type baculovirus-infected cells were disrupted, some nuclear structures and cytoplasmic vesicules remained but no cytoskeletal elements and particularly no MTs were present (Fig. 3a). In contrast, when infected cells were producing P18 (Fig. 3b) or P18 157m (Fig. 3c), the atypical microtubular network described above was the last remaining structure, suggesting a particularly high stability. Indeed, such a cytopathic effect was never observed with any wild-type baculovirus or other recombinant baculoviruses tested in our laboratory, whatever the multiplicity and the time of infection.
Figure 3.
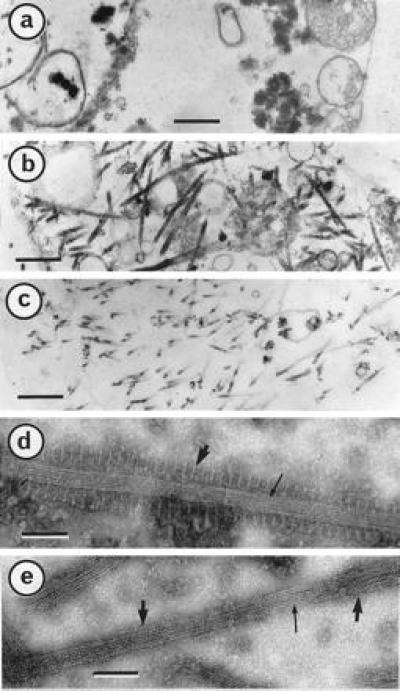
P18– and P18 157m–MT complexes are highly stable. Cells were infected for 5 days, harvested, fixed and embedded as described (15). Micrographs represent thin sections of Sf9 cells infected with wild-type baculovirus (a), recombinant producing P18 (b), and recombinant producing P18 157m (c). P18– and P18 157m–MT complexes (d and e, respectively) were extracted and submitted to MT disassembly-inducing factors as described in the text before negative staining with ammonium molybdate (15). [Bars = 500 nm (a–c) and 60 nm (d and e).]
We developed an electron microscopy-based assay to evaluate the resistance of the P18– or P18 157m-MT complexes, formed in Sf9 cells, to any one or a combination of MT depolymerization-inducing factors such as dilution, cold, and calcium. P18– or P18 157m-MT complexes were extracted from 5 × 106 Sf9 cells, infected for 2 days, in 1 ml AB under optimal conditions, including 1 mM GTP, elevated temperature, and stabilization with 20 μM Taxol. The complexes were trapped on an electron microscope grid and the frequency of their appearance—i.e., the number observed per microscopic field—was assigned a reference value of 100. Infected cells, alternatively, were disrupted in AB with varying MT-depolymerizing factors in the absence of Taxol and trapped and observed in the same way. The frequency of appearance of the complexes was scored as a value relative to the reference—i.e., between 0 and 100. The various treatments applied to the P18– and P18 157m-MT complexes are summarized in Table 1.
Table 1.
Stability of P18– and P18 157m–MT complexes
| Dilution* | Cold† | Calcium‡ | Combination§ | |
|---|---|---|---|---|
| P18–MT | ||||
| complexes observed | ||||
| 30 min | 60 | 65 | 55 | 55 |
| 6 hr | 50 | 50 | 45 | 40 |
| P18 157m–MT | ||||
| complexes observed | ||||
| 30 min | 100 | 100 | 100 | 100 |
| 6 hr | 100 | 100 | 100 | 100 |
All values are noted as relative to the value of reference as described in the text.
Infected cells were disrupted in 10 ml AB buffer for either 30 min or 6 hr and centrifuged at 10,000 × g for 5 min. Pellets were resuspended in 1 ml AB buffer before being trapped and observed.
Infected cells were disrupted in 1 ml AB buffer and maintained on ice 30 min and 6 hr before being trapped and observed.
Infected cells were disrupted in 1 ml AB buffer containing 5 mM CaCl2 for either 30 min or 6 hr before being trapped and observed.
