Abstract
The retroviral integrase superfamily (RISF) comprises numerous important nucleic acid-processing enzymes, including transposases, integrases and various nucleases. These enzymes are involved in a wide range of processes such as transposition, replication and repair of DNA, homologous recombination, and RNA-mediated gene silencing. Two out of the four enzymes that are encoded by the human immunodeficiency virus—RNase H1 and integrase—are members of this superfamily. RISF enzymes act on various substrates, and yet show remarkable mechanistic and structural similarities. All share a common fold of the catalytic core and the active site, which is composed primarily of carboxylate residues. Here, I present RISF proteins from a structural perspective, describing the individual members and the common and divergent elements of their structures, as well as the mechanistic insights gained from the structures of RNase H1 enzyme complexes with RNA/DNA hybrids.
Keywords: Argonaute, integrase, protein structure, RNase H, transposase
Glossary
DDE aspartate, aspartate, glutamate catalytic triad
HIV human immunodeficiency virus
MID middle domain (in Argonautes)
PAZ Piwi/Argonaute/Zwille (protein domain)
PIWI protein domain homologous to piwi proteins (encoded by the ‘P-element induced wimpy testis' class of genes in Drosophila)
Prp8 precursor RNA-processing protein 8
RAG1 recombination-activating gene 1
RISC RNA-induced silencing complex
SN2 bimolecular nucleophilic substitution
Tn3 transposon 3
Tn5 transposon 5
V(D)J variable (V), diversity (D) and joining (J) immunoglobulin and T-cell receptor gene segments
Introduction
RNase H1 was the first retroviral integrase superfamily (RISF) enzyme for which a three-dimensional structure was solved (Table 1; Katayanagi et al, 1990; Yang et al, 1990). Since then, the characteristic fold of the catalytic cores of RISF proteins has been known as the ‘RNase H fold'. RNases H bind to RNA/DNA hybrids in a sequence-nonspecific manner and degrade the RNA strand. Two groups of RNases H have now been identified: type 1 (RNase H1 or HI) and type 2 (RNase H2 or HII) enzymes. RNase H1 enzymes are present in all forms of life from bacteria to animals, as well as in retroviruses in which they constitute a domain of reverse transcriptases. RNase H1-null mice die during embryonic development because the enzyme is essential for mitochondrial DNA replication (Cerritelli et al, 2003). RNase H1 enzymes have also been implicated in the removal of the RNA primers that are used to start the synthesis of Okazaki fragments during DNA replication (Kogoma & Foster, 1998), although they are not essential for this process. The RNase H activity of retroviral reverse transcriptases—in particular that of HIV—is essential for viral replication, where it has a crucial role in the conversion of viral genomic RNA into double-stranded DNA, which is subsequently integrated into the host genome (Schultz & Champoux, 2008). The structures of viral, bacterial and human RNase H1 enzymes have now been solved, including those in complex with RNA/DNA hybrid substrates (Table 1; Nowotny et al, 2005; Nowotny et al, 2007).
Table 1.
Representative structures of members of the retroviral integrase superfamily
| Protein | Structure | References |
|---|---|---|
| RNase H1 | Escherichia coli RNase H1 | Katayanagi et al, 1990; Yang et al, 1990 |
| Bacillus halodurans RNase H1 catalytic domain in complex with RNA/DNA | Nowotny et al, 2005 | |
| Human RNase H1 catalytic domain in complex with RNA/DNA | Nowotny et al, 2007 | |
| RNase H2 | Archaeoglobus fulgidus RNase H2 | Chapados et al, 2001 |
| Methanococcus jannaschii RNase H2 | Lai et al, 2000 | |
| RuvC | E. coli RuvC | Ariyoshi et al, 1994 |
| Ydc2 | Ceschini et al, 2001 | |
| DDE transposase | Bacteriophage MuA | Rice & Mizuuchi, 1995 |
| E. coli Tn5 complex with DNA | Davies et al, 2000 | |
| Hermes transposase from Musca domestica | Hickman et al, 2005 | |
| Integrase | Human immunodeficiency virus integrase | Dyda et al, 1994 |
| Argonaute | Pyrococcus furiosus Argonaute | Song et al, 2004 |
| Thermus thermophilus Argonaute complex with single-stranded DNA | Wang et al, 2008 | |
| UvrC | UvrC, carboxy-terminal domain | Karakas et al, 2007 |
| Prp8 | Carboxy-terminal domain of human and yeast Prp8 | Pena et al, 2008; Yang et al, 2008 |
DDE, aspartate, aspartate, glutamate catalytic triad; MuA, Mu transposase; Prp8, precursor RNA-processing protein 8; Tn5, transposon 5; Ydc2, yeast mitochondrial RuvC homologue.
