Abstract
The production of functional myosin heavy chains in many eukaryotic organisms requires the function of proteins containing UCS domains (UNC-45/CRO1/She4), which bind to the myosin head domain and stimulate its folding. UCS proteins are essential for myosin-related functions such as muscle formation, RNA localization and cytokinesis. Here, we show that the Schizosaccharomyces pombe UCS protein Rng3 associates with polysomes, suggesting that UCS proteins might assist myosin folding cotranslationally. To identify Rng3 cotranslational targets systematically, we purified Rng3-associated RNAs and used DNA microarrays to identify the transcripts. Rng3 copurified with only seven transcripts (around 0.1% of S. pombe genes), including all five messenger RNAs encoding myosin heavy chains. These results suggest that every myosin heavy chain in S. pombe is a cotranslational target of Rng3. Furthermore, our data suggest that microarray-based approaches allow the genome-wide identification of cotranslational chaperone targets, and thus pave the way for the dissection of translation-linked chaperone networks.
Keywords: Schizosaccharomyces pombe, protein folding, chaperone-mediated folding, RIp-chip, DNA microarrays
Introduction
Newly synthesized myosin heavy chains require the assistance of chaperones for their folding and assembly (reviewed by Kim et al, 2008). For example, folding of striated muscle myosin needs the chaperonin-containing t-complex polypeptide 1 (TCP-1), as well as chaperones of the heat-shock protein 70 (Hsp70) and Hsp90 families (Srikakulam & Winkelmann, 1999, 2004). In addition to general chaperones, the production of myosin proteins requires the function of specialized factors such as proteins containing UCS domains (UNC-45/CRO1/She4; Barral et al, 1998). Proteins of this family are necessary for many myosin-related functions such as muscle formation, cytokinesis and RNA localization (reviewed by Hutagalung et al, 2002). UCS proteins are present in animals (a single member in Caenorhabditis elegans and Drosophila melanogaster, and two paralogues in vertebrates) and fungi (She4 in Saccharomyces cerevisiae and Rng3 in Schizosaccharomyces pombe; Kim et al, 2008). UNC-45 binds directly to the head of muscle myosin heavy chains and prevents thermal aggregation of myosin, consistent with a role as a chaperone (Barral et al, 2002; Liu et al, 2008). UNC-45 might also recruit additional chaperones, as it associates stoichiometrically with Hsp90 (Barral et al, 2002). Consistently, both isoforms of mouse UNC45 enhance the folding of in vitro synthesized smooth muscle myosin in an HSP90-dependent manner (Barral et al, 2002; Liu et al, 2008). UCS proteins also associate with non-muscle myosins; for example, the S. cerevisiae UCS protein She4 interacts with non-conventional class I and V myosins (Toi et al, 2003; Wesche et al, 2003), and the S. pombe Rng3 interacts physically and genetically with the class II myosin Myo2 (Wong et al, 2000). The rng3 gene is essential for viability and its inactivation results in strong defects in cytokinesis (Wong et al, 2000). Rng3 can activate Myo2 motility in vitro, suggesting that UCS proteins can regulate myosin function independently of folding (Lord & Pollard, 2004; Lord et al, 2008).
In addition to the chaperones required for the refolding of stress-denatured proteins, eukaryotic cells contain a specialized network of chaperones that assist with the folding of newly synthesized proteins. These chaperones associate with nascent chains as they are produced by the ribosome. Chaperone-mediated cotranslational folding is much more widespread in eukaryotic cells than in prokaryotic organisms. It is thought that the development of a ribosome-associated chaperone network allowed eukaryotic cells to evolve complex multidomain proteins (Craig et al, 2003; Young et al, 2004; Rospert & Chacinska, 2006).
