Abstract
By screening a fetal brain two-hybrid library with the death domain of the p75 neurotrophin receptor (NTR), we identified the Sall2 transcription factor as a novel interacting protein. Sall2 is a unique member of the Sall gene family, which is believed to be a tumour suppressor. Here, we show that Sall2 contains a p75NTR interaction domain not found in other Sall proteins and that p75NTR/Sall2 complexes co-immunoprecipitate from brain lysates. NGF dissociates p75NTR/Sall2 complexes and activates TrkA, which has an obligate function in the nuclear translocation of Sall2. NGF also increases Sall2 expression and this is mediated by p75NTR, but may not require TrkA. Depletion of Sall2 from cells decreases the expression and activity of p21WAF1/CIP1, as well as the ability of NGF to induce growth arrest and the development of neurites. Overexpression of Sall2 activates p21WAF1/CIP1, induces growth arrest, and promotes neurite outgrowth independently of NGF. These data establish Sall2 as a link between NTRs and transcriptional events that regulate the growth and development of neuronal cells.
Keywords: neurite outgrowth, p21WAF1/CIP1, p75NTR, Sall2, TrkA
Introduction
Neurotrophins (NT) are a family of secreted polypeptides that regulate the development, viability, differentiation and function of cells in the nervous system (Chao, 2003). The family includes nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), NT-3 and NT-4/5. The pleiotropic actions of the NTs are mediated by binding to members of two structurally unrelated families of receptors, the tropomyosin receptor tyrosine kinases (TrkA, TrkB and TrkC) and the p75 NT receptor (p75NTR), a member of the tumour necrosis factor receptor (TNFR) superfamily (Teng and Hempstead, 2004; Reichardt, 2006). All NTs bind the p75NTR receptor with similar affinities, but selectively interact with the Trk family members.
Trk receptors are single transmembrane receptor tyrosine kinases. NGF binding to TrkA induces receptor dimerization and the phosphorylation of tyrosine residues that function as docking sites for adaptor proteins. Once associated with adaptor proteins, TrkA can activate signalling pathways that are important to its cellular activities (Teng and Hempstead, 2004; Reichardt, 2006). Genetic and biochemical evidence supports a role for Grb2, Shc, FRS-2/SNT and Gab-1 as adaptor proteins used by TrkA to engage the Ras pathway that activates mitogen-activated protein kinases, and the PI 3-kinase pathway that activates Akt (Teng and Hempstead, 2004; Reichardt, 2006). NT signalling through Trk receptors regulates cell survival, proliferation, the fate of neural precursors, and axon and dendrite growth (Teng and Hempstead, 2004; Reichardt, 2006).
The p75NTR receptor is a type I membrane protein composed of an extracellular domain that contains four cysteine-rich regions, and an intracellular domain related to FAS and the type I TNFR (TNFR1), as it contains a death domain (Teng and Hempstead, 2004; Reichardt, 2006). p75NTR is a pan-NTR that binds BDNF, NT-3 and NT-4, as well as NGF. The activities of p75NTR are mediated by its interaction with an array of intracellular proteins. Among these, NRIF (Casademunt et al, 1999), NRAGE (Salehi et al, 2000) and NADE (Mukai et al, 2000) contribute to apoptosis, RhoA and SC-1 affect neurite elongation and induce growth arrest (Chittka and Chao, 1999; Chittka et al, 2004), and TRAF6 (Khursigara et al, 1999; Ye et al, 1999) and RIP (Khursigara et al, 2001) activate NF-κB. JNK kinase, NF-κB and ceramide are downstream components of the transduction mechanism used by p75NTR to elicit cellular responses. Studies with isolated cells, and the phenotype of mice deficient in p75NTR (Lee et al, 1992; von Schack et al, 2001; Naumann et al, 2002), show that the receptor promotes context-dependent apoptosis, cell survival, neurite outgrowth or myelination (Dechant and Barde, 2002; Teng and Hempstead, 2004; Reichardt, 2006).