RNase H2 enzymes have different substrate specificity and biochemical properties (Ohtani et al, 1999). Most importantly, type 2 enzymes cleave preferentially at the 5′ end of RNA in chimeric DNA–RNA–DNA/DNA hybrids. They can hydrolyse this substrate even if it contains only a single ribonucleotide, whereas type 1 enzymes require at least four ribonucleotides (Ohtani et al, 1999). Therefore, RNase H2 enzymes are thought to be involved in the removal of single ribonucleotides that have been misincorporated into DNA (Rydberg & Game, 2002). Mutations of the human enzyme lead to Aicardi–Goutières syndrome, which is an autosomal recessive genetic disorder that severely affects the nervous system and has symptoms that are reminiscent of those caused by in utero viral infection (Crow et al, 2006). Eukaryotic RNase H2 enzymes contain three subunits ( Jeong et al, 2004), whereas bacterial and archaeal RNase H2 enzymes are monomeric (Chapados et al, 2001; Lai et al, 2000).
Bacterial RuvC resolvase is another member of the RISF. It cleaves the four-way DNA structures called Holliday junctions that are intermediates of the homologous-recombination process (Bennett et al, 1993). RuvC is a dimeric enzyme that cleaves the Holliday junction at two strands of the same polarity (Dunderdale et al, 1991). Substrate binding does not depend on the DNA sequence, but the cleavage is specific and occurs only at an (A/T)TT↓(G/C) cognate sequence (Shida et al, 1996).
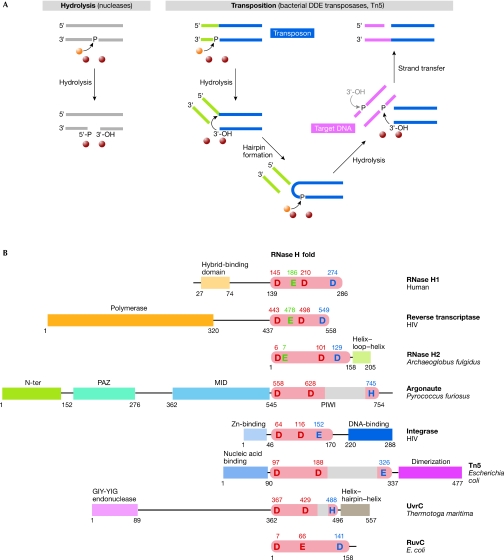
The transposases of the DDE family are also members of the RISF (Polard & Chandler, 1995), and catalyse the movement of DNA fragments called transposons from one location to another within or across genomes using a single active site. This multi-step reaction begins with the hydrolysis of both transposon ends, which generates free 3′-OH groups; the strategies that are used to produce this intermediate vary (Curcio & Derbyshire, 2003). Transposases such as MuA and Tn3 nick the transposon ends and join them with the target sequence forming a branched intermediate, which is later resolved by DNA replication. Another strategy that is often used by bacterial transposases such as Tn5 is the formation of hairpins on the ends of the transposon (Fig 1A). In this case, the transposon end is initially nicked in a hydrolysis reaction and the liberated 3′-OH groups perform a nucleophilic attack on the phosphate of the other strand, thereby forming a hairpin. The hairpin is resolved by a repetition of the first hydrolysis step and the product is a linear excised transposon with free 3′-OH groups, which is then joined with the target DNA by a nucleophilic attack of the 3′-OH groups on a phosphate group of the target DNA.
Figure 1.
Examples of reactions catalysed by members of the retroviral integrase superfamily and domain representation of selected members. (A) Schematic representation of two reactions catalysed by the RISF: nucleic-acid hydrolysis and a transposition reaction. The processing of only one end of the transposon is shown for simplicity. The attacking 3′-OH on the other end of the transposon is shown in grey. Metal ions and the water molecule are shown as purple and orange spheres, respectively. (B) RISF members. The insertions in the RNase H fold are shown as grey boxes. The functions or names of other domains are given and the positions of the active-site residues are indicated. The red residues are the two spatially conserved carboxylates, and the blue residue is the last and more variant residue. RNase H-specific glutamates are shown in green. DDE, aspartate, aspartate, glutamate catalytic triad; HIV, human immunodeficiency virus; RISC, RNA-induced silencing complex; RISF, retroviral integrase superfamily; Tn5, transposon 5; Zn, zinc.