Little is known about how UCS proteins function in vivo; in particular, it is not known whether they influence the folding of myosin heads in a cotranslational or post-translational manner. We used the S. pombe UCS protein Rng3 as a model to investigate whether UCS proteins function cotranslationally. We employed a microarray-based method to systematically identify RNAs associated with Rng3, and found that Rng3 specifically copurifies with all RNAs encoding myosin heavy chains in S. pombe. Rng3 was present on polysomes, and the interactions between Rng3 and myosin-encoding transcripts were sensitive to conditions that disassemble ribosomes. Our data support the idea that Rng3 assists myosin folding in a cotranslational manner.
Results
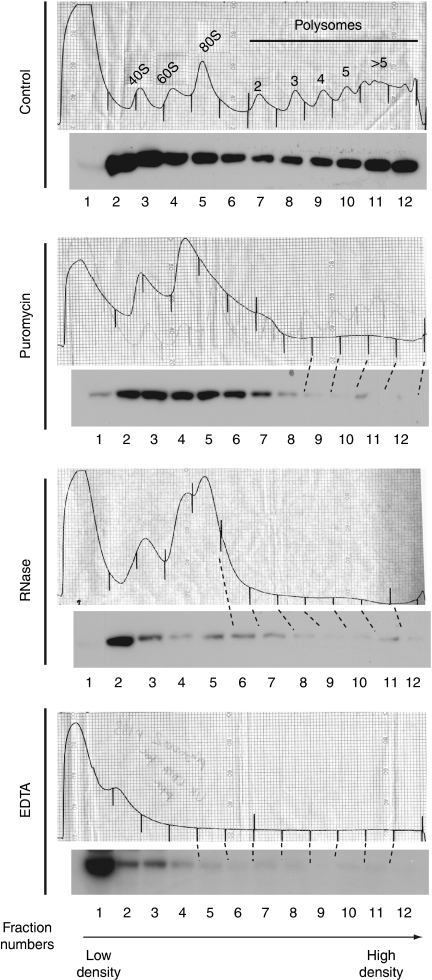
Chaperones that assist folding cotranslationally are associated with polysomes (Craig et al, 2003; Young et al, 2004; Rospert & Chacinska, 2006). Therefore, we investigated whether Rng3 was present in polysomal fractions. To detect Rng3, we constructed a strain in which endogenous Rng3 was marked at the carboxyl terminus with a TAP (tandem affinity purification) tag. Rng3-TAP-expressing cells had normal morphologies and grew with standard generation times, showing that the tagged protein was functional. We isolated polysomes by fractionating cell lysates using ultracentrifugation on sucrose gradients and followed the distribution of Rng3-TAP by immunoblotting. Although some Rng3 was present in the low-density fractions that corresponded to soluble proteins (Fig 1), a substantial amount of Rng3 was detected in the higher density fractions, which were enriched in polysomes (Fig 1). Disassembly of the polysomes using EDTA, puromycin or RNase (Fig 1) resulted in a shift of Rng3 towards the low-density fractions, showing that the presence of Rng3-TAP in the higher density fractions was caused by the association with polysomes. We considered the possibility that the association of Rng3 with ribosomes might reflect binding to its own RNA; however, proteins not expected to associate with ribosomes accumulate in the soluble fractions in similar experiments (data not shown), ruling out this explanation.
Figure 1.
Rng3 associates with polysomes. Extracts were fractionated using ultracentrifugation through sucrose gradients and Rng3-TAP was detected by Western blot. The top of the gradient is shown on the left. The position of the ribosomes was determined from the absorbance at 254 nm and confirmed by the distribution of the large subunit ribosomal proteins Rpl7 (data not shown). Ribosomes were stabilized by treatment of cells with cycloheximide, or disassembled by the addition of EDTA, RNase or puromycin to the extracts.