Infected cells were disrupted in 10 ml AB buffer containing 5 mM CaCl2 and kept on ice for either 30 min or 6 hr before being trapped and observed.
Comparable amounts of P18–MT complexes resisted cold-, dilution-, and calcium-induced MT disassembly as well as a combination of all three, the relative values varying between 40 and 65. Resistant MT portions were those tightly associated with P18 paracrystals. They remained intact following a 6-hr treatment combining all three depolymerizing factors (Fig. 3d). In contrast, all portions of MT not directly in contact with a P18 paracrystal were no longer observable after such treatments.
P18 157m–MT complexes were totally resistant to cold-, dilution-, and calcium-induced MT disassembly as well as to a combination of all three for over 6 hr (Fig. 3e and Table 1).
P18 Binds to Preformed MTs at High Strength.
MTs from uninfected and infected Sf9 cells were Taxol-purified as indicated in Materials and Methods. In the case of infected cells, the very small amount of P18 and P18 157m soluble in AB buffer copurified with the MTs (data not shown).
As mentioned earlier, P18 solubility is slightly increased in the presence of 500 mM NaCl, we therefore assessed whether P18 would bind to and cosediment with MTs at high ionic strength. Because tubulin does not polymerize at high salt concentrations, preformed Taxol-treated Sf9 MTs (200 μg) were mixed with 10 ml salt-soluble P18 extract (see Materials and Methods) and gently stirred at room temperature for 20 min before centrifugation. Fig. 4a, lane 3, shows a large amount of P18 cosedimented with tubulin, suggesting that P18 binds to Taxol-treated MTs in these experimental conditions. Electron microscopic observations of the pellet contents revealed MT clusters covered with newly formed P18 paracrystals, thus confirming that P18 bound MTs at high ionic strength (Fig. 4c).
Figure 4.
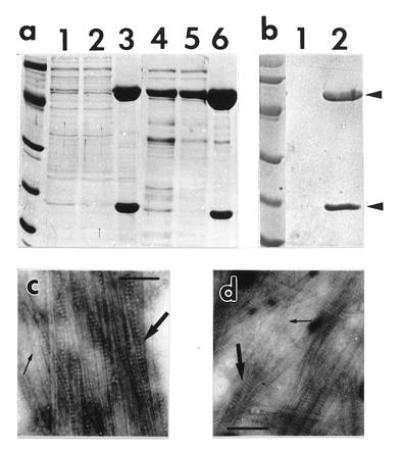
Cosedimentation of P18 and MTs at elevated ionic strength. Samples were fractionated in a SDS/12% polyacrylamide gel and stained with Coomassie blue R250 (a) or Ponceau Red (Sigma) after transfer onto a nitrocellulose membrane (b). (a) Samples were processed as described in the text, and 10 μl were loaded in each lane. A salt-soluble P18 extract mixed with Sf9 preformed MTs was loaded in lane 1, and the supernatant and pellet (resuspended in 200 μl AB) from a 15,000 × g, 20-min centrifugation through a 15% sucrose cushion made in AB were loaded in lanes 2 and 3, respectively. P18 was also cosedimented with pig brain MTs. Samples in lanes 4, 5, and 6 were prepared in the same conditions as in lanes 1, 2, and 3, except for slight modifications indicated in the text. A control experiment (b) was carried out omitting the addition of preformed MTs in the salt-soluble P18 extract, and the corresponding pellet was loaded in lane 1; lane 2 is as in lane 3 in a. P18 and tubulin are indicated with arrows. Molecular weight reference scales are phosphorylase b (94 kDa), albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa). Proteins of pellets loaded in lanes 3 and 6 in a were trapped on an electron microscope grid, negatively stained and observed (c and d, respectively). P18 paracrystals and MTs are indicated with large and small arrows. (Bars = 150 nm.)