The structure of the bacteriophage MuA transposase was the first to show that some transposases contain the RNase H fold (Rice & Mizuuchi, 1995); since then, several other structures of prokaryotic and eukaryotic transposases have been reported (Table 1). The related RAG1 protein—responsible for reshuffling the V(D)J segments during antigen–receptor gene assembly—is also thought to belong to the RISF on the basis of secondary-structure prediction and the identification of active-site residues (Kim et al, 1999; Landree et al, 1999).
Integrases catalyse the insertion of reverse-transcribed retroviral DNA into the host genome (Chiu & Davies, 2004). This reaction usually involves two steps: 3′-end processing and strand transfer. For example, in the case of HIV-1 integrase, the end processing consists of the removal of a terminal GT dinucleotide to produce a 5′ overhang and a 3′ end with free OH group. In the strand-transfer reaction, the target DNA—the genomic DNA of the host cell—is cleaved and the 3′ ends of the DNA that will become integrated are joined with the target. Integrases were found to be related to RNases H only when the structure of the HIV integrase was solved (Table 1; Dyda et al, 1994).
A recent addition to the RISF is Argonaute, the nuclease component of the RISC, which is a complex responsible for gene silencing by small-interfering RNAs (Mello & Conte, 2004). The RISC also contains a 20–24 nucleotide RNA that acts to select and capture a complementary messenger RNA target, which, if the complementarity is perfect, is then cleaved by Argonaute. The first crystal structure of Argonaute revealed that one of its domains—known as the PIWI domain—contains an RNase H-like segment (Table 1; Song et al, 2004). This structure, in combination with biochemical data, confirmed that Argonaute is the nuclease component of the RISC (Rivas et al, 2005).
UvrC is one of the crucial elements of the nucleotide-excision DNA repair pathway in bacteria. This pathway removes a wide range of DNA lesions (Truglio et al, 2006), and the role of UvrC is to cleave the DNA on both sides of the damage so that the fragment harbouring the lesion can be removed by a helicase. Each cleavage is carried out by one of two nuclease domains in UvrC, one of which is related to homing endonucleases and the other of which was recently shown by protein crystallography to adopt an RNase H fold (Table 1; Karakas et al, 2007).
Prp8 is the largest protein component of the spliceosome and is considered to be its main regulator. Recent structures of a domain from its carboxy-terminal region showed that it adopts the RNase H fold (Table 1; Pena et al, 2008; Yang et al, 2008). Prp8 is not a typical member of the RISF, however, as the carboxylate-rich active site is not conserved in its RNase H-like domain and there is no evidence of metal-ion binding to this domain. Whether the RNase H-like domain of Prp8 has catalytic activity remains to be seen.
The RNase H fold
In RISF members, the RNase H-like domain is usually linked to other functional domains that are responsible for nucleic-acid binding, protein–protein interactions or additional enzymatic activities (Fig 1B). Some RISF proteins function as dimers when required by their substrate. One such example is RuvC, which, as mentioned earlier, cleaves the two strands of the Holliday junctions. Also dimeric are the Tn5 transposase and integrases, which process the two ends of transposons and reverse-transcribed DNA, respectively. The modes of dimerization are different; for example, RuvC dimerizes through interactions that are mediated by two helices located on the side of its central β-sheet that is opposite to the active site (Ariyoshi et al, 1994). The Tn5 dimer contains two DNA molecules representing the transposon ends (Davies et al, 2000). Each DNA interacts with both protein molecules, and this bridging by nucleic acid is mainly responsible for dimer stabilization. Smaller contributions are made by the interactions between the C-terminal domains of Tn5. For the Hermes transposase, a hexameric active form has been proposed based on modelling and biochemical studies, and on electron-microscopy imaging (Hickman et al, 2005).
