Abstract
The structural characterization of de novo designed metalloproteins together with determination of chemical reactivity can provide a detailed understanding of the relationship between protein structure and functional properties. Toward this goal, we have prepared a series of cyclic peptides that bind to water-soluble metalloporphyrins (FeIII and CoIII). Neutral and positively charged histidine-containing peptides bind with a high affinity, whereas anionic peptides bind only weakly to the negatively charged metalloporphyrin. Additionally, it was found that the peptide becomes helical only in the presence of the metalloporphyrin. CD experiments confirm that the metalloporphyrin binds specific cyclic peptides with high affinity and with isodichroic behavior. Thermal unfolding experiments show that the complex has ”native-like” properties. Finally, NMR spectroscopy produced well dispersed spectra and experimental restraints that provide a high-resolution solution structure of the complexed peptide.
One approach to understanding the folding and function of proteins is to attempt their design from first principles (1-3). Such design, however, requires not only incorporation of nonpolar interactions, but also inclusion of specific hydrogen bonds, disulfide bridges, ion pairs, and metal chelation. Without these interactions, proteins form compact folding intermediates referred to as molten globules (4). Molten-globule structures are compact and possess a high degree of secondary structure, but they are also highly dynamic, with both internal and external side chains rapidly interconverting among a large family of rotamers. Analysis of protein structures, on the other hand, reveals that the interior hydrophobic core is well packed and rigid. Therefore, two challenges occur in the design of ”native-like” proteins. By using positive design (3, 5-7), one can take into account issues such as hydrophobicity and the helix- or sheet-forming propensity of amino acids. By using negative design [a term developed to describe the selective introduction of unique stabilizing interactions (2, 3)], one can introduce features that stabilize one specific fold, while, at the same time, they destabilize alternative topologies. By using this dual strategy, one can increase the free-energy gap between the native and molten-globule states.
With the challenge of design making rapid progress, several groups have begun to look at the incorporation of functional cofactors in hopes of producing de novo catalytic proteins (8-14). Many enzymes require mono- and dinuclear metal ions (15), metal clusters (ferredoxin and nitrogenase) (16), heme (cytochromes and globins) (17), and organic cofactors (flavin- and quinone-based enzymes) (18). These cofactors are important, especially when processes such as oxidation/reduction or substrate binding and activation are required (15).
Studies of natural enzymes have led to considerable understanding of their structure and function. Reduction of these complicated systems to minimal models, however, will allow us to test our understanding of their functional properties and to explore environmental and therapeutic applications. The creation of new biocatalysts is a sizable challenge. The problem has been approached by a variety of methods, including de novo (”rational”) design (2, 3, 12, 14, 19), directed evolution of new catalytic processes (20-22), production of catalytic antibodies (23), and the use of phage display to identify proteins that bind to transition-state analogs (24-27). Rational design, which we use here, allows us to start with a simplified initial model that can be characterized at each stage in detail and refined accordingly.
Another step toward the creation de novo of catalytic proteins is the acquisition of high-resolution structures of the synthetic proteins. Although several such structures exist (28-31), very few have a cofactor bound to the polypeptide (1). With the confirmation of these structures in hand, it should then be possible to design features that can bind and activate small substrate molecules, such as dioxygen and hydrocarbons.
We have described the design and characterization of 15-mer, tweezers, and cyclic heme-binding peptides (32-34). In this work, we present the solution structure of a metalloporphyrin-peptide complex, designed from first principles. This complex is a unique example of a high-resolution solution structure of a metalloporphyrin cofactor bound to a designed peptide. What is perhaps most interesting about this system is that the peptide is unfolded in the absence of the porphyrin. In the presence of the porphyrin, however, the peptide is folded and rigid, with only a single, well defined conformation in solution.
Materials and Methods
UV Spectroscopy. UV-binding studies were done on a Hitachi U-3300 UV spectrometer (Hitachi, Tokyo) in a thermostated cell holder. For studies comparing charge, all measurements were made in 50 mM KH2PO4, (pH 6.8-7.5). Binding constants were determined as reported earlier (32).
CD Spectroscopy. All CD spectra were measured on a Jasco J-700 spectrapolarimeter (Jasco, Easton, MD) at 4°C. Unless otherwise specified, all measurements were made on samples whose concentration was 2-10 μM (pH 7.0, 2 mM potassium phosphate/5 mM potassium chloride). Analyses of thermal unfolding data were done with established methods (35). The chemical unfolding data were best-fit by using the nonlinear least-squares algorithms in SIGMA-PLOT to a two-state denaturation function (36).
NMR Spectroscopy. All NMR spectra were acquired at 4°C by using samples that were 3 mM in the peptide complexes of [CoIII(coproporphyrinate I)]3- (pH 7.2, 50 mM potassium phosphate/10 mM potassium chloride). All spectra were acquired in 90% H2O/10% D2O to suppress the exchange of backbone amide protons with deuterons. Suppression of the water signal was accomplished by selective excitation of the water signal during the recycling delay. NOESY spectra were acquired by using standard methods (37-39). Spectra were acquired by using mixing times ranging from 10 to 250 ms. For total correlation spectroscopy experiments (40, 41), the spin-lock power was set to 49 and mixing times to 70 ms. All other parameters were identical with those in NOESY experiments. Double-quantum-filtered COSY experiments (42) were acquired at very high digital resolution, typically with 512-1,024 increments in the F1 dimension.