Next we investigated whether the P18–MT complexes could be obtained at high ionic strength with tubulin from a mammal’s brain (Fig. 4a). Pure pig brain tubulin (4 mg tubulin/ml) was polymerized in AB buffer containing 1 mM GTP and 20 μM Taxol at 37°C for 1 hr. Taxol-treated MTs (2 mg) were diluted in 10 ml of salt-soluble P18 extract (in AB buffer/500 mM NaCl) and gently stirred at room temperature for 20 min. Fig. 4a, lane 6, demonstrates that a large proportion of tubulin sedimented together with P18 upon centrifugation. As with Sf9 MTs, the pellet content was observed by electron microscopy as MT aggregates embedded in P18 paracrystals (Fig. 4d). In control experiments, salt-soluble P18 extracts were processed similarly in the absence of preformed MTs; no P18 was pelleted upon centrifugation (Fig. 4b).
P18 Colocalizes in Vivo with Plant Cell MTs.
Transfection of tobacco protoplasts with transcripts of either pHST2-14, pHST218, or pHST218m was successful (rates of infection ranged from 10% to 30% of the cells as estimated by immunofluorescence) and both P18 and P18 157m could easily be detected by Western blots (data not shown).
Indirect immunofluorescence double labeling was used to visualize both P18 and tubulin as green and red fluorescence, respectively, in membrane ghosts from infected protoplasts transfected with pHST218 transcripts. Consistent with the results in insect cells, P18 distribution appeared as a filamentous network (Fig. 5b) exactly matching the tubulin distribution (Fig. 5a), thus demonstrating that P18 colocalizes in vivo with plant cell MTs. Similar results were obtained with protoplasts inoculated with pHST218m transcripts, while no green fluorescence could be detected in membrane ghosts from protoplasts inoculated with pHST2-14 which encodes neither P18 nor P18 157m (not shown).
Figure 5.
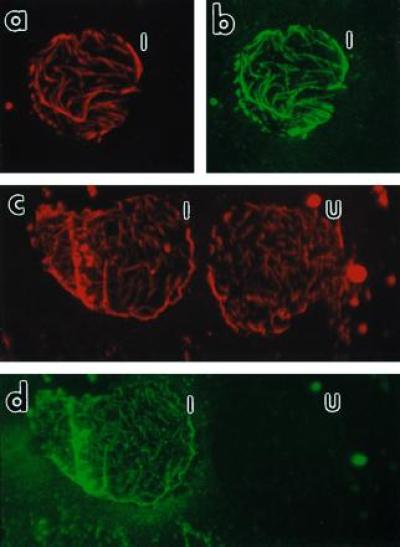
Fluorescence micrographs of Nicotiana tabacum cv. Xanthi membrane ghosts. Membrane ghosts were double labeled as indicated for visualizing both MTs (a and c) and P18 (b and d). The same membrane ghost from an infected protoplast producing P18 is shown in a with a rhodamine filter cube for visualizing the MTs and in b with a fluorescein filter cube for visualizing P18. After Taxol treatment (see text), MTs of two membrane ghosts are shown as a rhodamine fluorescence in c and the presence of P18 in only one of these cells is shown by fluorescein in d. I, infected cell; U, uninfected cell. (×400 for a–d.)
MT networks similar to that shown in Fig. 5a were very rarely observed in cells which did not produce P18 (or P18 157m) and those observed were usually greatly degraded. This indicates that P18 might also stabilize plant MTs in vivo. In contrast, when Taxol was added at a 10 mM final concentration to the protoplasts suspension 4 hr before immunofluorescence staining, the MT network was always observable in membrane ghosts whether the cell produced P18 or not. Fig. 5c shows the MT network in membrane ghosts originating from two Taxol-treated cells, only one of which was producing P18 as demonstrated by the very specific fluorescein labeling in Fig. 5d.
