Abstract
Qualitative and quantitative changes in mitochondrial DNA (mtDNA) have been shown to be common causes of inherited neurodegenerative and muscular diseases, and have also been implicated in ageing. These diseases can be caused by primary mtDNA mutations, or by defects in nuclear-encoded mtDNA maintenance proteins that cause secondary mtDNA mutagenesis or instability. Furthermore, it has been proposed that mtDNA copy number affects cellular tolerance to environmental stress. However, the mechanisms that regulate mtDNA copy number and the tissue-specific consequences of mtDNA mutations are largely unknown. As post-mitotic tissues differ greatly from proliferating cultured cells in their need for mtDNA maintenance, and as most mitochondrial diseases affect post-mitotic cell types, the mouse is an important model in which to study mtDNA defects. Here, we review recently developed mouse models, and their contribution to our knowledge of mtDNA maintenance and its role in disease.
Keywords: mitochondria, mitochondrial DNA, mouse model
Glossary
8-OHdG 8-hydroxydeoxyguanosine
ANT1 adenine nucleotide translocator; heart, muscle and brain-specific isoform
ATP adenosine triphosphate
cDNA complementary DNA
COI mitochondrially encoded cytochrome c oxidase 1
dNTP deoxyribonucleotide triphosphate
dTTP deoxythymidine triphosphate
HSP40 heat-shock protein 40 mitochondrial chaperone
ND6 mitochondrially encoded NADH dehydrogenase 6
p53R2 ribonucleotide reductase subunit
POLG mitochondrial DNA polymerase γ
RNaseH1 ribonuclease H1
RRM2B ribonucleotide reductase subunit M2B
TFAM mitochondrial transcription factor A
TK2 thymidine kinase 2
tRNA transfer RNA
Introduction
Mitochondrial dysfunction is a major cause of inherited metabolic diseases, and mitochondrial DNA (mtDNA) mutations have an estimated prevalence of 1:200 and carrier frequencies of more than 1:100 for nuclear-encoded gene mutations in Western populations (Elliott et al, 2008; Hakonen et al, 2005; Hudson & Chinnery, 2006). Furthermore, mitochondrial dysfunction has an important role in age-related pathologies such as cancer, type II diabetes and neurodegeneration, and possibly even in normal ageing. This exceptionally wide clinical spectrum can be attributed to the range of mitochondrial functions, which extend from oxidative ATP production and synthesis of iron–sulphur clusters, haem, amino acids, steroid hormones and neurotransmitters, to the regulation of cytoplasmic calcium levels and key events in apoptosis (Chan, 2006). However, most of the known mitochondrial disorders are caused primarily by a dysfunctional respiratory chain (DiMauro & Schon, 2003; Shoubridge, 2001; Zeviani & DiDonato, 2004). Thirteen key catalytic subunits of oxidative phosphorylation (OXPHOS) complexes are encoded by the mitochondrial genome (Anderson et al, 1981), whereas the rest of the approximately 80 OXPHOS proteins, as well as all other mitochondrial proteins, are encoded by nuclear genes. The existence of mtDNA is completely controlled by the nucleus, which encodes all of the proteins that are required for mtDNA maintenance (Fig 1). Mutations in these nuclear genes cause autosomal inherited secondary mtDNA instability.
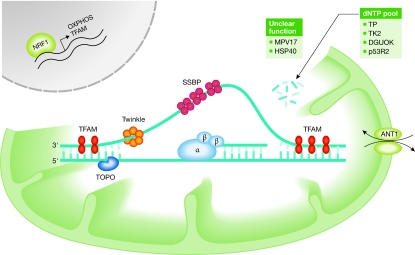
Figure 1.
Essential mitochondrial DNA maintenance proteins. All mtDNA maintenance proteins are encoded by nuclear genes, which are transcriptionally regulated by factors such as NRF1. Some proteins are involved directly in mtDNA replication and some provide nucleotides for DNA synthesis. The specific function of some of these proteins is still unclear but their absence induces mtDNA defects. ANT1, adenine nucleotide translocator 1; DGUOK, deoxyguanosine kinase; HSP40, heat-shock protein 40 mitochondrial chaperone; MPV17, mitochondrial inner membrane protein; mtDNA, mitochondrial DNA; NRF1, nuclear respiratory factor 1; OXPHOS, oxidative phosphorylation; p53R2, ribonucleotide reductase subunit; SSBP, single-stranded DNA-binding protein; TFAM, mitochondrial transcription factor A; TK2, thymidine kinase 2; TOPO, topoisomerase; TP, thymidine phosphorylase.
