Abstract
The mouse double minute 2 (MDM2)–p53 interaction regulates the activity of p53 and is a potential target for human cancer therapy. Here, we report that RYBP (RING1- and YY1-binding protein), a member of the polycomb group (PcG), interacts with MDM2 and decreases MDM2-mediated p53 ubiquitination, leading to stabilization of p53 and an increase in p53 activity. RYBP induces cell-cycle arrest and is involved in the p53 response to DNA damage. Expression of RYBP is decreased in human cancer tissues compared with adjacent normal tissues. These results show that RYBP is a new regulator of the MDM2–p53 loop and that it has tumour suppressor activity.
Keywords: MDM2, p53, RYBP, ubiquitination, human cancer
Introduction
Tumour suppressor p53 helps to maintain genomic integrity, regulates the cell-stress response, and controls human cancer development and progression. The loss of normal functional p53 can result in malignancies. In response to cellular stress, p53 stabilization and activation lead to cell-cycle arrest, DNA repair, apoptosis and/or senescence (Harris & Levine, 2005). Approximately half of all human malignancies carry mutant p53, and many tumours containing wild-type p53 have abnormalities in p53 regulators such as mouse double minute 2 (MDM2; Vogelstein et al, 2000).
The MDM2 oncogene is amplified and/or overexpressed in a wide range of human cancers (reviewed by Rayburn et al, 2005). MDM2 inhibits the activities of p53, and promotes its ubiquitination and degradation (Haupt et al, 1997). In turn, the transcription of MDM2 is activated by p53. This MDM2–p53 feedback loop is one of the main mechanisms of p53 regulation, and it has been considered a target for human cancer therapy and prevention (Poyurovsky & Prives, 2006). The molecular mechanisms responsible for regulating the MDM2–p53 feedback loop are not fully understood; however, we and others have identified several regulators of the MDM2–p53 loop (Zhang et al, 2003, 2004; Dai et al, 2004; Chen et al, 2007; He & Sun, 2007; Sun et al, 2008; Zhang & Zhang, 2008). Here, we identified RYBP (RING1 and YY1 binding protein), a polycomb group (PcG) complex member, as a new modulator of the MDM2–p53 interaction.
Results
RYBP binds to MDM2
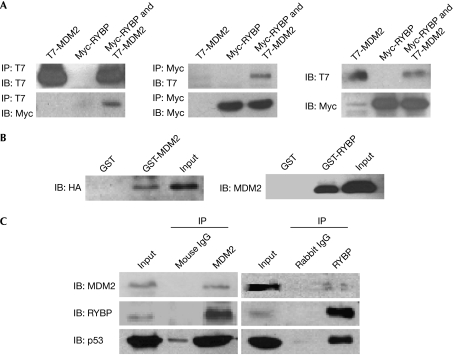
A yeast two-hybrid assay identified several proteins that interact with MDM2, including ribosomal protein (RP) L11, L23 and S7, which have been shown previously to bind to MDM2 (Zhang et al, 2003; Dai et al, 2004; Chen et al, 2007). We also identified three clones encoding a protein corresponding to RYBP. To determine whether MDM2 and RYBP interact in mammalian cells, T7-MDM2 and Myc-RYBP were co-transfected into COS7 (Fig 1A) and A549 (supplementary Fig S1A online) cells. T7-MDM2 co-precipitated with Myc-RYBP in cells with both MDM2 and RYBP, but not in cells with either protein alone. Assays performed in vitro showed that MDM2 pulls down RYBP and vice versa (Fig 1B), suggesting an interaction between MDM2 and RYBP. This was confirmed by reciprocal co-immunoprecipitation with endogenous MDM2 and RYBP in U2OS cells (Fig 1C). In addition, we found that two RYBP fragments (residues 1–73 and 144–228) were able to bind to MDM2, and the RYBP-binding domain of MDM2 was mapped to residues 180–298 (supplementary Fig S1B–E online).
Figure 1.