The death domain of p75NTR is distinct from those in TNFR1 and FAS, especially in the placement of the first of six alpha helices (Teng and Hempstead, 2004). The death domain of p75NTR associates with a unique array of proteins, including RhoA, RIP-2, and Bex1, and through these interactions permits the receptor to affect neurite outgrowth, survival, and cell cycle arrest, respectively (Yamashita et al, 1999; Khursigara et al, 2001; Gehler et al, 2004; Vilar et al, 2006). Using yeast two-hybrid assays, we showed earlier that the death domain in the type 1 TNF receptor is a motif that fosters protein–protein interactions (Song et al, 1994), leading us to hypothesize that the death domain in p75NTR might interact with yet-to-be identified proteins. Here, we used the death domain of p75NTR as a bait in a two-hybrid screen. This resulted in the identification of the Sall2 transcription factor as a novel p75NTR-interacting protein.
Results
Sall2 binds the death domain of p75NTR
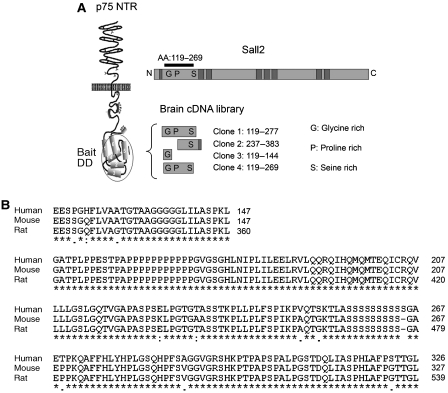
Only a few p75NTR-binding proteins directly associate with its death domain, prompting us to search for proteins that interact with this region of the receptor. By screening a human fetal brain two-hybrid library, we identified four human clones that bound p75NTR, each of which encoded amino-acid sequences of the Sall2 transcription factor (Figure 1A). This discovery was particularly interesting, as little is known of the functions of Sall2. Analysis of the Sall2 clones showed that each encodes a portion of the N-terminal domain of Sall2. Taken together, the clones encompass amino acids 119–269 of Sall2, which contains glycine-, proline-, and serine-rich sequences characteristic of transcription factors (Figure 1A). Human Sall2 is homologous to mouse and rat Sall2 (90% identity). Sequence alignment shows that the domain that interacts with p75NTR is highly conserved among human, mouse and rat Sall2 (96%; Figure 1B). Sall2 is not highly homologous with human Sall1, Sall3 and Sall4 (∼30%), and the domain in Sall2 that interacts with the p75NTR receptor is not in human Sall1, Sall3 or Sall4 (∼11% identity; Supplementary Figure 1A). The minimal sequences in the Sall2 clones that interact with p75NTR (clone 2, amino acids 237–269; clone 3, amino acids 119–144) are also not in Sall1, Sall3 or Sall4 (Supplementary Figure 1B).
Figure 1.
Sall2 binds the death domain of p75NTR. (A) The death domain (amino acids 324–399) of p75NTR was used to screen a human fetal brain library. Four interacting clones encompass sequences in a 150-amino-acid region in the N-terminal domain of Sall2. The relationship of the clones to full-length Sall2 (shown as a bar) and the death domain (DD) is illustrated. (B) Alignment of primary amino-acid sequences of mouse, rat and human Sall2 using ClustalW. The region of human Sall2 identified by the two-hybrid screen is compared with the N-terminal regions of mouse and rat Sall2.
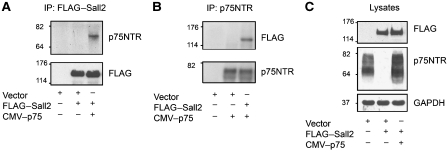
We used immunoprecipitation to test whether p75NTR and Sall2 interact in a mammalian system. In agreement with the two-hybrid results, p75NTR specifically co-immunoprecipitated with FLAG-tagged Sall2 from lysates of transfected 293 cells (Figure 2A–C). The interaction of the proteins was demonstrated by immunoprecipitation of FLAG-tagged Sall2 followed by western blotting for p75NTR (Figure 2A), or by immunoprecipitation of p75NTR followed by western blotting for FLAG-tagged Sall2 (Figure 2B).