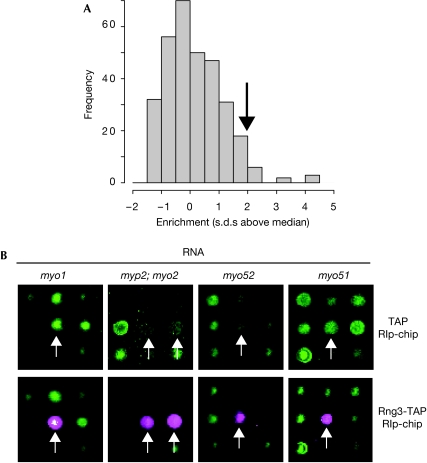
Ribosome-associated chaperones interact indirectly with the messenger RNA (mRNA) encoding proteins, whose folding they assist. Therefore, we reasoned that the identification of chaperone-associated transcripts could be used as a surrogate for the discovery of chaperone targets. To test this idea, we purified TAP-tagged Rng3 and used DNA microarrays to identify the transcripts present in the immunoprecipitate (RIp-chip, for ribonucleoprotein immunoprecipitation analysed with DNA chips; Fig 2; Table 1). Markedly, all five mRNA encoding myosin heavy chains encoded in the S. pombe genome were enriched in the immunoprecipitates. The results were highly consistent, as these were the five most enriched transcripts in four independent biological repeats. None of these transcripts was present in control immunoprecipitates from strains containing no tagged proteins, or expressing only TAP or unrelated TAP-tagged proteins, showing the specificity of the association (Fig 2; unpublished observations). We considered the possibility of cross-hybridization between the various myosin probes on our microarray system; however, the fact that the probes in our platform have been designed from regions with no homology between the different myosins (Lyne et al, 2003) rules out this potential artefact. Rng3 associated with myosin RNAs belonging to three classes: class I (myo1), class II (myo2 and myp2) and class V (myo51 and myo52; Win et al, 2002). These data suggest that every myosin heavy chain is an in vivo target of Rng3.
Figure 2.
The Rng3 chaperone binds to messenger RNAs encoding myosin heavy chains. (A) Histogram of RNA enrichment ratios from a Rng3-TAP RIp-chip experiment. The x axis represents log2-transformed enrichment ratios that have been median-centred and divided by their standard deviation (that is, the numbers on the axis represent standard deviations above or below the median), and the y axis shows the number of genes in each category. The arrow indicates the threshold used to define enriched RNAs. Only RNAs that were enriched above the threshold in four out of four independent experiments were selected (Table 1). (B) Sections of DNA microarrays hybridized with immunoprecipitated RNAs (magenta) and total RNA (green). The intensity of the signal is proportional to the amount of RNA. The arrows show the position of the myosin RNAs labelled above. TAP only (top row); Rng3-TAP (bottom row). RIp-chip, ribonucleoprotein immunoprecipitation analysed with DNA chips; TAP, tandem affinity purification.
Table 1.
Messenger RNAs associated with Rng3
| Common name | Systematic name | Description | Enrichment (s.d.s above median) |
|---|---|---|---|
| myo1 | SPBC146.13 | Type I myosin heavy chain | 5.37±0.98 |
| myo2 | SPCC645.05c | Type II myosin heavy chain | 5.24±0.90 |
| myp2 | SPAC4A8.05c | Type II myosin heavy chain | 4.28±0.67 |
| myo51 | SPBC2D10.14c | Type V myosin heavy chain | 4.93±1.06 |
| myo52 | SPCC1919.10c | Type V myosin heavy chain | 5.57±1.16 |
| sib1 | SPAC23G3.02c | Ferrichrome synthetase | 3.26±0.49 |
| spp42 | SPAC4F8.12c | Component of the U5 snRNP | 2.71±0.33 |
| Only mRNAs that passed the enrichment threshold in four independent biological repeats are shown. The five most enriched RNAs in every experiment were those of the five myosin heavy chains. The enrichment column shows the average enrichment in four independent experiments (measured as the number of standard deviations above the median enrichment of all mRNAs in the immunoprecipitation, see Methods) and the standard error of the mean over the four repeats. mRNA, messenger RNA; snRNP, small nuclear ribonucleoprotein. |
Apart from those mRNAs encoding myosin heavy chains, only the sib1 and spp42 transcripts passed the selection criteria for enrichment (see Methods). sib1 encodes ferrichrome synthetase and spp42 is a component of the U5 small nuclear ribonucleoprotein particle (snRNP) involved in splicing. Both are large proteins, with predicted molecular weights of 560 and 275 kDa, respectively. Some other mRNAs were consistently enriched in Rng3 immunoprecipitates, although below the enrichment threshold (see supplementary information online). The proteins they encode might represent additional Rng3 targets; however, given that their enrichment levels (including those of sib1 and spp42 RNAs) are much lower than those of myosins, the significance of their binding to Rng3 is unclear. Although myosin heavy chains seem to be the main targets of UCS proteins, a recent report has identified UNC45A as a putative regulator of the progesterone receptor (Chadli et al, 2006).