RYBP binds to MDM2. (A) COS7 cells (5 × 105 cells per well) were transfected with 10 μg of T7-MDM2, Myc-RYBP or both for 24 h. The lysates were immunoprecipitated (IP) with T7 (left panel) or Myc (middle panel) antibodies or directly immunoblotted (IB) with T7 and Myc antibodies (right panel). (B) GST, GST-MDM2 or GST-RYBP proteins were incubated with in vitro-translated HA-RYBP or Myc-MDM2 at 4°C overnight. The bound proteins were resolved on SDS–PAGE and detected by HA (left panel) or MDM2 antibodies (right panel). (C) U2OS cells were lysed in NP-40 lysis buffer, and the lysates were immunoprecipitated with mouse IgG or MDM2 2A10 (left panel), rabbit IgG or rabbit RYBP antibodies (right panel) at 4°C overnight. Protein G beads were then added to the mixture and rotated for another 1 h at 4°C. After washing with NP-40 buffer, the precipitates were separated by SDS–PAGE and the target proteins were detected with MDM2 2A10, p53 DO-1 or RYBP antibodies. DO-1, antibody against p53; GST, glutathione S-transferase; HA, haemagglutinin; MDM2, mouse double minute 2; RYBP, RING1- and YY1-binding protein; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
RYBP stabilizes p53 and MDM2
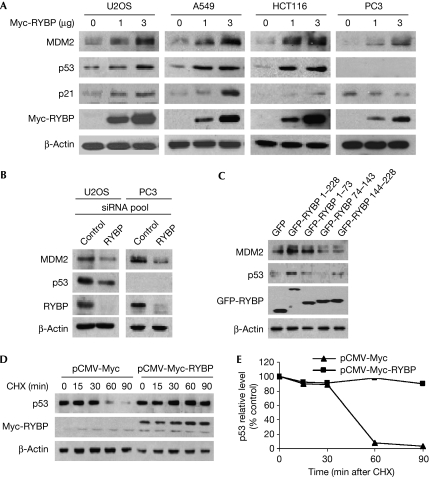
To examine the functional significance of the RYBP–MDM2 interaction, RYBP was overexpressed in U2OS, A549, HCT116 and PC3 cells. In a dose-dependent manner, ectopically expressed RYBP upregulated p53, MDM2 and p21WAF1 in U2OS, A549 and HCT116 cells (Fig 2A). In PC3 cells (p53 null), MDM2 was upregulated and p21WAF1 downregulated (Fig 2A, right panel). In U2OS cells, when RYBP was knocked down by a specific small interfering RNA (siRNA) pool, p53 and MDM2 were downregulated. MDM2 was also downregulated in PC3 cells when RYBP was knocked down (Fig 2B), confirming that RYBP directly stabilizes MDM2 independently of p53. To determine whether RYBP binding to MDM2 is necessary for the effects on p53, various deletion mutants of RYBP were transfected into U2OS cells. Wild-type RYBP increased the p53 protein level, while the two RYBP fragments (residues 1–73 or 144–228) able to bind to MDM2 had a less pronounced effect, and the RYBP mutant unable to bind to MDM2 failed to increase the stability of p53 (Fig 2C). This suggests that full-length RYBP binding to MDM2 is required to stabilize p53 efficiently. In a time-course study, overexpression of RYBP led to an increase in the half-lives of both p53 and MDM2 (Fig 2D,E; supplementary Fig S2A,B online), whereas knockdown of RYBP by siRNA led to a decrease in the stability of both MDM2 and p53 (supplementary Fig S2C online).
Figure 2.
RYBP stabilizes p53. (A) U2OS, A549, HCT116 and PC3 cells were transfected with a Myc-RYBP plasmid for 24 h. Immunoblotting was used to assess the MDM2, p53 and p21WAF1 protein levels. (B) U2OS or PC3 cells were transfected with 100 nM of control or RYBP siRNA pool for 72 h. MDM2, p53 and RYBP proteins in whole lysates were detected by immunoblotting. (C) U2OS cells were transfected with GFP vector, or deletion mutants of GFP-RYBP (4 μg) for 24 h. The lysates were cleared, and SDS–PAGE was used to detect MDM2 and p53. (D,E) A549 cells were transfected with a CMV vector or CMV-RYBP (4 μg) for 24 h followed by exposure to CHX (10 μg/ml) for the indicated times. (D) Cell lysates were collected and detected as above. (E) The intensity of the p53 bands was analysed by densitometry (Bio-Rad Model GS-670 Imaging Densitometer; Bio-Rad, Hercules, CA, USA). The relative densitometry at each time point is expressed as a percentage of the density at time 0 h after normalization to the corresponding β-actin level. CHX, cycloheximide; CMV, cytomegalovirus; GFP, green fluorescent protein; MDM2, mouse double minute 2; RYBP, RING1- and YY1-binding protein; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; siRNA, small interfering RNA.