Figure 2.
Sall2 interacts with p75NTR in cells. HEK 293 cells were transfected with p75NTR and FLAG–Sall2. (A) FLAG–Sall2 was immunoprecipitated from a cell lysate and a western blot was probed for p75NTR or FLAG (Sall2). (B) p75NTR was immunoprecipitated from a cell lysate and a western blot was probed for FLAG (Sall2) or p75NTR. (C) Overexpression of p75NTR and Sall2 in the lysates was confirmed by western blotting.
Sall2 in the brain
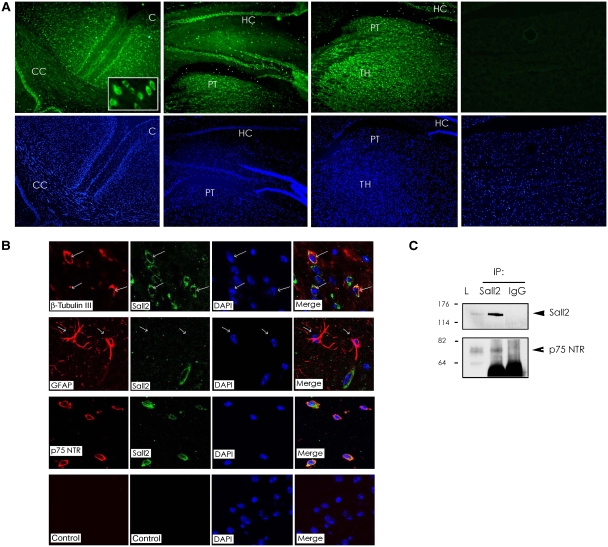
Previous work detected mRNA for Sall2 in mouse brain (Kohlhase et al, 2000), but the expression and distribution of the Sall2 protein in brain have not been characterized. This led us to test the distribution of Sall2 at the cellular level in mouse brain. We probed sections of mouse brain with an antibody that recognizes Sall2 by immunohistochemistry (Nielsen et al, 2003), and then used immunofluorescence microscopy to visualize Sall2 expression. At low (10 ×) magnification, Sall2 is present in most areas of the brain. As an example, we show Sall2 staining in the cortex, the hippocampus and the thalamus (Figure 3A), brain regions known to express p75NTR. Higher (40 ×) magnification showed that Sall2 resides mostly in the cytoplasm of brain cells (Figure 3A, insert), although nuclear staining was also observed. We next identified the cell type in mouse brain that expresses Sall2. Confocal microscopy indicated that neurons specifically contain Sall2. This conclusion was based on co-expression of β-tubulin III with Sall2. Astroglial cells, identified by glial fibrillary acidic protein (GFAP) immunoreactivity, did not contain Sall2 (Figure 3B).
Figure 3.
Relationship of Sall2 and p75NTR in the brain. (A) Immunofluorescence of coronal mouse brain sections probed with antibody to Sall2 at 10 × and 40 × (inset) magnification. C, cortex; CC, corpus callosum; HC, hippocampus; PT, protectum; TH, thalamus. DAPI-stained nuclei appear blue. (B) Confocal microscopy of coronal mouse brain sections probed with antibodies to: β-tubulin III (red) and Sall2 (green); GFAP (red) and Sall2 (green); p75NTR (red) and Sall2 (green); the negative control shows a confocal image without primary antibodies. (C) Endogenous Sall2 was immunoprecipitated from a mouse brain lysate (L) using a Sall2 polyclonal antibody. Rabbit IgG was a control for the immunoprecipitation. A western blot was probed for Sall2 or endogenous p75NTR.