Several key questions have yet to be answered in order to understand mtDNA-associated pathology (Sidebar A). If OXPHOS deficiency leads to insufficient ATP synthesis, why are there tens of different mtDNA diseases instead of just one? Mutations in different mitochondrial tRNA genes cause specific syndromes: tRNALeu mutations are involved in hearing loss, diabetes, cardiomyopathy and early strokes (Finsterer, 2007; Kobayashi et al, 1991); tRNALys mutations lead to myoclonic epilepsy (Shoffner et al, 1990); and tRNASer mutations typically cause deafness (Reid et al, 1994). This pathogenic variability cannot be explained on the basis of ATP deficiency. How does the level of heteroplasmy affect the disease outcome? Primary mtDNA mutations cause maternally inherited disorders, the severity of which is loosely correlated with the amount of mutant mtDNA—that is, the level of heteroplasmy. However, low levels of one mutation cause diabetes, whereas high levels of the same mutation cause stroke, indicating that subtle-to-severe dysfunction must be interpreted differently by different sets of cells. Does developmental and somatic mtDNA segregation affect disease outcomes? During oogenesis, the amount of mtDNA undergoes a bottleneck and then rapidly expands (Jenuth et al, 1996; Cree et al, 2008). Are some mutations actively selected away during development? Does the heteroplasmy level change somatically? Understanding this process would be the cornerstone of preimplantation diagnosis. In addition, what are the physiological consequences of mtDNA disorders? Which cells are susceptible at which stage of development? Why do families with the same nuclear mutation show wide variation in their disease severity? Can ‘nuclear background' affect the individual capacity for stress response or can mtDNA copy number explain variation in disease manifestations? What proteins are required for mtDNA maintenance? Many such proteins have been found because, when they are mutated, they lead to mitochondrial disease.
Sidebar A | In need of answers.
Why are mitochondrial DNA (mtDNA) diseases so phenotypically variable?
How does the level of mtDNA heteroplasmy affect the phenotypic outcome?
What are the mechanisms of mtDNA segregation?
What are the physiological consequences of mtDNA disorders?
Which set of proteins is required for mtDNA maintenance?
These fundamental questions about mtDNA biology remain mostly unsolved and will only be answered by using appropriate animal models. Therefore, the genetic manipulation of mtDNA in mice has become a crucial tool for understanding mitochondrial pathology (Table 1).
Table 1.
Manipulation of mitochondrial DNA or proteins involved in its maintenance in mice
| Mouse model | References |
|---|---|
| Transmitochondrial models | |
| Mito-mice | Inoue et al, 2000; Nakada et al, 2004 |
| Chloramphenicol resistance | Marchington et al, 1999; Sligh et al, 2000 |
| COI T6589C + ND6 13885insCdelT | Fan et al, 2008 |
| NZB+BALB | Jenuth et al, 1996; Battersby et al, 2003 |
| Restriction endonuclease targeted to mitochondria | |
| PstI, muscle-specific | Srivastava & Moraes, 2005 |
| Manipulation of mtDNA by modifying nuclear disease genes | |
| ANT1−/− | Graham et al, 1997 |
| TP−/−,UP−/− | Haraguchi et al, 2002 |
| Twinkle dup353–365 transgenic, Deletor | Tyynismaa et al, 2005 |
| POLGA−/− | Hance et al, 2005 |
| POLGA D181A heart-specific transgenic | Zhang et al, 2000 |
| POLGA, D257A knock-in, Mutator | Trifunovic et al, 2004; Kujoth et al, 2005 |
| POLGA D181A brain-specific transgenic | Kasahara et al, 2006 |
| POLGA Y955C heart-specific transgenic | Lewis et al, 2007 |
| TFAM−/− | Larsson et al, 1998 |
| TFAM−/− heart | Wang et al, 1999; Li et al, 2000 |
| TFAM−/− pancreatic β-cell | Silva et al, 2000 |
| TFAM−/− neocortex, neuronal | Sorensen et al, 2001 |
| TFAM−/− skeletal muscle | Wredenberg et al, 2002 |
| TFAM−/− dopamine neurons | Ekstrand et al, 2007 |
| RNaseH1−/− | Cerritelli et al, 2003 |
| RRM2B−/− | Kimura et al, 2003; Bourdon et al, 2007 |
| TK2−/− | Akman et al, 2008 |
| TK2 H126N knock-in | Zhou et al, 2008 |
| NRF1−/− | Huo & Scarpulla, 2001 |
| MPV17−/− | Weiher et al, 1990; Viscomi et al, 2008 |
| HSP40−/− | Hayashi et al, 2006 |
| Mice with increased mtDNA copy number | |
| TFAM overexpression | Ekstrand et al, 2004 |
| TFAM overexpression | Ikeuchi et al, 2005 |
| TK2 overexpression | Hosseini et al, 2007 |
| Twinkle overexpression | Tyynismaa et al, 2004 |
See supplementary Tables 1, 2, 3 online for further details. ANT1, adenine nucleotide translocator 1; BALB, inbred mouse strain; COI, mitochondrially encoded cytochrome c oxidase 1; del, deletion; dup, duplication; HSP40, heat-shock protein 40 mitochondrial chaperone; ins, insertion; MPV17, mitochondrial inner membrane protein; mtDNA, mitochondrial DNA; ND6, mitochondrially encoded NADH dehydrogenase 6; NRF1, nuclear respiratory factor 1; NZB, inbred mouse strain; POLGA, DNA polymerase-γ α subunit; PstI, restriction endonuclease; RNaseH1, ribonuclease H1; RRM2B, ribonucleotide reductase subunit M2B; TFAM, mitochondrial transcription factor A; TK2, thymidine kinase 2; TP, thymidine phosphorylase; UP, uridine phosphorylase.