RYBP inhibits MDM2-mediated p53 ubiquitination
Interaction assays in COS7 cells transfected with only p53 and RYBP indicated that there was no direct binding between p53 and RYBP, and p53/RYBP co-immunoprecipitation was seen only when MDM2 was added (supplementary Fig S3A online). In addition, RYBP precipitated p53 in COS7 cells co-transfected with wild-type MDM2, but not in cells with mutant MDM2 lacking the RYBP-binding domain (supplementary Fig S3B online).
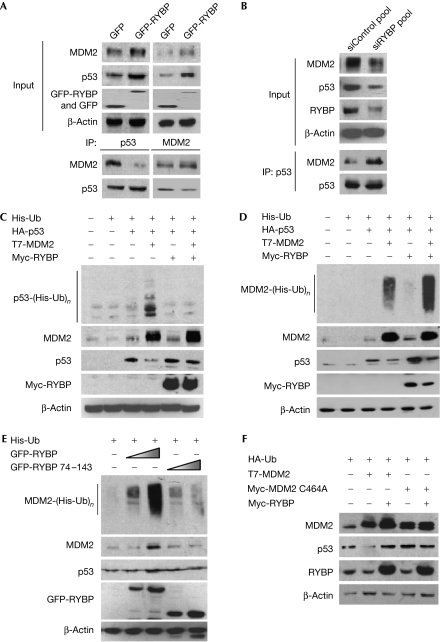
In U2OS cells, ectopically expressed RYBP decreased the interaction between MDM2 and p53 (Fig 3A), but knockdown of RYBP by siRNA increased the interaction (Fig 3B). These results were confirmed after MG132 was added under the same conditions (supplementary Fig S3C online). Furthermore, in COS7 cells co-transfected with T7-MDM2, haemagglutinin (HA)-p53 and green fluorescent protein (GFP)-RYBP, increasing the expression of RYBP decreased the interaction between MDM2 and p53, as measured by immunoprecipitation using a GFP antibody (supplementary Fig S3D online).
Figure 3.
RYBP inhibits MDM2-mediated ubiquitination of p53. (A) U2OS cells were transfected with plasmids expressing a GFP vector or GFP-RYBP (10 μg) for 24 h. The cell lysates were immunoprecipitated (IP) with p53 FL393 or MDM2 H221, and the MDM2 and p53 protein levels were determined using MDM2 2A10 or p53 DO-1. (B) U2OS cells were transfected with either the control or RYBP siRNA pools (100 nM) for 72 h. Immunoprecipitation was accomplished with the p53 FL393 antibody, and the corresponding proteins were detected by immunoblotting. (C,D) A549 cells were transfected with His-Ub, HA-p53, T7-MDM2 and Myc-RYBP as indicated in the figures. At 24 h after transfection, the cells were collected and divided into two fractions. One was used directly for Western blot analysis. The other was lysed with Ni-NTA lysis buffer, and the lysates were purified using Ni-NTA agarose beads and immunoblotted with corresponding antibodies. (E) A549 cells were transfected with His-Ub and with increasing doses of GFP-RYBP, or a mutant of GFP-RYBP lacking the MDM2-binding domain for 24 h. The same strategy as described in Fig. 4D was used to analyse the endogenous ubiquitination of MDM2. (F) U2OS cells were transfected with combinations of plasmids, including the MDM2 mutant lacking E3 ligase activity (C464A), as indicated, for 24 h. Target proteins in the whole-cell lysates were detected by immunoblotting. CHX, cycloheximide; DO-1, antibody against p53; GFP, green fluorescent protein; HA, haemagglutinin; MDM2, mouse double minute 2; Ni-NTA, nickel-nitrilotriacetic acid; RYBP, RING1- and YY1-binding protein; siRNA; small interfering RNA; Ub, ubiquitin.
To determine whether RYBP stabilizes p53 by inhibiting its ubiquitination, A549 cells were co-transfected with HA-ubiquitin (Ub) or His-Ub, T7-MDM2, HA-p53 and Myc-RYBP. MDM2 transfection led to an increase in the ubiquitination of p53 as measured by Western blotting, but when cells were co-transfected with RYBP, the level of ubiquitinated p53 decreased (supplementary Fig S3E online, left panel, compare lanes 4 and 6). The effect of RYBP on p53 ubiquitination was further shown by immunoprecipitation assays with a p53 antibody (supplementary Fig S3E online, middle panel) and by using nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Fig 3C). We also observed that the decrease in p53 ubiquitination occurred in an RYBP dose-dependent manner (supplementary Fig S3E online, right panel) and that the level of ubiquitinated p53 increased when RYBP was knocked down by siRNA in U2OS cells (supplementary Fig S3F online). Although co-transfection of RYBP increased the level of ubiquitinated exogenous and endogenous MDM2 in A549 cells in a dose-dependent manner (Fig 3D,E), this did not occur in cells transfected with a mutant RYBP that was unable to bind to MDM2 (Fig 3E).