UCS proteins might function by binding to myosin heads and recruiting chaperones of the Hsp90 family to stimulate myosin folding (Barral et al, 2002; Liu et al, 2008; Srikakulam et al, 2008). In S. pombe, Rng3 binds to the Hsp90 protein Swo1 (Mishra et al, 2005). Furthermore, rng3 and swo1 interact genetically, and some swo1 mutants show phenotypes similar to those of rng3 mutants (Mishra et al, 2005). To investigate whether Swo1 was also associated with myosin mRNAs, we TAP-tagged Swo1 as described above for Rng3 and used it for RIp-chip experiments. In contrast with Rng3, we did not detect binding to myosin or other mRNAs (data not shown). This lack of interaction could be attributed to the loss of binding during purification or might reflect the biochemical mode of action of the Rng3/Swo1 pair. For example, Rng3 could associate with myosin nascent chains and recruit Swo1 post-translationally.
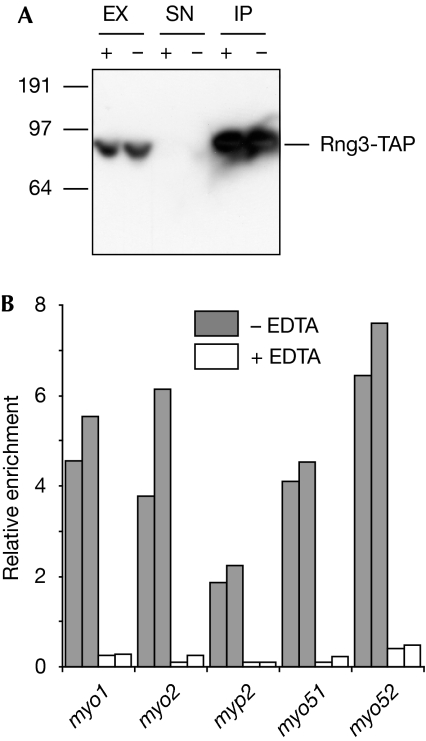
If the association of Rng3 with myosin mRNAs reflected the binding of Rng3 to nascent myosin chains on polysomes, it should be sensitive to conditions that disrupt ribosomes. To test this idea, we carried out RIp-chip experiments with Rng3-TAP in the presence or absence of EDTA. Although Rng3 was efficiently precipitated both in the presence and absence of EDTA (Fig 3A), EDTA treatment led to a strong reduction in the enrichment of myosin mRNAs in the Rng3 immunoprecipitate (Fig 3B), supporting the idea that Rng3 interacts with myosin proteins cotranslationally. To address directly the question of whether the association between Rng3 and myosin RNAs occurs on polysomes, we carried out two sequential immunoprecipitations from cells expressing Rng3-TAP. We purified Rng3-TAP together with associated RNAs, and then used antibodies against ribosomal RNA to isolate ribosomes from the Rng3-containing fraction; as a negative control, we carried out the second immunoprecipitation with an unrelated antibody. Myosin-encoding RNAs were still enriched in the ribosomal immunoprecipitate, but were not detectable in the negative control (Table 2). These results show that the myosin RNAs that copurify with Rng3 are also associated with ribosomes.