Structure Calculations. Interproton distance restraints were derived from NOESY experiments with 10-, 20-, 50-, 100-, and 250-ms mixing times acquired on a Varian Innova spectrometer operating at 600 MHz. It was found that, with a mixing time of 100 ms, the nuclear Overhauser effect (NOE) intensities were still in the linear region of the build-up curve, so a two-spin approximation was applied for the generation of distance constraints (31). Peaks involving nonexchanging protons were scaled to the largest Hβ-Hβ NOE cross-peak intensity, assuming a 1.8-Å separation. Peaks with exchangeable amide protons were scaled to the largest HN(i)-HN(i+1) cross-peak intensity, assuming a distance of 2.8 Å, as in an ideal α-helix. For those cross-peaks involving methyl groups, a pseudoatom approximation was used and the NOE distance was increased 0.5 Å. Finally, the upper-bound limit for each distance constraint was increased 10%. Backbone φ-angle restraints, derived from 3JNH-CH coupling constants, were determined by using the method of Kim and Prestegard (43). Restraints were restricted to -60 ± 40 for coupling constants <7 Hz and to -120 ± 50 for coupling constants >8 Hz.
Three-dimensional structure calculations were done by using the program INSIGHT/DISCOVER (Accelrys, San Diego) with a modified consistent valence force field. An NOE-restrained, simulated annealing protocol developed by Nilges and coworkers (44-46) was used for structure calculations by starting with a randomized set of structures, heating the molecule to 2,000 K, and then slowly cooling the molecule over the course of 45 ps to 50 K. Force constants for bonds were set to 25 kcal/(mol·rad2), whereas the constants for angles were set to 100 kcal/(mol·rad2). Of the 50 structures, 75% converged to acceptable structures. Of the acceptable structures, 14 were deemed exceptional. All of these structures displayed no violations >0.5 Å or >5°.
Results and Discussion
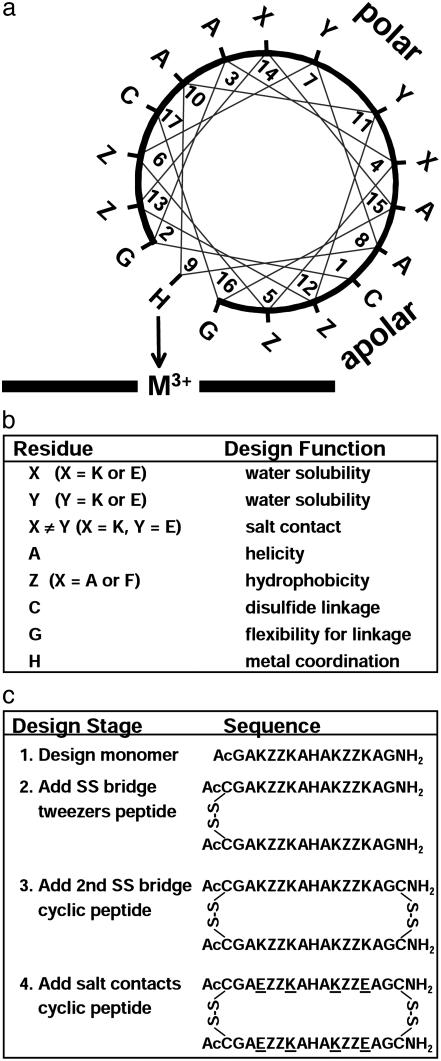
Design of the Cyclic (Cy) Peptides. The design of the primary sequence for Cy-AA-KK (Fig. 1; the sequences of Cy-AA-KK, Cy-AA-EE, and Cy-AA-EK are given in the legend) has been described (34). Our design approach is illustrated in Fig. 1b. Cy-AA-KK contains all lysines on its polar face (and hence is positively charged), whereas Cy-AA-EE has only glutamates on its polar face (and hence is negatively charged). The sequence of Cy-AA-EK is similar to Cy-AA-KK, except that the lysines in positions 4 and 14 were changed to glutamates with the intention of introducing salt-bridge contacts. Cysteines are at the termini of each of the monomeric peptides to permit cyclization. The oxidative cyclization of the peptide was carried out by using the orthogonal protecting groups described (34).
Fig. 1.