To determine whether the effects of RYBP on p53 are related to the inhibition of the E3 ligase activity of MDM2, wild-type or mutant MDM2 without E3 ligase activity (C464A) was co-transfected with or without RYBP into U2OS cells. Transfection with RYBP prevented p53 downregulation in cells transfected with wild-type MDM2, but when cells were transfected with the mutant MDM2 (C464A), there were no apparent changes in the expression or stability of p53 (Fig 3F). This indicates that, in unstressed cells, RYBP probably regulates the stability of p53 by forming a RYBP–MDM2–p53 complex that prevents the association between MDM2 and p53, thereby inhibiting the E3 ligase of MDM2 from ubiquitinating p53. RYBP also stabilizes native MDM2 while simultaneously increasing ubiquitinated MDM2. Thus, it might be possible that RYBP stabilizes MDM2 by inhibiting the proteasome-mediated degradation of ubiquitinated MDM2.
RYBP regulates the activity of p53
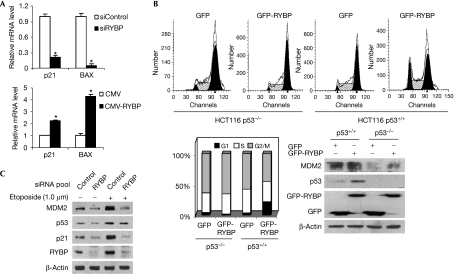
The effects of RYBP on the transactivational activity of p53 were evaluated using quantitative reverse transcription–PCR (qRT–PCR). As shown in Fig 4A, knockdown of RYBP by siRNA inhibited cyclin-dependent kinase inhibitor 1A (CDKN1A; p21) and Bcl2-associated X protein (BAX) messenger RNA (mRNA) levels by 4.7- and 20.0-fold respectively; overexpression of RYBP induced their mRNA expression levels by 2.3- and 4.3-fold, respectively. In addition, when U2OS cells were transiently co-transfected with vectors expressing RYBP and the p21WAF1 promoter luciferase reporter, the luciferase activity was increased by RYBP in a dose-dependent manner (supplementary Fig S4A online). Similar results were obtained in MCF7 cells with wild-type p53, but not in MCF7 cells with p53 knockdown (supplementary Fig S4B online). Transfection with RYBP also resulted in G1 arrest in HCT116 p53+/+ cells, but not in HCT116 p53−/− cells (Fig 4B).
Figure 4.
RYPB regulates the activity of p53. (A) U2OS cells were transfected with siControl or siRYBP pool (100 nM) for 72 h, or transfected with a CMV or CMV-RYBP vector (4 μg) for 24 h. The total RNA was then extracted and qRT–PCR was carried out to detect the relative messenger RNA (mRNA) levels of CDKN1A (p21) and BAX; *P<0.05. (B) HCT116 cells with or without p53 were transfected with a vector expressing either GFP or GFP-RYBP for 24 h, and then the GFP-positive populations were examined for DNA content (upper panels). A plot of the percentages of each phase of the cell cycle in various cells following RYBP treatment is included in the lower left panel. Analyses of protein lysates from the same transfected cells are shown in the lower right panel. (C) U2OS cells were exposed either to a control or to an RYBP siRNA pool (100 nM) for 48 h. The cells were then exposed to 1.0 μM etoposide for an additional 24 h, and the expression of target proteins was examined by immunoblotting. BAX, Bcl2-associated X protein; CDKN1A, cyclin-dependent kinase inhibitor 1A; CMV, cytomegalovirus; GFP, green fluorescent protein; qRT–PCR, quantitative reverse transcription–PCR; RYBP, RING1- and YY1-binding protein; si, small interfering.
When U2OS cells were exposed to the chemotherapeutic agent doxorubicin (0–100 nM) or etoposide (0–10 μM), there were dose-dependent increases in RYBP, together with p53, p21WAF1 and MDM2 (supplementary Fig S4C,D online, left panels). In cells exposed to doxorubicin (100 nM) or etoposide (1.0 μM) for various times (0–24 h), there were time-dependent increases in RYBP, together with p53, p21WAF1 and MDM2 (supplementary Fig S4C,D online, right panels). Furthermore, when U2OS cells were exposed to a siRNA pool against RYBP, the increases in p53, p21WAF1 and MDM2 following etoposide exposure were inhibited (Fig 4C), showing that RYBP is involved in the p53 response to DNA damage.