DISCUSSION
In contrast to animal viruses, very little is known about the relations between the host cell cytoskeleton and plant viruses. So far, plant virus cell-to-cell movement is the only function for which such relations have been documented. An early report, based on electron microscopic observations, suggested the participation of host MTs in the cell-to-cell movement of comoviruses (26). Furthermore, the beet yellows closterovirus 65-kDa protein, possibly involved in the cell-to-cell movement, binds to MTs in vitro (27), and the movement protein of tobacco mosaic virus was recently demonstrated to colocalize with the MT network in infected plant cells (28). In the present report, we describe the interaction between the CaMV ATF and the cellular cytoskeleton via an association with MTs, thus indicating that tubulin or MTs may also play a role in aphid transmission. For experimental reasons, the characterization of P18–MTs interaction was performed in insect cells, though the colocalization of P18 and plant cell MTs, in vivo, was verified and is discussed later.
When expressed in insect cells, the CaMV ATF and a derivative mutant colocalized with and decorated MTs in situ. Both the P18– and P18 157m–MT complexes, formed in vivo, appeared to be highly stable. These complexes were resistant to dilution-, cold-, and calcium-induced MT disassembly as well as to a combination of all three. Margolis and Job (29) reviewed the effect of various MT-binding proteins on the stability of the polymer. Structural MAPs are known to be factors affecting MT stability, but there are only a few polypeptides that are reported to confer a particularly high stability, comparable to that reported here for P18– and P18 157m–MT complexes. Stable tubule only polypeptides (30), myelin basic protein (31), and histone H1 (32) were reported to induce MT resistance to cold, dilution, and calcium as well as to a combination of all three. Hence, P18 and P18 157m might be considered as new candidates in the group of factors conferring “super stability” to MTs (33).
The interaction between MTs and an extensive variety of associated proteins is believed to be of an electrostatic nature and is disrupted by conditions of high ionic strength (20). Currently, histone H1 is the only described MT-binding protein that is not decisively released from MTs by salt treatment, and no experiments were reported that test whether the attachment of histone H1 onto MTs could occur at high ionic strength (32). Here we report that the P18–MT binding can occur at high ionic strength. In this respect, the behavior of P18 seems to be unique among the MT-binding proteins described so far, suggesting either that P18 has a particularly high affinity for MTs or that the interaction is not of an electrostatic nature.
Because all described MAPs are removed from MTs at high ionic strength, we can reasonably assume that they are not mediating an indirect attachment of P18 onto MTs. Furthermore, P18 and tubulin were the only major polypeptides concentrated in the pellet of the cosedimenting experiments (Fig. 4), thus suggesting that other proteins are not involved in the interaction and that P18 might bind directly to MTs. However, we cannot completely rule out the possibility of an additional component mediating the P18–MT binding. The development of a P18 purification protocol would allow a definitive answer.
The suitability of the baculovirus/insect cell system for studying the interaction between a heterologous protein and MTs has previously been demonstrated (34). Nevertheless, we decided to verify that P18 was also capable of binding to MTs from various origins, especially from plants.
CaMV isolates such as CM4-184, which exhibits a nearly complete deletion of gene II, replicate and spread perfectly in host plants. Therefore, it is obvious that P18-MT binding is involved neither in the replication cycle nor in the cell-to-cell movement, and hence the presumptive role of this binding is in the aphid transmission process. P18 has never been reported to be associated with MTs in infected plant cells. Rather, it is present in the plant cell cytoplasm as electron–lucent inclusion bodies (35), the matrices of which consist of P18 paracrystal clusters (15). Extensive electron microscopic observation of CaMV-infected leaves and P18 inclusion bodies, purified in the presence of Taxol, failed to demonstrated a P18–MT colocalization. Hence, we concluded that if P18-MT interactions occur in CaMV-infected cells, they probably do so under tight regulation. This may explain why such property has so far been overlooked and it also prompted us to assess P18–MT binding in plant cells using a system where no putative inhibition due to other CaMV gene products could occur.