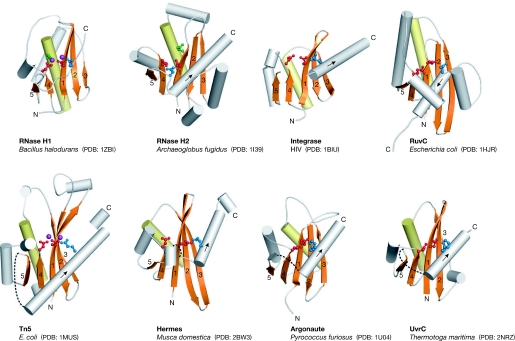
Although there is no detectable homology between the members of the RISF, the structure of their catalytic RNase H-like domains is remarkably conserved (Yang & Steitz, 1995). The central and most invariant element of this domain is a five stranded β-sheet (Fig 2). The first three strands are anti-parallel and usually run without any insertions; the two exceptions are RNase H2, which has two α-helices between strands 2 and 3, and Tn5, which has a short helix between strands 1 and 2. The shorter fourth and fifth strands run parallel to the first strand. In addition to the central β-sheet, the fold contains α-helices of variable position and arrangement. The most conserved α-helix is located after strand 3. It is adjacent to one face of the β-sheet and runs across it, probably stabilizing and reinforcing the central β-sheet (Fig 2). The RNase H fold can be disrupted by the insertions of various structures, which most often occur after strand 5 and before the last catalytic residue (Figs 1A,2). In UvrC, Tn5 transposase and Argonaute, this insertion predominantly consists of β-strands, and a large insertion in Hermes contains only α-helices.
Figure 2.
Catalytic core structures. The central β-sheet (with numbered strands) and the conserved α-helix are shown in orange and yellow, respectively. More divergent parts of the fold are shown in grey. The active-site residues are shown in ball-and-stick representation and are colour coded as described in Fig 1B. The two metal ions observed in the Tn5 and RNase H1 structures are shown in purple. The sites of insertions into the RNase H fold are shown as dashed lines. The direction of the last helix is indicated with an arrow. HIV, human immunodeficiency virus; PDB, Protein Data Bank code; Tn5, transposon 5.
Catalysis and the active sites
The enzymes of the RISF catalyse two general types of reaction: the hydrolysis of the phosphate group of a nucleic acid that leads to the formation of products containing 5′-phosphate and 3′-OH groups; and strand-transfer reactions in which a 3′-OH group of one DNA molecule attacks a phosphate group of another DNA molecule to join the two (Fig 1A). Stereochemical studies show that reactions catalysed by RISF members occur by a one-step SN2-like mechanism that includes the generation of a pentacovalent intermediate and the inversion of the phosphate stereo configuration (Kennedy et al, 2000; Krakowiak et al, 2002).
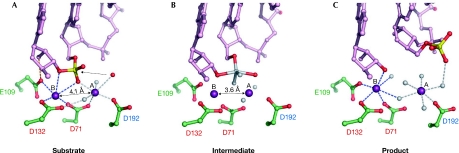
Divalent metal ions are essential for catalysis; the preferred ion of RISF enzymes is Mg2+, but Mn2+ also supports catalysis. Ca2+ inhibits hydrolysis, but can support strand transfer in transposition (Savilahti et al, 1995). The crystal structures solved in the presence of nucleic acid—Tn5 (Davies et al, 2000) and RNase H1 enzymes (Nowotny et al, 2005, 2007)—point to the involvement of two ions and a general two-metal ion mechanism (Steitz & Steitz, 1993; Yang et al, 2006). In this mechanism, metal ions are located on two sides of the scissile phosphate: the A-site Mg2+ coordinates, positions and activates the nucleophile; and the B-site Mg2+ stabilizes the transition state and the leaving group (Fig 3). The active sites of RISF members are composed predominantly of negatively charged carboxylate residues that coordinate the metal ions. Two residues—invariantly aspartates, except in the case of RuvC in which one is replaced by a glutamate—are particularly important in this process and are well conserved. The position of these two residues relative to the rest of the fold is also conserved: the first is in the middle of the first β-strand and the second is at the end of the fourth strand, adjacent to strand 1 (Figs 1B,2). The first carboxylate is the only one that coordinates directly both metal ions and is located at the heart of the active site. The second residue coordinates metal ion B, and in some structures metal ion A, through a water molecule (Fig 3; Nowotny et al, 2005).
Figure 3.