RYBP is downregulated in human cancers
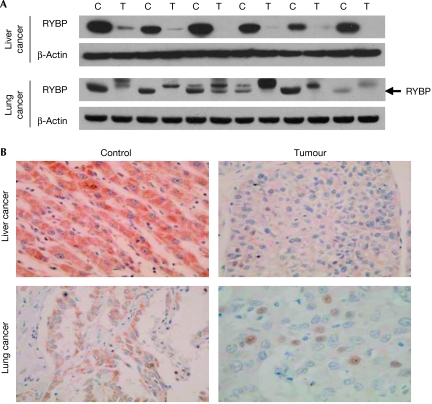
To show the clinical relevance of RYBP, we evaluated its expression in human liver and lung cancers, and matched adjacent non-malignant tissues. As determined by Western blotting, the cancer tissues had lower or undetectable RYBP protein levels, or might have had unknown post-translational modifications to the protein, compared with their corresponding control tissues (Fig 5A). Further immunohistochemical analyses indicated that RYBP was downregulated in tumour tissues, compared with their matched control tissues (Fig 5B).
Figure 5.
RYBP is downregulated in cancer tissues compared with matched normal tissues. (A) Proteins were extracted from adjacent normal and malignant liver and lung tissues, and immunoblotting was used to detect the expression of RYBP and β-actin. (B) Immunohistochemical analyses were performed to determine the expression of RYBP in tumour and normal adjacent tissue samples. Representative pictures from liver and lung tissues were selected to show the alterations in the expression of RYBP. C, control tissue; RYBP, RING1- and YY1-binding protein; T, tumor tissue.
Discussion
The regulation of p53 has been a major focus for human cancer research (Poyurovsky & Prives, 2006). The p53 protein is primarily regulated at the post-translational level through the ubiquitination–proteasome system, and its ubiquitination is largely induced by MDM2 (Jones et al, 1995; Kubbutat et al, 1997). This study represents our continuing efforts in determining the mechanisms responsible for the regulation of the MDM2–p53 interaction. Herein, we show that RYBP is a new regulator of the MDM2–p53 loop, and provide evidence that it functions as a tumour suppressor.
Although several studies have indicated that RYBP is a ubiquitin-binding protein (Arrigoni et al, 2006) that induces apoptosis (Zheng et al, 2001; Danen-van Oorschot et al, 2004; Novak & Phillips, 2008), its precise function remains unclear. Here, we used human tissue samples to show that RYBP is downregulated or modified in tumour tissues compared with corresponding control tissues. We also showed an increase in RYBP and p53 following exposure to chemotherapeutic agents. Our studies suggest that the activation of p53 following etoposide-induced DNA damage requires RYBP.
We speculate that RYBP binding to MDM2 results in changes in the conformation of MDM2, leading to an alteration of its interaction with p53, precluding its function in the ubiquitination and degradation of p53. In addition, we suggest that there is a distinct role for the ternary complex of RYBP, MDM2 and p53, beyond its main activity in inhibiting the MDM2–p53 interaction. As RYBP binds to MDM2 but not to p53, it is likely that the ternary complex acts as a reservoir of both p53 and MDM2, maintaining a basal level of the proteins. Following cellular stress such as DNA damage, the ternary complex might undergo conformational changes, allowing p53 to be released—to respond to the stress—while limiting the free MDM2 pool, thereby reducing the rate of p53 degradation. In addition, the ternary complex might be required for the functions of both RYBP and p53 in regulating gene expression (García et al, 1999). These hypotheses should be tested in future studies.
We cannot exclude other effects of RYBP on MDM2. Our results indicate that RYBP stabilizes MDM2, independently of p53. In the absence of p53, MDM2 is able to repress transcription when tethered to a reporter gene promoter (Minsky & Oren, 2004). Therefore, RYBP and MDM2 might form a protein complex in which RYBP recruits MDM2 to its target gene promoters. Furthermore, as the PcG complex is necessary for the repression of important developmental genes, and as RYBP, MDM2 and p53 are all involved in apoptosis, future studies should investigate the functional consequences of the link between the PcG complex, RYBP, MDM2 and p53 in mammalian development and carcinogenesis, as well as their implications in disease prevention and treatment.