(a) Helix-wheel representation depicting the designed peptide complexed with the metalloporphyrin on one face. The 60° tilt of the bound peptides (from NMR solution structure) shows that residues 5 and 12 are in contact with the porphyrin. The peptide abbreviations are Cy-ZZ-XY, where residues 4 and 14 are X, residues 7 and 11 are Y, and residues 5, 6, 12, and 13 are Z, as shown. (b) Design strategy and intended function of specific residues. (c) Sequence of design (32-34) resulting in the current cyclic peptides. Cy-AA-KK [—C(Ac)GAKAAKAHAKAAKAGC(NH2)—]2 is design stage 3 with lysines (K) on the polar face. Cy-AA-EE [—C(Ac)GAEAAEAHAEAAEAGC(NH2)—]2 also is at design stage 3 but with glutamates (E) on the polar face. Cy-AA-EK [—C(Ac)GAEAAKAHAKAAEAGC(NH2)—]2 is at design stage 4 with X = glutamate and Y = lysine; it thus provides salt bridges between lysines and glutamates on the polar face. Cy-FF-KK [—C(Ac)GAKFFKAHAKFFKAGC(NH2)—]2 is the same as Cy-AA-KK, except Z = phenylalanine (F) in place of alanine (A). SS, disulfide link.
Starting from our earlier work with monomeric 15-mer peptides (32-34), we have iterated systematically toward a peptide that would display ”native-like” properties. We have described the effect of disulfide addition on the binding affinity and secondary structure (34). We have now extended this work to the effect of charge and salt bridges on the 3D structure of the complex.
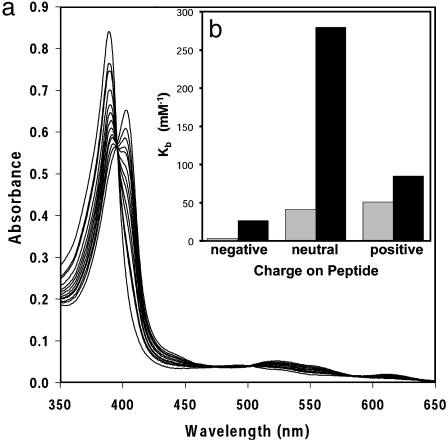
Solution Binding Studies. We have examined the effect of peptide charge on the binding affinity to MIII(coproporphyrinate I)3-. Coproporphyrin I has four carboxylates on the periphery of the macrocycle, so its MIII complex has a net charge of -3 at pH 7. A spectrophotometric titration, shown in Fig. 2a, displays the binding of the free peptide to the metalloporphyrin. The data indicate that the high-spin metalloporphyrin is converted into a low-spin complex, typical of bishistidine-ligated hemes (17). Isosbestic behavior confirms that the only species in solution are the free and complexed (six-coordinate) metalloporphyrin. Three peptides were synthesized: Cy-AA-KK (positively charged), Cy-AA-EE (negatively charged), and Cy-AA-EK (overall neutral). Fig. 2b shows a comparison of the binding of Fe-porphyrin complexes at pH 6.7 and 7.6.
Fig. 2.
(a) Spectrophotometric titration of [FeIII(coproporphyrinate I)]3- with Cy-AA-KK at 21°C. Clean isosbestic behavior was observed in all titrations, implying that only free porphyrin and the complex are present in solution. (b) Summary of binding affinities for the negatively charged (Cy-AA-EE), neutral (Cy-AA-EK), and positively charged (Cy-AA-KK) peptides to [FeIII-(coproporphyrinate I)]3- at pH 6.7 ( ) and 7.6 (▪). The measured binding constants were ≈0.03, 40, and 50 mM-1 at pH 6.7 and 25, 279, and 83 mM-1 at pH 7.6 for Cy-AA-EE, Cy-AA-EK, and Cy-AA-KK, respectively. Cy-FF-KK (data not shown) has Kb = 390 mM-1 at pH 7.6. For comparison, histidine has Kb = 0.008 mM-2. Estimated errors are less than ±10%.
) and 7.6 (▪). The measured binding constants were ≈0.03, 40, and 50 mM-1 at pH 6.7 and 25, 279, and 83 mM-1 at pH 7.6 for Cy-AA-EE, Cy-AA-EK, and Cy-AA-KK, respectively. Cy-FF-KK (data not shown) has Kb = 390 mM-1 at pH 7.6. For comparison, histidine has Kb = 0.008 mM-2. Estimated errors are less than ±10%.
The preorganization provided by the cyclization of the peptides with disulfide bonds has a dramatic impact on heme binding compared with free histidine. Despite the different stoichiometries (2:1 histidine/metalloporphyrin vs. 1:1 cyclic peptides/metalloporphyrin, as determined from Hill plots of the titration data), it is still possible to roughly compare the binding properties. At pH 7.6, the ratio of free to bound [FeIII(coproporphyrin I)]+ will be 1.0 for Cy-AA-EK (the cyclic peptide with the strongest binding; Fig. 2b) at a peptide concentration of 3.6 μM. By way of comparison with histidine itself, a free-to-bound heme ratio of 1.0 requires a histidine concentration of 5.6 mM, a three order of magnitude difference.