The reaction catalysed by RNase H1 enzymes. (A) Substrate or pre-reactive state. The RNA is shown in pink ball-and-stick representation with the scissile phosphate shown in red and yellow, the attacking nucleophile is shown as a red sphere and the two Mg2+ ions as purple spheres. The contact between Glu (E)109 and the 2′-OH is shown as an orange line. The numbers of the active-site carboxylates are given and are colour-coded as described in Figs 1B and 2. (B) The transition state model based on a structure with a non-phosphorylated nick at the active site. The observed oxygen atoms are shown in red and the modelled transition state atoms are shown in grey. The dashed line indicates the bond that will be broken in the reaction. Note that the distance between the metal ions is shorter than in (A). (C) Product complex. After the reaction is complete, and the 5′-phosphate and 3′-OH are generated, the phosphate group is displaced from the active site. The representations are based on the structures of Bacillus halodurans RNase H1 enzyme in complex with RNA/DNA hybrids as follows: (A) partly inactive D192N mutant, Protein Data Bank (PDB) code ; (B) D192N mutant, PDB ; and (C) E188A mutant, PDB .
For most RISF proteins, mutations in the active site do not inhibit substrate binding, and in some cases they can even enhance it (Nowotny et al, 2005). However, even conservative mutations of either of the two crucial carboxylates to amides render RISF enzymes inactive (Chapados et al, 2001; Ichiyanagi et al, 1998; Kanaya, 1998; Peterson & Reznikoff, 2003). For Argonaute, only aspartate to alanine mutations have been reported, and they completely abolished catalytic activity (Rivas et al, 2005). These mutational data further emphasize the essential role of the two carboxylates in catalysis.
The third residue of the active site—the most C-terminal of the three—coordinates metal ion A. This residue is comparatively variable and can be an aspartate (in RNases H and RuvC), a glutamate (in Tn5, Hermes and integrases) or a histidine (in UvrC). In many Argonautes this residue is a histidine, but there are also active Argonautes with a lysine or aspartate in this position ( Joshua-Tor, 2006; Wang et al, 2008). In the RISF, this residue is always located in a less conserved part of the core structure, after strand 5 of the central β-sheet. This region usually forms an α-helix that is adjacent to the β-sheet and runs roughly from strand 4 towards strand 3 (Fig 2). However, in RuvC, the last residue is located in a helix that runs in the opposite direction, and in RNase H2 enzymes it is located in a loop before the C-terminal α-helix. This last active-site residue is also relatively tolerant to mutation; for example, in RNase H1 enzymes it can be replaced by asparagine or histidine without significant loss of activity (Kanaya, 1998), and in RNase H2 an aspartic acid to asparagine substitution leads to only a partial loss of activity of the enzyme (Chapados et al, 2001).
In RNases H, there is an additional active-site residue, which in type 1 enzymes is a glutamate that coordinates metal ion B and forms contacts with the 2′-OH group of the ribonucleotide adjacent to the scissile phosphate (Fig 3A). It probably acts as an additional specificity check, relaying the information about the presence of the 2′-OH in the substrate to the active site (Nowotny et al, 2005). A similarly positioned glutamate is also present in RNase H2 enzymes; however, it comes from a different part of the fold and whether it has a similar function in the recognition of 2′-OH groups remains to be seen. Nevertheless, this glutamate is essential for the activity of both types of RNase H (Chapados et al, 2001; Kanaya, 1998).
Substrate binding and catalysis
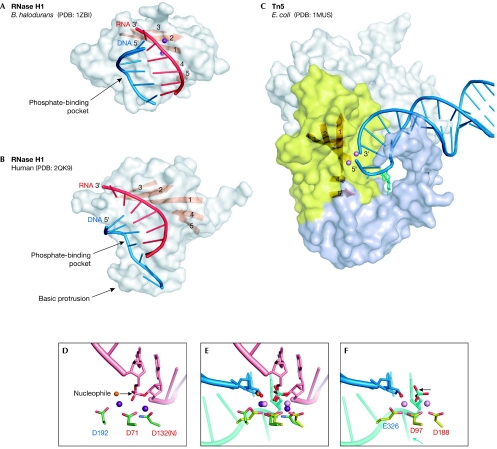
The structures of two RISF members in complex with nucleic acids have been described. The first structure to be solved was that of the Tn5 transposase with DNA forming a nicked hairpin, which represents the complex formed after the complete cleavage of the transposon from the flanking DNA (Davies et al, 2000). The other known structures are those of bacterial and human RNase H1 enzymes bound to RNA/DNA hybrids (Nowotny et al, 2005; Nowotny et al, 2007). In all cases, the nucleic acids were seen to interact with the active site; however, the positions of the nucleic acids relative to the RNase H fold are different (Fig 4). The Tn5 structures have been reviewed elsewhere (Steiniger-White et al, 2004); therefore, I focus here on the RNase H1 enzyme structures.