Direct manipulation of mtDNA
The generation of heteroplasmic mice with pathological mtDNA mutations would be an important tool for understanding mtDNA segregation and the tissue specificity of disease manifestations. However, engineering mtDNA has proved challenging owing to the multicopy nature of the mitochondrial genome: hundreds to thousands of mtDNAs exist within one cell. Furthermore, the transfection of plasmids or modified mtDNA into animal mitochondria has not been successful (McGregor et al, 2001). Supplementary Table 1 online summarizes the indirect approaches that have been used to introduce primary mtDNA mutations into mice.
Ageing-related somatic mtDNA mutagenesis was used to generate heteroplasmic mice: synaptosomes with mutant mtDNA were isolated and fused to cells lacking mtDNA, and enucleated cytoplasts were fused to fertilized pronuclear-state embryos (Inoue et al, 2000). The transmitochondrial ‘Mito-mice' carried a heteroplasmic single mtDNA deletion in high amounts, and presented respiratory-chain deficiency in the heart, skeletal muscle and kidneys. The mice developed severe anaemia, myopathy, cardiomyopathy, deafness and renal failure, and died by 6 months of age (Inoue et al, 2000; Nakada et al, 2004). Surprisingly, the deleted mtDNA was transmitted through the maternal line to the offspring, although single mtDNA deletions are not usually inherited. The transmission of the defect through the germline might have been possible through mtDNA duplications. The inherited nature of the trait allowed the maintenance of the mouse line and emphasized the importance of Mito-mice in studying mitochondrial disease pathogenesis. In humans, early-onset Pearson syndrome is caused by a single heteroplasmic mtDNA deletion that leads to anaemia, pancreatic and renal insufficiency, and mitochondrial myopathy, closely mimicking the phenotype of Mito-mice.
Heteroplasmic mice have also been generated by cytoplasmic fusion between two normal inbred mouse strains, generating mice with variable degrees of heteroplasmy (Jenuth et al, 1996). These mice did not show pathology, but there was consistent directional selection for one mtDNA genotype in some tissues (Battersby et al, 2003). Tissue-specific single factors mediated this selection (Battersby et al, 2005); the identification of the responsible proteins will be important to understand the basis of tissue specificity and somatic segregation. A similar strategy was used to introduce a chloramphenicol-resistance mutation into mtDNA (Marchington et al, 1999; Sligh et al, 2000), and to generate mice that carried two heteroplasmic point mutations in the ND6 and COI genes of mtDNA, which led to mitochondrial myopathy and cardiomyopathy (Fan et al, 2008). These results hold promise for obtaining models of mtDNA point mutations that are relevant in human diseases such as those that involve tRNA mutations.
Engineering nuclear genes to target mtDNA
The manipulation of nuclear mtDNA maintenance genes has been revealed as a successful way in which to affect mtDNA stability indirectly (supplementary Table 1 online). The mtDNA replication machinery—especially the DNA polymerase POLG (Kaguni, 2004; Graziewicz et al, 2006) and the replicative helicase Twinkle (Spelbrink et al, 2001; Korhonen et al, 2004)—has been an effective target to induce mtDNA mutagenesis (Fig 2).