Figure 3.
The Rng3–myosin messenger RNA association is sensitive to conditions that destabilize polysomes. (A) Western blots of samples from Rng3-TAP immunoprecipitation experiments from mock-treated (−) or EDTA-treated extracts (+). Here, 1.5% of the input and 5% of the IP were loaded. (B) Enrichment of myosin RNAs in Rng3-TAP IPs (normalized to the levels of the small nuclear RNA U3) from mock- (grey) or EDTA-treated extracts (white). Data from two independent experiments are shown. EX, whole-cell extract; IP, immunoprecipitate; SN, supernatant after immunoprecipitation; TAP, tandem affinity purification.
Table 2.
Messenger RNAs associated with Rng3 and with ribosomes
| Common name | Systematic name | Enrichment |
|---|---|---|
| myo51 | SPBC2D10.14c | 4.17 (4.62, 3.71) |
| myo2 | SPCC645.05c | 3.13 (2.63, 3.61) |
| SPAC1250.02 | 2.93 (2.95, 2.92) | |
| myo1 | SPBC146.13c | 2.74 (3.06, 2.42) |
| SPAC56F8.06c*** | 2.22 (2.10, 2.36) | |
| rng2 | SPAC4F8.13c | 2.04 (2.00, 2.08) |
| myo3: myp2 | SPAC4A8.05c | 2.04 (2.63, 1.36) |
| myo52 | SPCC1919.10c | 2.02 (1.84, 2.20) |
| SPCC794.02*** | 2.00 (1.85, 2.15) | |
| SPCC18.12c*** | 1.47 (1.20, 1.72) | |
| Fractions containing immunopurified Rng3 were used for immunoprecipitation with antibodies against a ribosomal RNA, and the immunoprecipitates were analysed using DNA microarrays. The enrichment levels were normalized to those of the small nuclear RNA U3 (SPNCRNA.03) in the immunoprecipitate. The 10 most enriched mRNAs are shown. mRNAs encoding myosin heavy chains are shown in bold. Those RNAs marked with stars are enriched in control immunoprecipitations (cells not expressing TAP-tagged proteins) and are likely to represent false positives. None of the myosin RNAs was enriched in control experiments in which the second immunoprecipitation was carried out with anti-Myc antibodies. The enrichment column shows the average enrichment and the numbers in parentheses show the values for each of the two independent experiments. Probes for ribosomal RNAs on the microarrays showed strong signals in the immunoprecipitation with ribosomal RNA antibodies, but were not detectable above background in the negative control (data not shown). mRNA, messenger RNA; TAP, tandem affinity purification. | ||
As Rng3 is associated with polysomes, we considered the possibility that Rng3 directly influences the translation of myosin proteins. To test this idea, we carried out genome-wide polysomal profiling of wild-type and rng3 mutant cells by comparing the number of ribosomes associated with each transcript, which can be used as a surrogate for translational efficiency. We used sucrose gradients to separate RNAs according to the number of associated ribosomes (supplementary Fig S1A online), and directly compared the corresponding fractions from rng3 and wild-type extracts. Changes in translation would alter the number of ribosomes associated with specific transcripts, and result in a redistribution of the RNAs across the various fractions. We did not observe any changes in the distribution of the five myosin-encoding RNAs (supplementary Fig S1B online), indicating that Rng3 does not regulate myosin translation. Consistent with a specific role in protein folding, myosin protein levels were not altered in rng3 mutants (supplementary Fig S2 online).