Consistent with the interaction of Sall2 with p75NTR shown in Figures 1 and 2, Sall2 colocalized with p75NTR at the cell surface in mouse brain (Figure 3B). The demonstration that Sall2 is expressed by neurons, and partly resides in the same cellular compartment as p75NTR, prompted us to test for association of the endogenous proteins. p75NTR and Sall2 co-immunoprecipitated and therefore interact under physiological conditions in the brain (Figure 3C).
Control experiments confirmed the identity of the protein recognized by the antibody to Sall2. Western blotting showed that the Sall2 antibody recognizes a similar protein in lysates of mouse brain, human HCT116 colon cancer cells and rat PC12 cells (Supplementary Figure 2A, lanes 1–4). Pretreatment of anti-Sall2 with the peptide used to generate the antibody (Supplementary Figure 2A, lanes 5–8) or treatment of cells with Sall2-specific siRNA (Supplementary Figure 3A and B; Materials and methods) resulted in loss of the protein identified as Sall2 on western blots and by immunofluorescence, confirming its identify.
NGF induces re-localization of Sall2
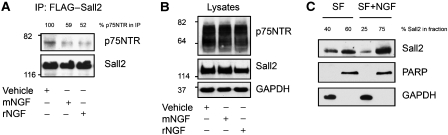
The fact that Sall2 is a transcription factor begs the question of how it could bind to a cell surface receptor and then function. To study this, we first investigated how NGF affects interaction of p75NTR with Sall2 by immunoprecipitation of p75NTR/Sall2 complexes from cells incubated in the absence or presence of NGF. We found that Sall2 and p75NTR associate constitutively, and 2.5S NGF or recombinant human NGF diminishes the amount of p75NTR/Sall2 immunoprecipitated from cells by more than 40% (Figure 4A), suggesting that NGF dissociates the complex.
Figure 4.
NGF induces translocation of Sall2 into the nucleus. (A) NGF diminishes interaction of Sall2 with p75NTR. HEK 293 cells were transfected with p75NTR and FLAG–Sall2. After 24 h, cells were cultured in serum-free medium for 24 h and then NGF was added. Cells were incubated in the absence or presence of NGF (100 ng/ml) for 15 min. FLAG–Sall2 was immunoprecipitated from cell lysates and western blots were probed with antibodies to p75NTR or FLAG. The percentage of p75NTR in the Sall2 complex (immunoprecipitate) is given above the blot and is the average of results from three independent experiments. (B) Overexpression of p75NTR and Sall2 in the lysates was confirmed by immunoblotting (right). (C) Localization of Sall2 in PC12 cells. Cells were incubated in the absence or presence of NGF for 1 h, after which cell fractions were isolated using NE-PER nuclear and cytoplasmic extraction reagents from Pierce Inc. Fractions were characterized by immunoblotting for Sall2, GAPDH and PARP, and a representative figure is shown. The percentage of Sall2 in fractions is the average of results from three independent experiments.
Next, we isolated nuclear and cytoplasmic fractions from control and NGF-treated PC12 cells to evaluate the effect of NGF on endogenous Sall2 localization. PC12 cells were used here because they exhibit neuronal properties similar to those of sympathetic neurons (Greene and Tischler, 1976), and are a useful model for studying the functions of NTRs. Equal amounts of each fraction were assayed for the expression level of Sall2 by western blotting. We found that under serum-free conditions, Sall2 resides in the cytoplasm and nucleus (Figure 4C). NGF decreased the fraction of Sall2 in the cytoplasm (from 40 to 25%), and increased the fraction of Sall2 in the nucleus (from 60 to 75%). To confirm the purity of the cell fractions, GAPDH served as a cytoplasmic marker, and PARP served as a nuclear marker. Our experiments show that NGF dissociates the p75NTR/Sall2 complex and induces Sall2 to move into the nucleus.