Figure 4.
Nucleic-acid complexes of Bacillus halodurans and human RNase H1 enzymes, and Escherichia coli Tn5 transposase. (A) B. halodurans RNase H1. The protein is shown in surface representation with strands of the central β-sheet shown as an orange ribbon. The portion of the nucleic acid that interacts with the protein is shown in cartoon representation with the RNA shown in red and the DNA in blue. The two metal ions at the active site are shown as purple spheres. (B) Human RNase H1. (C) Tn5 transposase in complex with a resolved DNA hairpin (blue) corresponding to the transposon end. Only one subunit of the dimer is shown. The insertion in the RNase H fold is shown in light blue. The nucleotide that is flipped out to form the DNA hairpin and is stabilized by the interactions with the insertion domain is shown in cyan stick representation. The amino-terminal domain that interacts with the nucleic acids and the carboxy-terminal dimerization domain are shown in grey. Two Mn2+ ions are shown as pink spheres. (D) Close-up view of the active site of B. halodurans RNase H1. Active-site carboxylates are shown in green and are labelled using the same colour coding as in Figs 1B and 2. The RNA strand is shown in pink with the two nucleotides joined by the scissile phosphate shown in stick representation. Two Mg2+ ions are shown as purple spheres. The attacking nucleophile is shown as an orange sphere and the direction of the attack is indicated with an arrow. (E) Superposition of the active sites of RNase H1 anf Tn5 transposase (coloured as in (D) and (F)) based on the positions of C-α atoms of the active-site carboxylates. Note that although they were not included in the superposition, the phosphate groups, metal ions and attacking nucleophiles (water in B. halodurans RNase H1 and 3′-OH in E.coli Tn5) occupy similar positions. After the non-transferred strand (cyan) is removed from the Tn5 active site, this superposition might represent the configuration of the transposon end (3′-OH group) attacking the target DNA mimicked by the RNA strand (pink) from the B. halodurans RNase H1 structure. (F) Active site of Tn5 transposase. The DNA is shown in blue (transferred strand) and cyan (non-transferred strand). The Mn2+ ions are shown as pink spheres. The 3′-OH group is indicated with a red sphere. The direction of the postulated last nucleophilic attack by a water molecule to generate this 3′-OH is indicated with an arrow. The flipped-out base is indicated with a cyan arrow. PDB, Protein Data Bank code; Tn5, transposon 5.
Both bacterial and human RNase H1 enzymes contain two grooves on their surface (Fig 4A,B), each of which accommodates one strand of the RNA/DNA hybrid. The RNA-strand groove contains the active site, and the RNA is specifically recognized by interactions with 2′-OH groups. The key to DNA recognition is the conformation of the nucleic acid: one of the phosphate groups of the non-cleaved strand binds to a tight phosphate-binding pocket on the surface of the protein, and this interaction requires the nucleic acid to adopt a B-form conformation. As only DNA can adopt such a conformation, the non-cleaved strand must be DNA. In human RNase H1 enzymes, there is an additional element, known as basic protrusion, which introduces further deformations to the DNA strand and can accommodate only 2′-deoxynucleotides, which leads to more stringent discrimination against RNA.
Two metal ions have been observed at the active sites of both bacterial and human RNase H1 enzymes (Fig 3A), which are coordinated not only by carboxylates from the active site but also by the backbone of the RNA. The presence of the correct nucleic acid is essential for the binding of both metal ions, which is probably the reason why often only one metal ion has been observed in the apo structures. This interdependence of metal ion and nucleic-acid binding ensures that the catalysis occurs only when the correct substrate is bound, thereby enhancing the specificity of the enzyme. In Bacillus halodurans RNase H1 structures—solved at high resolution—the metal ion A coordinates a water molecule that is clearly positioned as the nucleophile attacking the phosphorus of the scissile phosphate (Fig 3A). Such a configuration of the active site indicates that the catalysis occurs through a two-metal ion mechanism. Subsequent structures of B. halodurans RNase H1–RNA/DNA complexes allowed the reconstruction of the course of the catalytic reaction. The structure in complex with a non-phosphorylated nick at the active site was used to model the transition state, and the structure with a 5′-phosphorylated nick at the active site showed its configuration after the completion of the reaction (Fig 3B,C; Nowotny & Yang, 2006). On the basis of these structures, it has been proposed that the two metal ions move closer together to promote formation of the transition state. After the completion of the hydrolysis, the 5′-phosphate is displaced from the active site.