Because the metalloporphyrin has a substantial overall negative charge (-3), electrostatic interactions play a major role in affecting the binding constants. At pH 6.7, the positively charged (Cy-AA-KK) and neutral (Cy-AA-EK) peptides bind to the metalloporphyrin with similar affinities and much more strongly than the negatively charged peptide (Fig. 2b). We expected Cy-AA-KK (positively charged) to bind most tightly; competition between the metal center and protons for binding to the imidazole, however, may affect the binding affinity at pH 6.7; alternatively, the weakening of the salt bridges in Cy-AA-EK at lower pH values might also be expected to affect the relative affinities. In comparison, the negatively charged peptide (Cy-AA-EE) is bound relatively weakly. As an aside, during prolonged periods (several hours) at pH <6.7 only, the heme complex of this negatively charged peptide appears to precipitate (i.e., all heme absorption bands decrease).
Salt bridges and hydrophobic interactions are important positive design elements in the creation of metalloporphyrin-binding peptides. At pH 7.6, when all the lysines are still protonated and the glutamates are deprotonated, the neutral peptide has the highest affinity. This finding probably reflects the effects of the salt bridges of Cy-AA-EK that stabilize helix formation and hence increase binding to the metalloporphyrin. Salt bridges can stabilize proteins by up to 1 kcal/mol each (3). Hydrophobic contacts also affect the binding constants. The alanine residues shown by the NMR structure to be in contact with the heme face were replaced by phenylalanine (Cy-FF-KK). This replacement increases binding 5-fold compared with Cy-AA-KK, at pH 7.6, presumably because of increased hydrophobic interactions.
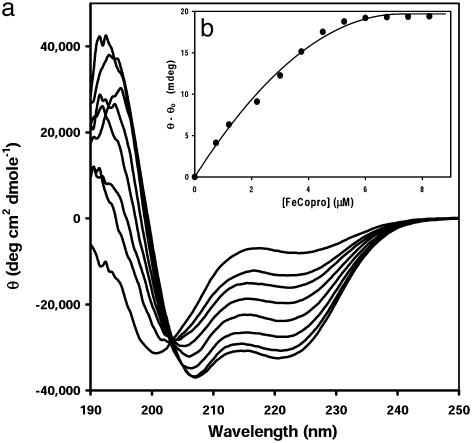
Studies of Secondary Structure and Stability. Both Cy-AA-KK and Cy-AA-EK bind the Fe and the Co system with good isodichroic behavior (Fig. 3a). This finding was not true for the tweezers or 15-mer peptides, as reported earlier (34). As expected, Cy-AA-EE does not assume any secondary structure. The observation of a random-coil to helix (Zimm-Bragg) type of transition in Cy-AA-KK and Cy-AA-EK is consistent with the metalloporphyrin binding to the unfolded peptide (Fig. 3), with the resulting complex assembling into one conformation.
Fig. 3.
(a) CD titration of Cy-AA-KK with [FeIII(coproporphyrinate I)]3- at 4°C. Aliquots of a metalloporphyrin solution (0.1 equivalent) were added until no change occurred in the negative signal at 222 nm. For clarity, only the spectra of the first seven aliquots are shown. Note that an isodichroic point occurs at ≈204 nm, again implying two-state solution behavior. (b) Binding isotherm showing saturation at 1 equivalent to bound porphyrin ([porphyrin] = 7 μM).
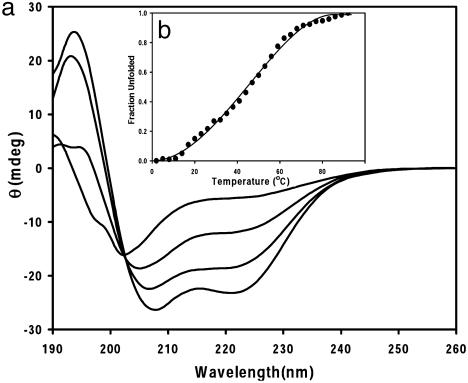
One characteristic of small, single-domain globular proteins is that they show a single, cooperative, thermal unfolding transition (47). The thermal unfolding (Fig. 4) of Cy-AA-KK bound to the slow-exchanging [CoIII(coproporphyrinate I)]3- is similar to that of small globular proteins, and similar data were obtained for Cy-AA-EK. In contrast, peptides containing phenylalanine residues at positions 4, 5, 11, and 12 gave a nonsigmoidal unfolding curve, possibly due to the high number of nonspecific interactions between the porphyrin and the peptide, which create an large ensemble of conformations. Table 1 lists the melting temperature and the van't Hoff enthalpies of the systems analyzed. Small, single-domain globular proteins typically have unfolding enthalpies (ΔHm) on the order of ≈40 kcal/mol (5, 6). The enthalpies that we have measured are somewhat lower than that (≈10-15 kcal/mol), as expected, because our complexes have much less buried hydrophobic surface area. The Tm of the cyclic peptides varies from 47 to 55°C. The alanine-based peptides appear to be half-unfolded at 47°C, which is slightly smaller than many small globular proteins, emphasizing the importance of working with them at low temperatures. The phenylalanine-based derivatives have higher Tm values, which might be due to π-π interactions or hydrophobic stabilization.
Fig. 4.
(a) Thermal unfolding for the complex formed between [CoIII(coproporphyrinate I)]3- and Cy-AA-KK (similar results were seen for the complex with Cy-AA-EK). Again, the isodichroic point confirms the two-state unfolding of the peptide. (b) Cooperative, two-state unfolding of the same complex monitored at 222 nm, with a melting temperature of 47°C.