Methods
Plasmids and reagents. The yeast two-hybrid assay was described previously (Chen et al, 2007). The amplified RYBP sequence was cloned into pGADT7 and pCMV-3Tag vectors. The preparation and sources of other plasmids, reagents, qRT–PCR primers and conditions are described in the supplementary information online.
Immunoprecipitation and immunoblotting. Plasmids were transfected with Lipofectin® into cells as indicated. Cells were collected at the indicated times post-transfection, and lysed in NP-40 lysis buffer containing a protease inhibitor mixture from Sigma (St Louis, MO, USA). Cell lysates were used for immunoblotting as described previously (Zhang & Zhang, 2008). Immunoprecipitation was conducted using the indicated antibodies. Beads were washed, and bound proteins were detected by immunoblotting as reported previously (Zhang & Zhang, 2008).
Ni-NTA ubiquitination assay. Ni-NTA ubiquitination assays were conducted as described previously with minor modifications (Zhang et al, 2004).
Generation of GST fusion proteins and pull-down assays. Binding between the MDM2 protein and RYBP in vitro was assessed using glutathione S-transferase (GST) fusion proteins. The bound proteins were captured by glutathione-agarose beads and resolved using SDS–polyacrylamide gel electrophoresis (Zhang & Zhang, 2008).
Cell-cycle distribution assay. The cell-cycle distribution assay was accomplished as described previously (Zhang & Zhang, 2008).
Analysis of RYBP levels in human cancer tissues and controls. This study was approved by the Institutional Review Board of Nantong Hospital. Fresh surgical specimens of hepatocellular (n=6) and lung (n=6) cancer tissues and adjacent non-cancerous tissues were obtained from Nantong Cancer Hospital, Nantong, China. All the diagnoses were histologically confirmed. Western blot analyses with fresh tissues, as well as immunohistochemical analyses of formalin-fixed paraffin-embedded sections, were conducted as described in the supplementary information online.
Supplementary Information is available at EMBO Reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
This study was supported by the National Institutes of Health (NIH) grants R01 CA112029 and R01 CA121211. E.R.R. was supported by a Department of Defense (DoD) grant W81XWH-06-1-0063 and a T32 fellowship from the NIH/University of Alabama at Birmingham (UAB) Gene Therapy Center (CA075930). H.W. was supported by the Ministry of Science and Technology of China (2007CB947100) and the National Natural Science Foundation of China (30870513). We thank Dr Z. Zhang for helpful discussions, Q. Ba, L. Xiang, S. Wu, M. Li, Y. Liu and X. Li for excellent technical assistance.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N (2006) The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett 580: 6233–6241 [DOI] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R (2007) Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26: 5029–5037 [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24: 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen-van Oorschot AA, Voskamp P, Seelen MC, van Miltenburg MH, Bolk MW, Tait SW, Boesen-de Cock JG, Rohn JL, Borst J, Noteborn MH (2004) Human death effector domain-associated factor interacts with the viral apoptosis agonist Apoptin and exerts tumor-preferential cell killing. Cell Death Differ 11: 564–573 [DOI] [PubMed] [Google Scholar]
- García E, Marcos-Gutiérrez C, del Mar Lorente M, Moreno JC, Vidal M (1999) RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J 18: 3404–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- He H, Sun Y (2007) Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene 26: 2707–2716 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A (1995) Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208 [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M (2004) The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 16: 631–639 [DOI] [PubMed] [Google Scholar]
- Novak RL, Phillips AC (2008) Adenoviral-mediated Rybp expression promotes tumor cell-specific apoptosis. Cancer Gene Ther 15: 713–722 [DOI] [PubMed] [Google Scholar]
- Poyurovsky MV, Prives C (2006) Unleashing the power of p53: lessons from mice and men. Genes Dev 20: 125–131 [DOI] [PubMed] [Google Scholar]
- Rayburn E, Zhang R, He J, Wang H (2005) MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets 5: 27–41 [DOI] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H (2008) Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem 283: 12387–12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408: 307–310 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23: 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang R (2008) Proteasome activator PA28γ regulates p53 by enhancing its MDM2-mediated degradation. EMBO J 27: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang H, Li M, Agrawal S, Chen X, Zhang R (2004) MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J Biol Chem 279: 16000–16006 [DOI] [PubMed] [Google Scholar]
- Zheng L, Schickling O, Peter ME, Lenardo MJ (2001) The death effector domain-associated factor plays distinct regulatory roles in the nucleus and cytoplasm. J Biol Chem 276: 31945–31952 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information