Discussion
UCS proteins might function cotranslationally
We have used the S. pombe UCS protein Rng3 as a model to study the mode of action of UCS chaperones and to develop new approaches to dissect chaperone networks at the genome-wide level. We have shown that Rng3 is present in polysomes and specifically associates with mRNAs encoding all S. pombe myosin heavy chains. This is the first indication that UCS proteins might act cotranslationally in vivo. Although the functional significance of these interactions remains to be established, we propose that they reflect Rng3-assisted cotranslational folding of myosin nascent chains. This phenomenon might be advantageous for the cell under certain conditions; for example, partially folded myosin proteins might act as dominant-negative mutants and be toxic for the cell. Studies in C. elegans have shown that very small quantities of a dominant-negative form of myosin (just 2% of the amount of wild-type protein) are sufficient to block the assembly of thick filaments (Bejsovec & Anderson, 1988). Coupling translation and folding would prevent the release of partly folded, potently toxic myosins to the cytoplasm.
Rng3 function and myosin turnover
Myosin protein levels are reduced in the absence of UCS proteins in C. elegans (Barral et al, 2002; Landsverk et al, 2007) and S. cerevisiae (Lord et al, 2008), probably because denatured myosin proteins are ubiquitinated and degraded. In S. pombe, two studies have suggested that Myo2 levels are decreased when Rng3 is inactivated (Mishra et al, 2005; Lord et al, 2008); however, we did not detect strong changes in the four myosins that we investigated (including Myo2). We think that these apparent discrepancies can be explained by experimental differences. First, we measured the levels of wild-type Myo2, whereas Mishra et al investigated a mutant form of Myo2 (myo2-E1). Second, Lord et al compared the yields of purification of overexpressed Myo2 in wild-type and rng3 mutants, but did not examine Myo2 levels directly. The decreased yields that they observed in rng3 mutants might have been caused by changes in Myo2 that affected the purification process (such as altered solubility), rather than by altered myosin protein levels. Third, we looked at levels of TAP-tagged proteins, whereas the other two studies used antibodies to detect untagged Myo2. It is possible that the addition of TAP affected the stability of the proteins; given this, we do not think that our data are directly comparable to published reports on Myo2 levels. Finally, we cannot rule out that newly synthesized myosin proteins are less stable in rng3 mutants, but we were unable to detect any changes. Because rng3 is essential, we inactivated it using a thermosensitive mutation. We took samples after 4 h at the restrictive temperature, at which point cells already showed a strong phenotype. If myosin proteins were very stable, a decrease in myosin half-lives would not be observable within this time frame. Although there are no published estimates for the turnover of S. pombe myosin proteins, S. cerevisiae Myo5 has a half-life of 5 h (Lord et al, 2008). It is also possible that myosin misfolding does not lead to enhanced degradation; for example, although inactivation of Hsp90 blocks the folding of myosin motor domains in reticulocyte lysates, it does not affect their synthesis or stability (Liu et al, 2008). Therefore, our results are compatible with a role for Rng3 in cotranslational folding.
Global study of translation-linked chaperone networks
Eukaryotic cells contain numerous chaperones and co-chaperones that assist de novo folding of proteins cotranslationally (Craig et al, 2003; Young et al, 2004; Rospert & Chacinska, 2006); however, little is known about the spectrum of their targets. The genome-wide identification of chaperone substrates is not straightforward and involves laborious systematic mapping of physical and genetic interactions between chaperones and other proteins (McClellan et al, 2007). Furthermore, as these approaches rely on protein–protein interactions, they cannot be used to distinguish between cotranslational and post-translational targets. We have used for the first time a microarray-based approach to systematically discover mRNAs associated with a chaperone. Our results show remarkable specificity: only seven transcripts copurified with Rng3 (around 0.1% of all S. pombe protein-coding genes), including all five RNAs that encode myosin heavy chains. These data support the idea that RIp-chip can be used to identify chaperone cotranslational targets in a straightforward manner. The systematic application of this approach to chaperones and co-chaperones will give an insight into how these proteins recognize their targets and will help to understand the organization of translation-linked chaperone networks at the genome-wide level.