Table 1.
Thermodynamic properties of cyclic peptide complexes of [CoIII(coproporphyrinate I)]3-
| Parameter | Cy-AA-KK | Cy-AA-EK | Cy-FF-KK |
|---|---|---|---|
| Tm, °C* | 47 ± 3 | 47 ± 3 | 52 ± 3 |
| ΔSm, cal·mol-1·deg-1† | 31 ± 5 | 47 ± 5 | 34 ± 5 |
| ΔHm, kcal·mol-1‡ | 9.9 ± 2 | 15.0 ± 2 | 11.1 ± 3 |
| ΔG°unfolding, kcal·mol-1§ | 2.9 ± 0.24 | 3.3 ± 0.26 | —¶ |
| m, cal·mol-1·M-1∥ | 598 ± 47 | 735 ± 54 | —¶ |
Melting temperature. This is the temperature when ΔG = 0 and the molecule is half-unfolded
Entropy of unfolding (1 cal = 4.184 J)
van't Hoff enthalpy
Free energy of unfolding in urea
Not measured because of the dynamic nature of the complex and limited sample quantities
Molar cosolvation term
Urea-induced unfolding of the alanine peptides was also investigated. The free energies (Table 1) are a bit lower than the average value of 4 kcal/mol seen for globular proteins. However, these systems are miniature proteins without the extensive hydrophobic core seen in globular proteins. The most stabilizing interactions seen in these systems are the Co-histidine bond, the hydrophobic interactions of the porphyrin face with the alanine residues, and electrostatic interactions (in Cy-AA-EK).
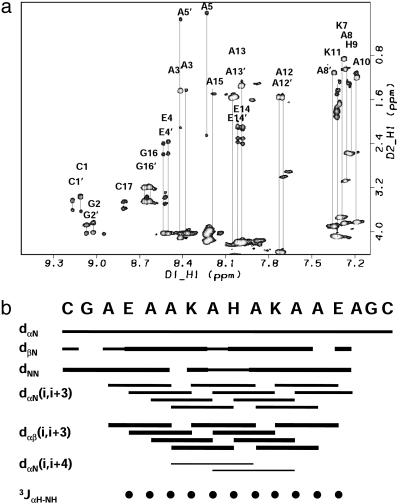
NMR Spectroscopy. Two-dimensional NMR spectra were well dispersed, consistent with conformational singularity (Fig. 5a). All the resonances for this molecule could be assigned by using standard methods after analysis of NOESY, total correlation spectroscopy, and double-quantum-filtered COSY data in 90% H2O/10% D2O (38). Analysis of the NOE cross-peak intensities reveals significant secondary structure in the molecule (Fig. 5b). Only one of the helices is shown for clarity because the other helix gives the same pattern. The data reveal a stronger intensity for the dNN cross-peaks relative to the dαN cross-peaks, which is a sign of helicity. Another strong pattern is the dαβ(i, i + 3) cross-peaks. Although it is difficult to distinguish between 310 and α-helices from NMR data, the presence of the dαβ(i, i + 3) cross-peaks as the most intense in the spectra, the presence of dαN(i, i + 4), and the absence of dαN(i, i + 2) cross-peaks provides further support for the α-helical structure. Several unique NOEs also exist between the metalloporphyrin and the peptide and the imidazole of the histidine and other residue side chains.
Fig. 5.
(a) Two-dimensional 1H-1H total correlation spectroscopy spectrum of the complex of [CoIII(coproporphyrinate I)]3- and Cy-AA-EK. The spectra show nice dispersion, characteristic of a structured system. (b) Summary of the sequential NOEs for the complex of [CoIII(coproporphyrinate I)]3- and Cy-AAEK. The intensities were grouped into three sets: weak, medium, and strong. The size of the bars reflects these intensities. NOEs were taken from a NOESY spectrum recorded with a mixing time of 100 ms. The three-bond 2JαNH coupling constants were taken from a high-resolution double-quantumfiltered COSY experiment and converted to φ-angle restraints by using the standard methods (36, 37). Only one of the helices is shown for clarity because the other helix gives the same pattern. The data reveal a stronger intensity for the dNN cross-peaks relative to the dαN cross-peaks, which is a sign of helicity. Another strong pattern is the dαβ(i, i + 3) cross-peaks. Although it is difficult to distinguish between 310 and α-helices from NMR data, the presence of the dαβ(i, i + 3) cross-peaks as the most intense in the spectra, the presence of dαN(i, i + 4), and the absence of dαN(i, i + 2) cross-peaks provide further support for the α-helical structure. Several unique NOEs also exist between the metalloporphyrin and the peptide and the imidazole of the histidine and other residue side chains.