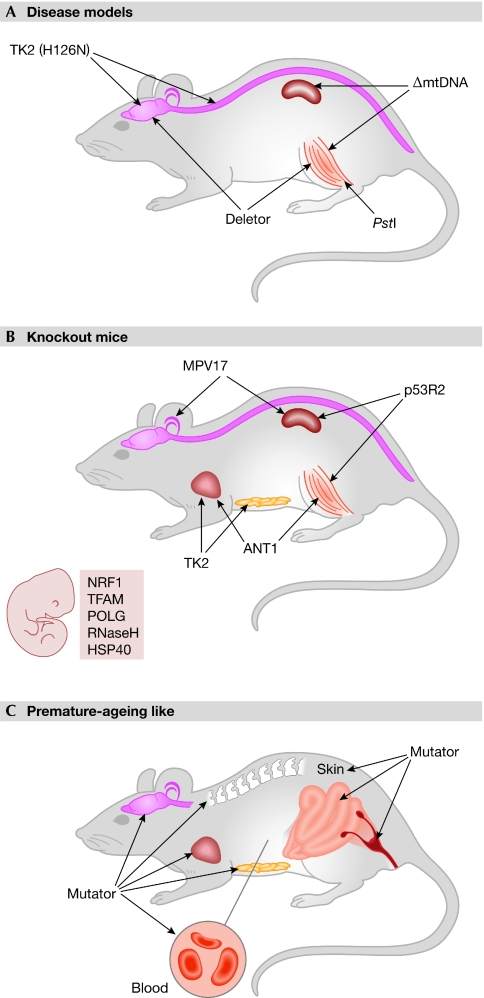
Figure 2.
Mouse models of mitochondrial DNA and its maintenance. (A) Main tissue manifestations of mtDNA disease models. (B) Crucial tissue manifestations of mice lacking mtDNA maintenance proteins. Mice in which mtDNA replication proteins are absent die during embryonic development, whereas those without proteins involved in maintaining the nucleotide pools present postnatal symptoms. (C) Various tissue manifestations of prematurely aged Mutator mice. ΔmtDNA, Mito-mice with single mtDNA deletion; ANT1, adenine nucleotide translocator; HSP40, heat-shock protein 40 mitochondrial chaperone; MPV17, mitochondrial inner membrane protein; mtDNA, mitochondrial DNA; NRF1, nuclear respiratory factor 1; p53R2, ribonucleotide reductase subunit; POLG, DNA polymerase γ; PstI, restriction endonuclease; RNaseH, ribonuclease H1; TFAM, mitochondrial transcription factor A; TK2, thymidine kinase 2.
Dominant mutations in Twinkle predispose individuals to human adult-onset progressive external ophthalmoplegia, which is associated with multiple mtDNA deletions (Suomalainen et al, 1997; Spelbrink et al, 2001). This disease is closely replicated in transgenic mice that ubiquitously express a mutant murine Twinkle cDNA—which carries a mutation that is homologous to a dominant patient mutation—in a 1:1 ratio with respect to endogenous Twinkle. This mouse suffers from late-onset (1 year of age) progressive respiratory-chain deficiency in the skeletal muscle, as well as in distinct neuronal populations such as cerebellar Purkinje cells and hippocampal neurons, with the accumulation of multiple mtDNA deletions (Tyynismaa et al, 2005), and was hence named ‘Deletor'. This phenotype could be replicated in several transgenic lines, whereas the overexpression of wild-type Twinkle caused no pathology (Tyynismaa et al, 2004). The lifespan and reproductive capabilities of the Deletor mouse were normal, and the distinct pathology makes it an attractive target for long-term therapeutic trials to treat adult-onset mitochondrial myopathy and neurodegeneration.
Human mutations of the mitochondrial replicative polymerase POLG have been shown to be a major cause of mitochondrial disease, and more than 2% of the population carries pathogenic POLG variants (Hakonen et al, 2005; Hudson & Chinnery, 2006). Knock-in inactivation of the POLG proof-reading exonuclease activity in mice results in efficient mtDNA mutagenesis (Trifunovic et al, 2004; Kujoth et al, 2005). These ‘Mutator' mice show signs of premature ageing, including weight loss, reduced subcutaneous fat content, hair loss, kyphosis, osteoporosis, anaemia, reduced fertility and an enlarged heart, and their median lifespan is shortened to 48 weeks (Fig 2C; Trifunovic et al, 2004; Kujoth et al, 2005). Mutator mice were the first experimental demonstration that extensive mtDNA mutagenesis can cause premature ageing-like symptoms, and will be important in revealing the contribution of mitochondria to various degenerative phenotypes. Studies on the Mutators have shown that coding-region mutations, which are potentially severely defective in function, are selected against during oocyte development, and an over-representation of tRNA mutations, which have a potentially milder functional effect, is maintained through the selection (Stewart et al, 2008). This intriguing finding could explain the abundance of tRNA mutations in human pathology. Furthermore, the Mutator might provide an excellent tool for creating mtDNA mutation-specific mouse lines by breeding the animals with wild-type strains, thereby maintaining and ‘diluting out' the original mtDNA mutations in the maternal line.
Overexpression of mutant POLG in mice caused cardiomyopathy when expressed in the heart and mood disorder-like behaviour with the neuronal promoter (Zhang et al, 2000; Kasahara et al, 2006; Lewis et al, 2007). However, these studies lacked the wild-type POLG overexpression control or reported only one transgenic line, leaving the contribution of insertional mutagenesis or protein overload to the phenotypes still to be excluded.