Methods
Protein detection. Expression of tagged proteins was verified by Western blot using peroxidase–anti-peroxidase-soluble complexes (Sigma, St Louis, MO, USA) to detect the protein A-binding domains of TAP. S. pombe ribosomal proteins Rpl701 and Rpl702 were detected with antibodies against the conserved C terminus of the human RPL7 protein (Bethyl, Montgomery, TX, USA) and tubulin with a mouse monoclonal antibody (Sigma).
Polysome profiling. Polysomes were fractionated by using sucrose gradients as described previously (Lackner et al, 2007). Protein samples were prepared as follows: fractions were mixed with 0.75 volumes of methanol and 1 volume of chloroform, centrifuged at 13,000 g for 5 min and the upper phase was discarded. The remaining sample was mixed with 1 volume of methanol and centrifuged again at 13,000 g for 5 min. After removing the supernatant, the pellet was air-dried and resuspended in sample buffer (Nupage; Invitrogen, San Diego, CA, USA). To prepare samples for RNA analysis, fractions were collected in tubes containing around 2 volumes of 8 M guanidium-HCl and pooled as described in the main text. This was followed by ethanol precipitation (with 1 volume of ethanol) and purification through RNeasy columns (Qiagen, Venlo, The Netherlands). RNA was eluted from the columns using 30 μl of water, and 12 μl of sample was used for labelling. For the EDTA-treated gradient, EDTA was added to the extract and to all buffers at a concentration of 25 mM. Puromycin treatment was carried out as described previously (Blobel & Sabatini, 1971) by adjusting the concentrations of KCl and MgCl2 in the buffer to 500 and 2 mM, respectively, and incubating the extracts at 30°C for 10 min in the presence of 1 mM puromycin. RNase treatment was carried out by incubation of the extract with 25 U/ml of RNase A and 1000 U/ml of RNase T1 (Ambion, Austin, TX, USA).
RIp-chip. The protocol for RIp-chip is based on Gerber et al, 2004 (supplementary information online for full details). Here, 10% of the cell extract was used to prepare reference RNA and the rest for immunoprecipitation. Immunoprecipitation was carried out using monoclonal antibodies against protein A (Sigma) coupled to magnetic beads (Invitrogen). After washing the beads, the immunoprecipitated RNA was purified using an RNAqueous-Micro kit (Ambion). Purification of immunoprecipitated RNA in this way resulted in a major improvement compared with the use of phenol–chloroform extraction and ethanol precipitation. Total RNA was purified with RNeasy columns (Qiagen). The amount of RNA in the immunoprecipitate was sufficient to allow direct labelling without amplification.
Labelling and microarray experiments. Total RNA purified from the cell extract was used as a reference in all experiments. Here, 20 μg of total RNA, all the RNA from the immunoprecipitate or around 40% of the sample for the genome-wide translational profiles were labelled using the SuperScript™ Plus Direct cDNA Labelling System (Invitrogen). Labelled cDNAs were hybridized with S. pombe DNA microarrays as described previously (Lyne et al, 2003). Microarrays were scanned with a GenePix 4000A microarray scanner and analysed with GenePix Pro 5.0 (Molecular Devices, Sunnyvale, CA, USA). Spots that did not show a minimum of 50% of pixels above the median background signal plus two standard deviations in both channels (or at least 90% in the channel of the immunoprecipitate) were removed. Median log10 ratios were used for the analysis. To allow comparison between different samples in the EDTA experiment, RNA enrichments were normalized to the levels of the small nuclear RNA U3 (SPNCRNA.03) in the immunoprecipitate. Selection of enriched RNAs in the RIp-chip experiments was carried out as described previously (Gerber et al, 2004), by choosing genes, the enrichment ratios of which were at least two standard deviations above the median enrichment of all genes (Fig 2A). Assuming a normal distribution of the enrichments, the expected fraction of false positives using this threshold with a single experiment would be around 0.05. However, as we only selected RNAs enriched in each of four experiments, this number is reduced to around 6.25 × 10−6. Rng3-TAP RIp-chip experiments were carried out four times; the test of EDTA sensitivity was performed in duplicate; the sequential immunoprecipitation experiment was performed twice and the negative control for this experiment was carried out once. All repeats were independent biological experiments and dyes were swapped when labelling replicates.