The tomato bushy stunt virus expression system yielded a significant amount of P18 in tobacco protoplasts. The double-immunofluorescence labeling of P18 and tubulin demonstrated that P18 colocalizes with the MT network. Although the preparation of protoplast membrane ghosts satisfactorily overcame the problem of the autofluorescence of chloroplasts, we observed some intact cells and in these, P18 was distributed similarly (data not shown). P18 never appeared to be stored in inclusion bodies indicating that accumulation of P18 in such structures is the result of more complex interactions with other CaMV gene product(s). In the absence of Taxol treatment, most of the MT network in cells not producing P18 was disrupted during the processing of samples, while it remained intact in cells producing P18. This suggests that, as for insect cell, plant cell MTs are stabilized when complexed with P18.
From the results reported here, we can assume that binding to tubulin and/or MTs from various origins is a property of the functional CaMV ATF. We have no definitive evidence demonstrating that the P18–MT interaction is actually involved in CaMV aphid transmission. However, one might speculate about such involvement in two different ways: (i) P18 may bind to tubulin-like receptor site(s) on the cuticule of the aphid feeding apparatus, and (ii) the aphid vector could inoculate P18 along with virus particles in the new host plant where an association of P18 with the cell cytoskeleton may be involved in the very early stages of the infection, such as a virion transport to the nucleus. In any case, the putative participation of tubulin and/or MTs in noncirculative virus vector transmission is a novel concept that opens new ways for investigating the molecular mechanisms of the activity of ATF type molecules.
Acknowledgments
We thank Joanne Hay and Thomas P. Pirone for critical reading of the manuscript, and we are grateful to Mark Mattson for fluorescence microscope facilities.
Footnotes
Abbreviations: CaMV, cauliflower mosaic virus; ATF, aphid transmission factor; MT, microtubule; MAP, MT-associated protein.
References
- 1.Shepherd R J, Wakeman R J, Romanko R R. Virology. 1968;36:150–152. doi: 10.1016/0042-6822(68)90127-x. [DOI] [PubMed] [Google Scholar]
- 2.Rothnie H M, Chapdelaine Y, Hohn T. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 3.Markham P G, Pinner M S, Raccah B, Hull R. Ann Appl Biol. 1987;111:571–587. [Google Scholar]
- 4.Pirone T P. Semin Virol. 1991;2:81–87. [Google Scholar]
- 5.Pirone T P, Blanc S. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 6.Lung M C Y, Pirone T P. Phytopathology. 1973;63:912–914. [Google Scholar]
- 7.Lung M C Y, Pirone T P. Virology. 1974;60:260–264. doi: 10.1016/0042-6822(74)90383-3. [DOI] [PubMed] [Google Scholar]
- 8.Woolston C J, Covey S N, Penswick J R, Davies D W. Gene. 1983;23:15–23. doi: 10.1016/0378-1119(83)90212-3. [DOI] [PubMed] [Google Scholar]
- 9.Woolston C J, Czaplewski L G, Markham P G, Goad A S, Hull R, Davies J W. Virology. 1987;160:246–251. [Google Scholar]
- 10.Armour S L, Melcher V, Pirone T P, Lyttle D G, Essenberg J M. Virology. 1983;129:25–30. doi: 10.1016/0042-6822(83)90392-6. [DOI] [PubMed] [Google Scholar]