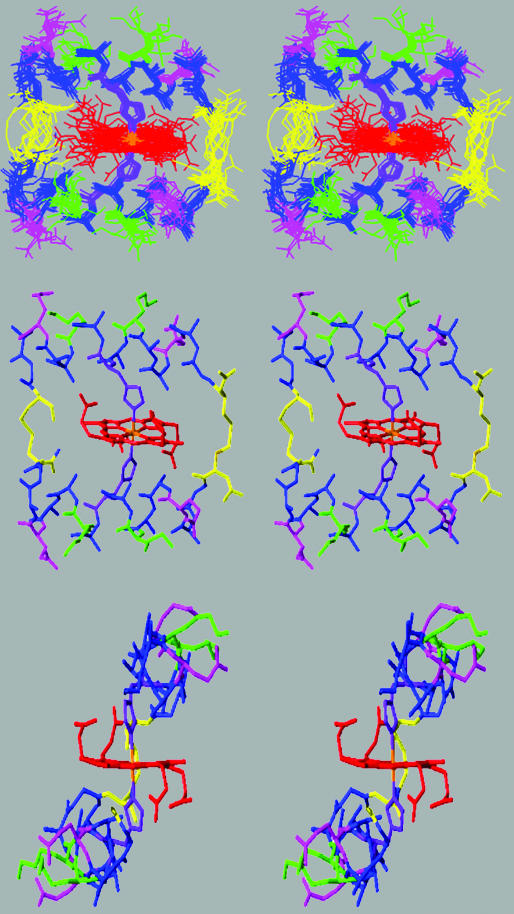
Structure Calculations. The quality of the NMR data permitted the use of NOE-restrained simulated annealing protocols for the calculation of the solution structure. By using the INSIGHTII/DISCOVER package, and a modified form of the consistent valence force field, 50 random structures were calculated from a total of 224 distance restraints (including 22 hydrogen bond restraints) and 30 φ-angle restraints (Table 2). Rigorous analysis of NOE cross-peaks gave a total of 202 distance restraints determined from peak volume measurements. Torsion-angle constraints were determined from analysis of double-quantum-filtered COSY spectral data. Displayed in Fig. 6 Top is a stereoview of the 14 best structures; topologically, the structure is similar to a leucine zipper. A representative low-energy structure is shown in Fig. 6 Middle. Strikingly, the structure shows that the two helices tilt to maximize close packing of the hydrophobic porphyrin π system and the hydrophobic residues of the peptide, so that the lysines are proximal to the carboxylates of the porphyrin. In this design, alanines were positioned on the side of the helix so that they could interact favorably with the hydrophobic porphyrin face. Of the 14 structures displayed, the overall backbone rms deviation is 0.9 Å for all nonhydrogen atoms (Table 2), 0.6 Å for the backbone atoms, and a very low 0.4 Å in the helical regions of the peptide. As expected, the turn regions, where the cysteine bridges have been placed, have more disorder and are presumably dynamic in solution.
Table 2. Structural statistics for a family of 14 calculated structures of the Cy-AA-EK complex of [CoIII(coproporphyinate -I)]3-.
| Experimental restraints | |
| Intraresidue (|i—j| = 0) | 64 |
| Short to medium (1 ≤ |i—j| ≤ 5 residues) | 131 |
| Long range (|i—j| > 5 residues) | 8 |
| Hydrogen bonds | 22 |
| ϕ-angles | 30 |
| Total | 255 |
| Deviations from experimental restraints | |
| Distance restraints | 0.07 |
| Dihedral angle | 2.7 |
| Deviations from ideal geometry | |
| Bonds, Å | 0.02 |
| Angles, ° | 1.87 |
| Impropers | 0.96 |
| rms deviations from the mean structure, Å | |
| Backbone atoms (residues 1—34: N, Cα, C) | 0.6 |
| Backbone atoms (residues 19—31: N, Cα, C) | 0.4 |
| All nonhydrogen atoms (residues 1—34) | 0.93 |
Of the 50 structures, 40 converged to acceptable structures. Of these, 14 structures were deemed exceptionally good (i.e., displayed no violations>0.5 Å or 5°). Distances were constrained in a square well potential by using force constants for bonds set to 25 kcal/(mol·rad2) and for angles set to 100 kcal/(mol·rad2). The structures were checked by using the NMRREFINE/PROSTAT module in the INSIGHT/DISCOVER package.
Fig. 6.
(Top) Overlay of the 14 best structures for the complex of [CoIII(coproporphyrinate I)]3- and Cy-AA-EK. Two stereo side views are shown for clarity. Structures were generated by using the insight/discover package. The color scheme is as follows: Ala and Gly, blue; Lys, green; Glu, pink; Cys, yellow; and His, purple. (Middle) Stereo view of the solution structure of [CoIII(coproporphyrinate I)]3- and Cy-AA-EK. (Bottom) As in b, rotated 90°. Coordinates for these structures are available in the Protein Data Bank at www.rcsb.org/pdb with PDB ID code 1PBZ.
Conclusions
The unifying theme in this work has shown that the metalloporphyrin plays a substantial role in the induction and stabilization of peptide structure. Furthermore, the described structures display native-like properties similar to small proteins. The broadening of the free energy gap between native and molten structures was possible only after incorporation of disulfide bridges and ionic contacts.