Eliminating mutant mtDNA
A potentially successful treatment strategy could shift the heteroplasmy to favour wild-type over mutant mtDNA. As a proof-of-principle for the enzymatic modification of mtDNA in vivo, PstI restriction endonuclease—which cuts mtDNA twice inducing double-stranded DNA breaks (DSBs)—was targeted to mouse skeletal muscle mitochondria (Srivastava & Moraes, 2005). Surprisingly, this led to mtDNA depletion and multiple large-scale mtDNA deletions, indicating that the 5′ ends of the DSBs had often been end-joined to the partly single-stranded D-loop, thereby creating large mtDNA deletions. The homologous D-loop region is also a common deletion breakpoint in humans and in the Deletor mouse (Zeviani et al, 1989; Tyynismaa et al, 2005). These similarities indicate that the formation of multiple mtDNA deletions might involve DSB repair. Between the ages of 6 and 7 months, the PstI mice developed a severe mitochondrial myopathy, but the PstI transgene was not transmitted to their offspring. Nonetheless, this experiment shows that mtDNA modifying enzymes can be introduced into animal tissues, indicating that point mutation-specific DNA-processing enzymes could be used for the sequence-specific degradation of mutant mtDNA.
No cell can live for long without mtDNA
MtDNA is completely dependent on nuclear genes: if an essential maintenance protein is defective, mtDNA is lost. The elucidation of the molecular background of mitochondrial DNA depletion syndrome (MDS)—a prevalent cause of respiratory-chain deficiency in childhood (Sarzi et al, 2007)—has uncovered several new proteins that are required for mtDNA maintenance.
The absence of proteins that directly regulate mtDNA copy number in mice leads to mtDNA loss and is not tolerated beyond mouse embryonic day 8.5 (Fig 2B), at which point mtDNA has probably been diluted too much from the ovum origin to be able to maintain life. At this developmental stage, cardiac function is required to enable oxygen supply because the embryo can no longer survive by oxygen diffusion. Essential mtDNA maintenance proteins—such as RNaseH1 and HSP40 (supplementary Table 2 online; Cerritelli et al, 2003; Hayashi et al, 2006)—have been found by studying knockout mice that die around embryonic day 8.5 and lack mtDNA.
TFAM is a histone-like packaging protein of mtDNA that is essential for mtDNA transcription and replication (Fisher & Clayton, 1988; Kanki et al, 2004; Kaufman et al, 2007). mtDNA levels closely follow TFAM levels, and TFAM knockout therefore leads to the progressive loss of mtDNA, severe OXPHOS defects and the death of the targeted cell type (supplementary Table 2 online; Larsson et al, 1998; Wang et al, 1999; Li et al, 2000; Silva et al, 2000; Sorensen et al, 2001; Wredenberg et al, 2002; Ekstrand et al, 2007). Conditional knockout studies have shown that different cell types vary considerably in their tolerance to reduced mtDNA levels. Tissue-specific TFAM knockouts are good models of respiratory-chain dysfunction at stages in which mtDNA is still present in reduced amounts.
Nucleosides and nucleotides for mtDNA maintenance
Molecular genetics studies of MDS have revealed an expanding group of proteins involved in mitochondrial nucleoside pool regulation (supplementary Table 2 online). These disorders are typically tissue specific, although this phenomenon is not currently understood. MtDNA maintenance requires both the mitochondrial dNTP salvage pathway and cytoplasmic de novo dNTP synthesis. TK2 is a mitochondrial deoxyribonucleoside kinase that is part of the salvage pathway. TK2 mutations manifest as early childhood-onset MDS with progressive myopathy and/or encephalopathy (Saada et al, 2001; Götz et al, 2008). Mice lacking functional TK2 showed rapid and progressive mtDNA depletion in all studied tissues, although only after postnatal day 12 (Akman et al, 2008; Zhou et al, 2008). The most severely affected tissue in the TK2 null mice was the brain rather than skeletal muscle, as is the case in human MDS. Defects in the de novo dNTP synthesis pathway also lead to a severe phenotype: mutations affecting the p53R2 subunit of the cytoplasmic ribonucleotide reductase, RRM2B, underlie a severe form of MDS that is associated with infantile myopathy, encephalopathy and nephropathy, which is fatal before 4 months of age (Bourdon et al, 2007). The RRM2B knockout mouse reproduces this phenotype relatively well, as it develops normally until weaning, but then experiences growth retardation and multiple organ failure leading to death 12–14 weeks after birth (Kimura et al, 2003). Lack of ANT1, which is the main regulator of adenine pools in post-mitotic tissues, does not compromise mouse lifespan or fertility, but leads to severe exercise intolerance, marked proliferation of mitochondria in the skeletal muscle and mtDNA rearrangements in the enlarged heart (Graham et al, 1997). The involvement of ANT1 in mtDNA maintenance has been verified in humans through the identification of dominant ANT1 mutations in familial progressive external ophthalmoplegia (Kaukonen et al, 2000), and recessive mutations in adult-onset myopathy and cardiomyopathy, both of which are associated with multiple mtDNA deletions (Palmieri et al, 2005).