- 11.Givord L, Xiong C, Giband M, Koenig I, Hohn T, Lebeurier G, Hirth L. EMBO J. 1984;3:1423–1427. doi: 10.1002/j.1460-2075.1984.tb01987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howarth A J, Gardner J C, Messing J, Shepherd R J. Virology. 1981;112:678–685. doi: 10.1016/0042-6822(81)90313-5. [DOI] [PubMed] [Google Scholar]
- 13.Espinoza A M, Usmany M, Pirone T P, Harvey M, Woolston C J, Medina V, Vlak J M, Hull R. Intervirology. 1992;34:1–12. doi: 10.1159/000150257. [DOI] [PubMed] [Google Scholar]
- 14.Blanc S, Cerutti M, Usmany M, Vlak J M, Hull R. Virology. 1993;192:643–650. doi: 10.1006/viro.1993.1080. [DOI] [PubMed] [Google Scholar]
- 15.Blanc S, Schmidt I, Kuhl G, Esperandieu P, Lebeurier G, Hull R, Cerutti M, Louis C. Virology. 1993;197:283–292. doi: 10.1006/viro.1993.1589. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt I, Blanc S, Kuhl G, Esperandieu P, Devauchelle G, Louis C, Cerutti M. Proc Natl Acad Sci USA. 1994;91:8885–8889. doi: 10.1073/pnas.91.19.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaabihi H, Ogliastro M, Martin M, Giraud C, Devauchelle G, Cerutti M. J Virol. 1993;67:2664–2671. doi: 10.1128/jvi.67.5.2664-2671.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delseny M, Hull R. Plasmid. 1983;9:31–41. doi: 10.1016/0147-619x(83)90029-x. [DOI] [PubMed] [Google Scholar]
- 19.Blanc S, Cerutti M, Chaabihi H, Louis C, Devauchelle G, Hull R. Virology. 1993;192:651–654. doi: 10.1006/viro.1993.1081. [DOI] [PubMed] [Google Scholar]
- 20.Vallee R B. J Cell Biol. 1982;92:435–442. doi: 10.1083/jcb.92.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheley S, Kosik K S, Paskevich P, Bakalis S, Bayley H. J Cell Sci. 1994;102:739–752. doi: 10.1242/jcs.102.4.739. [DOI] [PubMed] [Google Scholar]
- 22.Malekzadeh-Hemmat K, Gendry P, Launay J F. Cell Mol Biol. 1993;39:279–285. [PubMed] [Google Scholar]
- 23.Scholthof H B, Scholthof K-B G, Jackson A O. Annu Rev Phytopathol. 1996;34:299–323. doi: 10.1146/annurev.phyto.34.1.299. [DOI] [PubMed] [Google Scholar]
- 24.Luciano C S, Rhoads R E, Shaw J G. Plant Sci. 1987;51:295–303. [Google Scholar]
- 25.Marchant J H. Exp Cell Res. 1978;115:25–30. doi: 10.1016/0014-4827(78)90397-x. [DOI] [PubMed] [Google Scholar]
- 26.Kim K S, Fulton J P. Virology. 1975;64:560–565. doi: 10.1016/0042-6822(75)90134-8. [DOI] [PubMed] [Google Scholar]
- 27.Karasev A V, Kashina A S, Gelfand V I, Dolja V V. FEBS Lett. 1992;304:12–14. doi: 10.1016/0014-5793(92)80578-5. [DOI] [PubMed] [Google Scholar]
- 28.Heinlein M, Epel B L, Padgett H S, Beachy R N. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 29.Margolis R L, Job D. In: Microtubules. Hyams J, Lloyd C, editors. New York: Wiley–Liss; 1994. pp. 221–228. [Google Scholar]
- 30.Pirollet F, Rauch C T, Job D, Margolis R L. Biochemistry. 1988;28:835–841. doi: 10.1021/bi00428a064. [DOI] [PubMed] [Google Scholar]
- 31.Pirollet F, Derancourt J, Haiech J, Job D, Margolis R L. Biochemistry. 1992;31:8849–8855. doi: 10.1021/bi00152a022. [DOI] [PubMed] [Google Scholar]
- 32.Multigner L, Gagnon J, Van Dorsselaer A, Job D. Nature (London) 1992;360:33–39. doi: 10.1038/360033a0. [DOI] [PubMed] [Google Scholar]
- 33.Job D, Margolis R L. Biochemistry. 1984;23:3025–3031. doi: 10.1021/bi00308a028. [DOI] [PubMed] [Google Scholar]
- 34.Kosik K S, McConlogue L. Cell Motil Cytoskeleton. 1994;28:195–198. doi: 10.1002/cm.970280302. [DOI] [PubMed] [Google Scholar]
- 35.Espinoza A M, Medina V, Hull R, Markham P G. Virology. 1991;185:337–344. doi: 10.1016/0042-6822(91)90781-6. [DOI] [PubMed] [Google Scholar]