Stability studies reveal a cooperative denaturation of both the Cy-AA-KK and Cy-AA-EK complexes. Whereas the thermodynamic data show only a marginal increase in the stability of Cy-AA-EK over Cy-AA-KK, the NMR spectra of Cy-AA-EK were the best resolved. NMR, therefore, is a critical screen for native-like properties.
The use of the diamagnetic, slow-exchanging CoIII analogue of [FeIII(coproporphyrinate I)]3- in the characterization of the cyclic complexes permitted the solution of the high-resolution structure of the [CoIII(coproporphyrinate I)]3- complex of Cy-AA-EK. Notably, the structure shows an ≈60° helical tilt relative to the heme, which maximizes hydrophobic contacts and intramolecular ion pairing. Analysis of the structure suggested that a four-helix bundle could easily be engineered by extension of these sequences.
Possible biomedical implications exist for these studies. It has been well established that intrinsically unstructured proteins (i.e., prions) (48, 49) are key elements in neurodegenerative diseases. We find it most interesting that porphyrins (50, 51) and other cofactors (52) have been shown to interact strongly with these unstructured proteins and may have important pharmaceutical applications. The ability of porphyrins and metalloporphyrins to stabilize α-helical structures in peptides may have important implications for remediation of peptide aggregates in vivo.
Acknowledgments
This work was supported by National Institutes of Health Grant HL 25934, with instrument support, in part, from the W. M. Keck Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: NOE, nuclear Overhauser effect.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1PBZ).
References
- 1.Lombardi, A., Summa, C. M., Geremia, S., Randaccio, L., Pavone, V. & DeGrado, W. F. (2000) Proc. Natl. Acad. Sci. USA 97, 6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill, R. B., Raleigh, D. P., Lombardi, A. & DeGrado, W. F. (2000) Acc. Chem. Res. 33, 745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nastri, F., Lombardi, A. & Pavone, V. (2001) Chem. Rev. 101, 3165-3189. [DOI] [PubMed] [Google Scholar]
- 4.Ptitsyn, O. B. (1995) Trends Biochem. Sci. 20, 485. [Google Scholar]
- 5.DeGrado, W. F., Raleigh, D. P. & Handel, T. (1991) Curr. Opin. Struct. Biol. 1, 984-993. [Google Scholar]
- 6.Betz, S. F., Raleigh, D. P. & DeGrado, W. F. (1993) Curr. Opin. Struct. Biol. 3, 601-610. [Google Scholar]
- 7.Bryson, J. W., Betz, S. F., Lu, H. S., Suich, D. J., Zhou, H. X., O'Neil, K. T. & DeGrado, W. F. (1995) Science 270, 935-941. [DOI] [PubMed] [Google Scholar]
- 8.Gibney, B. R. & Dutton, P. L. (1999) Protein Sci. 8, 1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rojas, N. R. L., Kamtekar, S., Simons, C. T., McLean, J. E., Vogel, K. M., Spiro, T. G., Farid, R. S. & Hecht, M. H. (1997) Protein Sci. 6, 2512-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rau, H. K., Snigula, H., Struck, A., Robert, B., Scheer, H. & Haehnel, W. (2001) Eur. J. Biochem. 268, 3284-3295. [DOI] [PubMed] [Google Scholar]
- 11.Mutz, M. W., Case, M. A., Wishart, J. F., Ghadiri, M. R. & McLendon, G. L. (1999) J. Am. Chem. Soc. 121, 858-859. [Google Scholar]
- 12.Moffet, D. A., Certain, L. K., Smith, A. J., Kessel, A. J., Beckwith, K. A. & Hecht, M. H. (2000) J. Am. Chem. Soc. 122, 7612-7613. [Google Scholar]
- 13.Hellinga, H. W. (1998) Folding Des. 3, R1-R8. [DOI] [PubMed] [Google Scholar]
- 14.Benson, D. E., Wisz, M. S. & Hellinga, H. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6292-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, R. H., Kennepohl, P. & Solomon, E. I. (1996) Chem. Rev. 96, 2239-2314. [DOI] [PubMed] [Google Scholar]
- 16.Beinert, H., Holm, R. H. & Munck, E. (1997) Science 277, 653-659. [DOI] [PubMed] [Google Scholar]
- 17.Lever, A. B. P. & Gray, H. B., eds. (1983) Iron Porphyrins, Physical Bioinorganic Chemistry Series 1 (Addison-Wesley, Reading, MA), Parts 1 and 2.