On the basis of the available models, the proteins that are involved in mitochondrial dNTP pool regulation in various species seem to be conserved and, if inactivated, lead to mtDNA depletion. However, the tissue-specific need for these proteins seems to differ between species. The dNTP-knockout models vary from other mtDNA maintenance protein knockouts in that all of the former undergo normal development, despite the great need for DNA replication and dNTPs during embryonal life, and disease develops only after weaning, resembling the situation in humans. This points to an alternative, perhaps maternal, source of mitochondrial dNTPs during embryonal development, which is a possibility that deserves further study.
Is an increase in mtDNA copy number beneficial?
The copy number of mtDNA is tightly regulated; however, overexpression of human TFAM or murine Twinkle in mice has led to mtDNA increase (supplementary Table 3 online; Ekstrand et al, 2004; Tyynismaa et al, 2004; Ikeuchi et al, 2005). Twinkle helicase could be rate limiting for replication initiation and might affect mtDNA copy number through increased synthesis, whereas TFAM could stabilize mtDNA and increase its half life. Elevated levels of mtDNA seem not to be deleterious in either model; by contrast, mice overexpressing TFAM were found to have increased survival after myocardial infarction (Ikeuchi et al, 2005) as well as reversed age-dependent memory impairment (Hayashi et al, 2008), implying a beneficial effect of increased mtDNA levels. TK2 overexpression—that is, 300-fold increased TK2 activity—in the heart of transgenic mice led to a 40% increase in mtDNA copy number (Hosseini et al, 2007), indicating that heavy upregulation of the dNTP salvage pathway/dTTP pool can also affect the amount of mtDNA. Mice with increased mtDNA provide an intriguing model in which to study the cross-talk between energy metabolism and the environment, and to identify conditions in which having more mtDNA could be advantageous.
Need for extensive pathogenesis studies
The growing number of mtDNA mouse models has indicated an essential role of mtDNA maintenance proteins for viability. Knockouts of such proteins inevitably kill the targeted cell, and hence their physiological relevance for understanding mitochondrial dysfunction is low. Knockouts of dNTP pool regulators have provided important information about early-onset disorders, reflecting the marked consequences of loss-of-activity mutations for early postnatal life; perhaps surprisingly, none of those proteins seems to be essential for development. As mitochondrial proteins are usually well conserved among various species, mouse strains carrying homologous patient mutations can often be obtained. This strategy—either through knock-in techniques, transgenesis or the introduction of primary mtDNA mutations—has been relatively successful in replicating human diseases, as exemplified by the Mito-mice and the Deletor mice. However, even these true disease models have not been fully utilized to study pathogenesis or therapeutic-intervention strategies. Surprisingly, most models studied so far have not shown increased production of reactive oxygen species (Silva & Larsson, 2002; Kujoth et al, 2005; Trifunovic et al, 2005), which is generally thought to be a crucial mediator of mitochondrial pathology, thereby implying that an increase in reactive oxygen species is not a general consequence of respiratory-chain dysfunction. The exception is the POLGA Y955C heart-specific transgenic model (Lewis et al, 2007), which did show elevated 8-OHdG in mtDNA.
Conclusions
Much remains to be elucidated about mtDNA, its maintenance and its effect on disease; however, the existing mouse models will be useful for studies of pathogenesis and, in some cases, for testing potential treatments for mitochondrial disorders. However, more models carrying patient mutations would need to be generated in order to understand the phenotypic variation, tissue specificity and physiological consequences of mitochondrial diseases. The effect of ‘nuclear background' on phenotypic variability within one disorder could be studied by back-crossing disease models to various pure inbred backgrounds; if variation was found, the responsible genes could then be mapped. Intriguingly, mitochondrial dysfunction is increasingly associated with common disease entities such as neurodegeneration and metabolic syndrome, as well as ageing, which emphasizes the role of the mtDNA-mouse models in understanding the role of mitochondrial dysfunction in common pathologies.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary information

Henna Tyynismaa (left) & Anu Suomalainen
Acknowledgments
This work was supported by the European Molecular Biology Organization Young Investigator Programme, the Sigrid Juselius Foundation, the Academy of Finland, the University of Helsinki, the Helsinki University Central Hospital and the Helsinki Biomedical Graduate School.