The conservation of the RNase H1 active site and its geometry makes it possible to formulate predictions for the catalytic mechanism of other members of the RISF. A comparison of RNase H1 and Tn5 complex structures reveals that the same fold and two-metal ion catalysis can be used to carry out a multi-step reaction of DNA transposition (Nowotny et al, 2005). When the two structures are superimposed, the metal ions occupy similar positions, and the nucleophilic water in the RNase H1 structures can almost be superimposed with the 3′-OH at the transposon end, which is the nucleophile for the strand-transfer reaction (Fig 4D–F). This 3′-OH group is generated by the attack of a water molecule on the phosphate from the opposite side of the 3′-OH. It has been proposed that the symmetrically coordinated metal ions in the Tn5 transposase can alternately activate a water molecule and the 3′-OH in successive chemical reactions, and that the 3′-OH at the transposon end remains coordinated to the same metal ion throughout the course of transposition (Fig 1A; Nowotny et al, 2005). In the last step of the reaction, the 3′-OH attacks the target DNA. These analyses show that the findings from one RISF enzyme can potentially be used to illuminate the details of the reaction catalysed by another enzyme.
The molecular details of substrate binding by RISF proteins are more difficult to predict because nucleic-acid binding often triggers conformational changes of the protein. For example, the domains of Argonautes—in particular the MID and PAZ domains—are relatively mobile (Wang et al, 2008). Superposition of RNase H1–RNA/DNA complexes with Argonaute proteins confirms that the nucleic-acid duplex should be placed in the central cavity of Argonaute to interact with the RNase H-like active site. However, in the model, the nucleic acid clashes with other domains, and in the actual complex these clashes are probably alleviated by the movements of individual domains of Argonaute.
Prospects
The RISF is fascinating and has diverse functions in nucleic-acid metabolism. The structural studies of these enzymes provide an unparalleled insight into the molecular details of their mechanism of action. One clear goal for the near future is to obtain additional crystal structures of RISF members in complex with the nucleic acids that they act upon (Sidebar A). Together with biochemical data, these structures will provide mechanistic insights into the important biological processes that depend on members of this family of enzymes.
Sidebar A | In need of answers.
Structures in complex with nucleic acid are not available for most enzymes of the retroviral integrase superfamily. These structures would help to answer the following questions: How is the target DNA captured by integrases and transposases? How does the strand transfer occur? Which protein conformational changes accompany this process? For the crossover junction endodeoxyribonuclease RuvC, the details of sequence-specific Holliday junction recognition need to be elucidated.
New members of the retroviral integrase superfamily need to be identified through sequence alignment and structural studies. In particular, the structures of the RAG1 and endonuclease V proteins would resolve whether they belong to the retroviral integrase superfamily.

Marcin Nowotny
Acknowledgments
I apologize to all those whose work has not been cited owing to the space constraints. I thank W. Yang and J. Bujnicki for critical reading of the manuscript. This work was supported by a European Molecular Biology Organization Installation Grant and a Wellcome Trust Senior Research Fellowship.