- 18.Klinman, J. P. (1996) J. Biol. Chem. 271, 27189-27192. [DOI] [PubMed] [Google Scholar]
- 19.Summa, C. M., Lombardi, A., Lewis, M. & DeGrado, W. F. (1999) Curr. Opin. Struct. Biol. 9, 500-508. [DOI] [PubMed] [Google Scholar]
- 20.Kuchner, O. & Arnold, F. H. (1997) Trends Biotechnol. 15, 523-530. [DOI] [PubMed] [Google Scholar]
- 21.Arnold, F. H. (2001) Nature 409, 253-257. [DOI] [PubMed] [Google Scholar]
- 22.Arnold, F. H. (1998) Acc. Chem. Res. 31, 125-131. [Google Scholar]
- 23.Wade, H. & Scanlan, T. S. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 461-493. [DOI] [PubMed] [Google Scholar]
- 24.Demartis, S., Huber, A., Viti, F., Lozzi, L., Giovannoni, L., Neri, P., Winter, G. & Neri, D. (1999) J. Mol. Biol. 286, 617-633. [DOI] [PubMed] [Google Scholar]
- 25.Atwell, S. & Wells, J. A. (1999) Proc. Natl. Acad. Sci. USA 96, 9497-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, G. P. & Petrenko, V. A. (1997) Chem. Rev. 97, 391-410. [DOI] [PubMed] [Google Scholar]
- 27.Forrer, P., Jung, S. & Pluckthun, A. (1999) Curr. Opin. Struct. Biol. 9, 514-520. [DOI] [PubMed] [Google Scholar]
- 28.Hill, R. B. & DeGrado, W. F. (1998) J. Am. Chem. Soc. 120, 1138-1145. [Google Scholar]
- 29.Walsh, S. T. R., Cheng, H., Bryson, J. W., Roder, H. & DeGrado, W. F. (1999) Proc. Natl. Acad. Sci. USA 96, 5486-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Struthers, M. D., Cheng, R. P. & Imperiali, B. (1996) Science 271, 342-345. [DOI] [PubMed] [Google Scholar]
- 31.Skalicky, J. J., Gibney, B. R., Rabanal, F., Urbauer, R. J. B., Dutton, P. L. & Wand, A. J. (1999) J. Am. Chem. Soc. 121, 4941-4951. [Google Scholar]
- 32.Huffman, D. L., Rosenblatt, M. M. & Suslick, K. S. (1998) J. Am. Chem. Soc. 120, 6183-6184. [Google Scholar]
- 33.Huffman, D. L. & Suslick, K. S. (2000) Inorg. Chem. 39, 5418-5419. [DOI] [PubMed] [Google Scholar]
- 34.Rosenblatt, M. M., Huffman, D. H., Wang, X., Remmer, H. & Suslick, K. S. (2002) J. Am. Chem. Soc 124, 12394-12395. [DOI] [PubMed] [Google Scholar]
- 35.Pace, C. N. & Scholtz, J. M. (1997) in Protein Structure: A Practical Approach, ed. Creighton, T. E. (Oxford Univ. Press, New York), 2nd Ed., pp. 299-321.
- 36.Ghadiri, M. R., Soares, C. & Choi, C. (1992) J. Am. Chem. Soc. 114, 825-831. [Google Scholar]
- 37.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 38.Cavanagh, J., Fairbrother, W. J., Palmer, A. G. & Skelton, N. J. (1996) Protein NMR Spectroscopy: Principles and Applications (Academic, San Diego).
- 39.Bax, A. (1988) J. Magn. Reson. 77, 134-147. [Google Scholar]
- 40.Davis, D. G. & Bax, A. (1985) J. Am. Chem. Soc. 107, 2820-2821. [Google Scholar]
- 41.Bax, A. & Davis, D. G. (1985) J. Magn. Reson. 65, 355-360. [Google Scholar]
- 42.Griesinger, C., Sorensen, O. W. & Ernst, R. R. (1986) J. Chem. Phys. 85, 6837-6852. [Google Scholar]
- 43.Kim, Y. & Prestegard, J. H. (1990) Proteins Struct. Funct. Genet. 8, 377-385. [DOI] [PubMed] [Google Scholar]
- 44.Nilges, M., Clore, G. M. & Gronenborn, A. M. (1987) FEBS Lett. 219, 11-16. [Google Scholar]
- 45.Nilges, M., Gronenborn, A. M., Bruenger, A. T. & Clore, G. M. (1988) Protein Eng. 2, 27-38. [DOI] [PubMed] [Google Scholar]
- 46.Nilges, M., Clore, G. M. & Gronenborn, A. M. (1988) FEBS Lett. 239, 129-136. [DOI] [PubMed] [Google Scholar]
- 47.Privalov, P. L. & Gill, S. J. (1988) Adv. Protein Chem. 39, 191-234. [DOI] [PubMed] [Google Scholar]
- 48.Wright, P. E. & Dyson, H. J. (1999) J. Mol. Biol. 293, 321-331. [DOI] [PubMed] [Google Scholar]
- 49.Dyson, H. J. & Wright, P. E. (1998) Nat. Struct. Biol. 5, 499-503. [DOI] [PubMed] [Google Scholar]
- 50.Caughey, W. S., Raymond, L. D., Horiuchi, M. & Caughey, B. (1998) Proc. Natl. Acad. Sci. USA 95, 12117-12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Priola, S. A., Raines, A. & Caughey, W. S. (2000) Science 287, 1503-1506. [DOI] [PubMed] [Google Scholar]
- 52.Viles, J. H., Cohen, F. E., Prusiner, S. B., Goodin, D. B., Wright, P. E. & Dyson, H. J. (1999) Proc. Natl. Acad. Sci. USA 96, 2042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]