References
- Akman HO, Dorado B, Lopez LC, Garcia-Cazorla A, Vila MR, Tanabe LM, Dauer WT, Bonilla E, Tanji K, Hirano M (2008) Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum Mol Genet 17: 2433–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Loredo-Osti JC, Shoubridge EA (2003) Nuclear genetic control of mitochondrial DNA segregation. Nat Genet 33: 183–186 [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Redpath ME, Shoubridge EA (2005) Mitochondrial DNA segregation in hematopoietic lineages does not depend on MHC presentation of mitochondrially encoded peptides. Hum Mol Genet 14: 2587–2594 [DOI] [PubMed] [Google Scholar]
- Bourdon A et al. (2007) Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet 39: 776–780 [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell 11: 807–815 [DOI] [PubMed] [Google Scholar]
- Chan DC (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH, Chinnery PF (2008) A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet 40: 249–254 [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348: 2656–2668 [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG (2004) Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet 13: 935–944 [DOI] [PubMed] [Google Scholar]
- Ekstrand MI et al. (2007) Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc Natl Acad Sci USA 104: 1325–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF (2008) Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet 83: 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC (2008) A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319: 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J (2007) Genetic, pathogenetic, and phenotypic implications of the mitochondrial A3243G tRNALeu(UUR) mutation. Acta Neurol Scand 116: 1–14 [DOI] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol 8: 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz A, Isohanni P, Pihko H, Paetau A, Herva R, Saarenpää-Heikkilä O, Valanne L, Marjavaara S, Suomalainen A (2008) Thymidine kinase 2 defects can cause multi-tissue mtDNA depletion syndrome. Brain 131: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC (1997) A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet 16: 226–234 [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC (2006) DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev 106: 383–405 [DOI] [PubMed] [Google Scholar]
- Hakonen AH et al. (2005) Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am J Hum Genet 77: 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hance N, Ekstrand MI, Trifunovic A (2005) Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet 14: 1775–1783 [DOI] [PubMed] [Google Scholar]
- Haraguchi M et al. (2002) Targeted deletion of both thymidine phosphorylase and uridine phosphorylase and consequent disorders in mice. Mol Cell Biol 22: 5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Imanaka-Yoshida K, Yoshida T, Wood M, Fearns C, Tatake RJ, Lee JD (2006) A crucial role of mitochondrial Hsp40 in preventing dilated cardiomyopathy. Nat Med 12: 128–132 [DOI] [PubMed] [Google Scholar]
- Hayashi Y et al. (2008) Reverse of age-dependent memory impairment and mitochondrial DNA damage in microglia by an overexpression of human mitochondrial transcription factor a in mice. J Neurosci 28: 8624–8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SH et al. (2007) Targeted transgenic overexpression of mitochondrial thymidine kinase (TK2) alters mitochondrial DNA (mtDNA) and mitochondrial polypeptide abundance: transgenic TK2, mtDNA, and antiretrovirals. Am J Pathol 170: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Chinnery PF (2006) Mitochondrial DNA polymerase-gamma and human disease. Hum Mol Genet 15: R244–R252 [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC (2001) Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol 21: 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M et al. (2005) Overexpression of mitochondrial transcription factor A ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation 112: 683–690 [DOI] [PubMed] [Google Scholar]
- Inoue K, Nakada K, Ogura A, Isobe K, Goto Y, Nonaka I, Hayashi JI (2000) Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet 26: 176–181 [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA (1996) Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet 14: 146–151 [DOI] [PubMed] [Google Scholar]
- Kaguni LS (2004) DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem 73: 293–320 [DOI] [PubMed] [Google Scholar]
- Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D (2004) Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol 24: 9823–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara T, Kubota M, Miyauchi T, Noda Y, Mouri A, Nabeshima T, Kato T (2006) Mice with neuron-specific accumulation of mitochondrial DNA mutations show mood disorder-like phenotypes. Mol Psychiatry 11: 577–593 [DOI] [PubMed] [Google Scholar]
- Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA (2007) The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell 18: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttälä A, Zeviani M, Comi GP, Keränen S, Peltonen L, Suomalainen A (2000) Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289: 782–785 [DOI] [PubMed] [Google Scholar]
- Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H (2003) Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet 34: 440–445 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Momoi MY, Tominaga K, Shimoizumi H, Nihei K, Yanagisawa M, Kagawa Y, Ohta S (1991) Respiration-deficient cells are caused by a single point mutation in the mitochondrial tRNA-Leu (UUR) gene in mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes (MELAS). Am J Hum Genet 49: 590–599 [PMC free article] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M (2004) Reconstitution of a minimal mtDNA replisome in vitro. EMBO J 23: 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC et al. (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309: 481–484 [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18: 231–236 [DOI] [PubMed] [Google Scholar]