All microarray data have been deposited in ArrayExpress (accession numbers, E-TABM-516, E-TABM-564 and E-TABM-565).
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Information
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
Acknowledgments
We thank Jürg Bähler for providing support throughout this project. We also thank Jürg Bähler and Samuel Marguerat for comments on the paper, and Daniel Lackner and Stephen Watt for help with microarray experiments and polysome separation. Kathy Gould (Vanderbilt University Medical Center, Nashville) provided the rng3-65 mutant. This study was supported by a Medical Research Council New Investigator Award (G0501168) to J.M.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barral JM, Bauer CC, Ortiz I, Epstein HF (1998) Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol 143: 1215–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF (2002) Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295: 669–671 [DOI] [PubMed] [Google Scholar]
- Bejsovec A, Anderson P (1988) Myosin heavy-chain mutations that disrupt Caenorhabditis elegans thick filament assembly. Genes Dev 2: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Blobel G, Sabatini D (1971) Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci USA 68: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadli A, Graham JD, Abel MG, Jackson TA, Gordon DF, Wood WM, Felts SJ, Horwitz KB, Toft D (2006) GCUNC-45 is a novel regulator for the progesterone receptor/hsp90 chaperoning pathway. Mol Cell Biol 26: 1722–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Eisenman HC, Hundley HA (2003) Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol 6: 157–162 [DOI] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Landsverk ML, Price MG, Epstein HF (2002) The UCS family of myosin chaperones. J Cell Sci 115: 3983–3990 [DOI] [PubMed] [Google Scholar]
- Kim J, Lowe T, Hoppe T (2008) Protein quality control gets muscle into shape. Trends Cell Biol 18: 264–272 [DOI] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk ML, Li S, Hutagalung AH, Najafov A, Hoppe T, Barral JM, Epstein HF (2007) The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J Cell Biol 177: 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Srikakulam R, Winkelmann DA (2008) Unc45 activates Hsp90-dependent folding of the myosin motor domain. J Biol Chem 283: 13185–13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Pollard TD (2004) UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol 167: 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Sladewski TE, Pollard TD (2008) Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells. Proc Natl Acad Sci USA 105: 8014–8019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J (2007) Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131: 121–135 [DOI] [PubMed] [Google Scholar]
- Mishra M, D'Souza VM, Chang KC, Huang Y, Balasubramanian MK (2005) Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot Cell 4: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Chacinska A (2006) Distinct yet linked: chaperone networks in the eukaryotic cytosol. Genome Biol 7: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R, Winkelmann DA (1999) Myosin II folding is mediated by a molecular chaperonin. J Biol Chem 274: 27265–27273 [DOI] [PubMed] [Google Scholar]
- Srikakulam R, Winkelmann DA (2004) Chaperone-mediated folding and assembly of myosin in striated muscle. J Cell Sci 117: 641–652 [DOI] [PubMed] [Google Scholar]
- Srikakulam R, Liu L, Winkelmann DA (2008) Unc45b forms a cytosolic complex with Hsp90 and targets the unfolded myosin motor domain. PLoS ONE 3: e2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toi H, Fujimura-Kamada K, Irie K, Takai Y, Todo S, Tanaka K (2003) She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol Biol Cell 14: 2237–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche S, Arnold M, Jansen RP (2003) The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr Biol 13: 715–724 [DOI] [PubMed] [Google Scholar]
- Win TZ, Mulvihill DP, Hyams JS (2002) Take five: a myosin class act in fission yeast. Cell Motil Cytoskeleton 51: 53–56 [DOI] [PubMed] [Google Scholar]
- Wong KC, Naqvi NI, Iino Y, Yamamoto M, Balasubramanian MK (2000) Fission yeast Rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. J Cell Sci 113: 2421–2432 [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3