References
- Ariyoshi M, Vassylyev DG, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K (1994) Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell 78: 1063–1072 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Dunderdale HJ, West SC (1993) Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell 74: 1021–1031 [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell 11: 807–815 [DOI] [PubMed] [Google Scholar]
- Ceschini S, Keeley A, McAlister MS, Oram M, Phelan J, Pearl LH, Tsaneva IR, Barrett TE (2001) Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J 20: 6601–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapados BR, Chai Q, Hosfield DJ, Qiu J, Shen B, Tainer JA (2001) Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J Mol Biol 307: 541–556 [DOI] [PubMed] [Google Scholar]
- Chiu TK, Davies DR (2004) Structure and function of HIV-1 integrase. Curr Top Med Chem 4: 965–977 [DOI] [PubMed] [Google Scholar]
- Crow YJ et al. (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi–Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38: 910–916 [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM (2003) The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol 4: 865–877 [DOI] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85 [DOI] [PubMed] [Google Scholar]
- Dunderdale HJ, Benson FE, Parsons CA, Sharples GJ, Lloyd RG, West SC (1991) Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature 354: 506–510 [DOI] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F (2005) Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol 12: 715–721 [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K, Iwasaki H, Hishida T, Shinagawa H (1998) Mutational analysis on structure-function relationship of a holliday junction specific endonuclease RuvC. Genes Cells 3: 575–586 [DOI] [PubMed] [Google Scholar]
- Jeong HS, Backlund PS, Chen HC, Karavanov AA, Crouch RJ (2004) RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res 32: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua-Tor L (2006) The Argonautes. Cold Spring Harb Symp Quant Biol 71: 67–72 [DOI] [PubMed] [Google Scholar]
- Kanaya S (1998) Enzymatic activity and protein stability of E. coli ribonuclease HI. In Ribonucleases H, RJ Crouch, JJ Toulme (Eds), pp 1–38. Paris, France: INSERM
- Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C (2007) Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J 26: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T, Morikawa K (1990) Three-dimensional structure of ribonuclease H from E. coli. Nature 347: 306–309 [DOI] [PubMed] [Google Scholar]
- Kennedy AK, Haniford DB, Mizuuchi K (2000) Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. Cell 101: 295–305 [DOI] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA (1999) Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev 13: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Foster PL (1998) Physiological functions of E. coli RNase HI. In Ribonucleases H, RJ Crouch, JJ Toulme (Eds), pp 39–66. Paris, France: INSERM
- Krakowiak A, Owczarek A, Koziolkiewicz M, Stec WJ (2002) Stereochemical course of Escherichia coli RNase H. Chembiochem 3: 1242–1250 [DOI] [PubMed] [Google Scholar]
- Lai L, Yokota H, Hung LW, Kim R, Kim SH (2000) Crystal structure of archaeal RNase HII: a homologue of human major RNase H. Structure Fold Des 8: 897–904 [DOI] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB (1999) Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev 13: 3059–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Conte D (2004) Revealing the world of RNA interference. Nature 431: 338–342 [DOI] [PubMed] [Google Scholar]
- Nowotny M, Yang W (2006) Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J 25: 1924–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W (2007) Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell 28: 264–276 [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S (1999) Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry 38: 605–618 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Luhrmann R, Wahl MC (2008) Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson G, Reznikoff W (2003) Tn5 transposase active site mutations suggest position of donor backbone DNA in synaptic complex. J Biol Chem 278: 1904–1909 [DOI] [PubMed] [Google Scholar]
- Polard P, Chandler M (1995) Bacterial transposases and retroviral integrases. Mol Microbiol 15: 13–23 [DOI] [PubMed] [Google Scholar]
- Rice P, Mizuuchi K (1995) Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell 82: 209–220 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Rydberg B, Game J (2002) Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci USA 99: 16654–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savilahti H, Rice PA, Mizuuchi K (1995) The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J 14: 4893–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz SJ, Champoux JJ (2008) RNase H activity: structure, specificity, and function in reverse transcription. Virus Res 134: 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Iwasaki H, Saito A, Kyogoku Y, Shinagawa H (1996) Analysis of substrate specificity of the RuvC holliday junction resolvase with synthetic Holliday junctions. J Biol Chem 271: 26105–26109 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Steiniger-White M, Rayment I, Reznikoff WS (2004) Structure/function insights into Tn5 transposition. Curr Opin Struct Biol 14: 50–57 [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Croteau DL, Van Houten B, Kisker C (2006) Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev 106: 233–252 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ (2008) Structure of the guide-strand-containing Argonaute silencing complex. Nature 456: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R (2008) Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci USA 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hendrickson WA, Crouch RJ, Satow Y (1990) Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science 249: 1398–1405 [DOI] [PubMed] [Google Scholar]
- Yang W, Lee JY, Nowotny M (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell 22: 5–13 [DOI] [PubMed] [Google Scholar]
- Yang W, Steitz TA (1995) Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3: 131–134 [DOI] [PubMed] [Google Scholar]