- Lewis W et al. (2007) Decreased mtDNA, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest 87: 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, Larsson NG (2000) Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci USA 97: 3467–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchington DR, Barlow D, Poulton J (1999) Transmitochondrial mice carrying resistance to chloramphenicol on mitochondrial DNA: developing the first mouse model of mitochondrial DNA disease. Nat Med 5: 957–960 [DOI] [PubMed] [Google Scholar]
- McGregor A, Temperley R, Chrzanowska-Lightowlers ZM, Lightowlers RN (2001) Absence of expression from RNA internalised into electroporated mammalian mitochondria. Mol Genet Genomics 265: 721–729 [DOI] [PubMed] [Google Scholar]
- Nakada K, Sato A, Sone H, Kasahara A, Ikeda K, Kagawa Y, Yonekawa H, Hayashi J (2004) Accumulation of pathogenic ΔmtDNA induced deafness but not diabetic phenotypes in mito-mice. Biochem Biophys Res Commun 323: 175–184 [DOI] [PubMed] [Google Scholar]
- Palmieri L et al. (2005) Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet 14: 3079–3088 [DOI] [PubMed] [Google Scholar]
- Reid FM, Vernham GA, Jacobs HT (1994) A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum Mutat 3: 243–247 [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O (2001) Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet 29: 342–344 [DOI] [PubMed] [Google Scholar]
- Sarzi E, Bourdon A, Chretien D, Zarhrate M, Corcos J, Slama A, Cormier-Daire V, de Lonlay P, Munnich A, Rotig A (2007) Mitochondrial DNA depletion is a prevalent cause of multiple respiratory chain deficiency in childhood. J Pediatr 150: 531–534 [DOI] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61: 931–937 [DOI] [PubMed] [Google Scholar]
- Shoubridge EA (2001) Nuclear genetic defects of oxidative phosphorylation. Hum Mol Genet 10: 2277–2284 [DOI] [PubMed] [Google Scholar]
- Silva JP, Larsson NG (2002) Manipulation of mitochondrial DNA gene expression in the mouse. Biochim Biophys Acta 1555: 106–110 [DOI] [PubMed] [Google Scholar]
- Silva JP, Kohler M, Graff C, Oldfors A, Magnuson MA, Berggren PO, Larsson NG (2000) Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet 26: 336–340 [DOI] [PubMed] [Google Scholar]
- Sligh JE, Levy SE, Waymire KG, Allard P, Dillehay DL, Nusinowitz S, Heckenlively JR, MacGregor GR, Wallace DC (2000) Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc Natl Acad Sci USA 97: 14461–14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen L, Ekstrand M, Silva JP, Lindqvist E, Xu B, Rustin P, Olson L, Larsson NG (2001) Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci 21: 8082–8090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN et al. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet 28: 223–231 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Moraes CT (2005) Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet 14: 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG (2008) Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol 6: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A et al. (1997) Autosomal dominant progressive external ophthalmoplegia with multiple deletions of mtDNA: clinical, biochemical, and molecular genetic features of the 10q-linked disease. Neurology 48: 1244–1253 [DOI] [PubMed] [Google Scholar]
- Trifunovic A et al. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423 [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG (2005) Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci USA 102: 17993–17998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A (2004) Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet 13: 3219–3227 [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A (2005) Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci USA 102: 17687–17692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscomi C, Spinazzola A, Maggioni M, Fernandez-Vizarra E, Massa V, Pagano C, Vettor R, Mora M, Zeviani M (2008) Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum Mol Genet 18: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J et al. (1999) Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet 21: 133–137 [DOI] [PubMed] [Google Scholar]
- Weiher H, Noda T, Gray DA, Sharpe AH, Jaenisch R (1990) Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell 62: 425–434 [DOI] [PubMed] [Google Scholar]
- Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG (2002) Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci USA 99: 15066–15071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, DiDonato S (2004) Mitochondrial disorders. Brain 127: 2153–2172 [DOI] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, Dimauro S, DiDonato S (1989) An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature 339: 309–311 [DOI] [PubMed] [Google Scholar]
- Zhang D, Mott JL, Chang SW, Denniger G, Feng Z, Zassenhaus HP (2000) Construction of transgenic mice with tissue-specific acceleration of mitochondrial DNA mutagenesis. Genomics 69: 151–161 [DOI] [PubMed] [Google Scholar]
- Zhou X, Solaroli N, Bjerke M, Stewart JB, Rozell B, Johansson M, Karlsson A (2008) Progressive loss of mitochondrial DNA in thymidine kinase 2 deficient mice. Hum Mol Genet 17: 2329–2335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information