Endogenous Sall2 localization is affected by NGF but not by other NTs
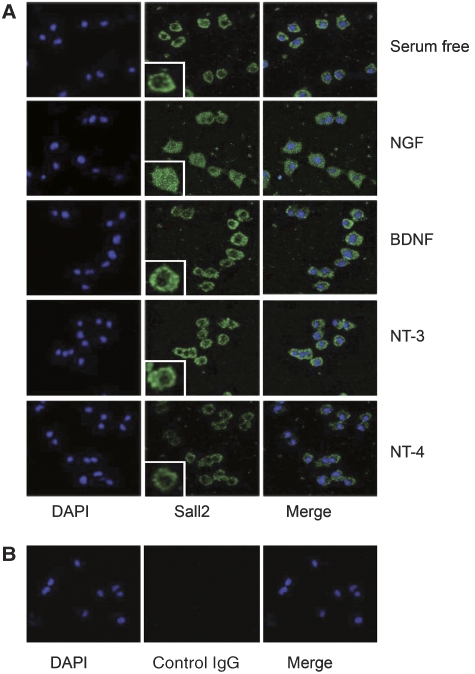
Confocal microscopy of Sall2 in PC12 cells yielded results consistent with observations made with fractionated PC12 cells (Figure 4C), showing that NGF induces redistribution of Sall2 into the nucleus (Figure 5). Quantification of the intensity of nuclear Sall2 staining showed a significant (33%) increase of nuclear Sall2 after NGF treatment (Figure 6B). As all NTs bind and activate p75NTR, we also examined the effect of BDNF, NT-3 and NT-4 on Sall2 localization. Movement of Sall2 into the nucleus was ligand specific because Sall2 localization was not affected by these NTs, but was affected by NGF (Figure 5).
Figure 5.
Localization of Sall2 is regulated by NGF, but not by other neurotrophins. PC12 cells were serum-starved and then incubated in the absence or presence of neurotrophins (100 ng/ml, 1 h). (A) Confocal microscopy characterized Sall2 localization after cells were fixed and stained with anti-Sall2 followed by an Alexa Fluor-conjugated secondary antibody. DAPI stained the nuclei, which appear blue. Inserts show magnified cells for each condition. (B) Confocal microscopy of a negative control in which rabbit IgG instead of Sall2 antibody was used for staining.
Figure 6.
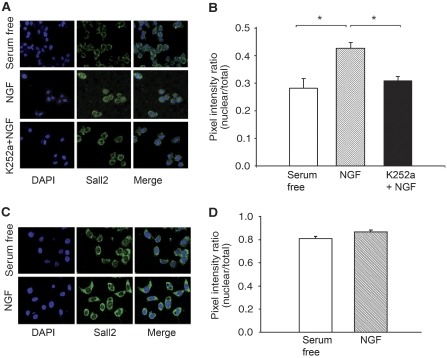
TrkA is necessary for entry of Sall2 into the nucleus. (A) PC12 cells were serum-starved for 24 h, incubated in the absence or presence of K252a for 30 min (10 nM) and then treated with NGF for 1 h. The cells were fixed, stained with anti-Sall2 and then with a secondary antibody coupled to Alexa Fluor 488. DAPI was used to visualize the cell nucleus. (B) Nuclear Sall2 increases after NGF treatment. The fluorescence intensity of Sall2 staining was quantified by calculating the ratio of nuclear pixel intensity × nuclear area/total pixel intensity × total cell area. Data are the mean±s.e.m. (n=35–40) from three independent experiments; *Student's t-test (P<0.005) for NGF versus control and for NGF versus K252a+NGF treatments. (C) PC12nnr5 cells were serum-starved for 24 h, incubated in the absence or presence of NGF (100 ng/ml) for 1 h, fixed, stained with anti-Sall2 and then with a secondary antibody coupled to Alexa Fluor 488. DAPI was used to visualize the cell nucleus. (D) Sall2 nuclear localization was quantified as in (B). Data are the mean±s.e.m. (n=35) from three independent experiments.
TrkA has a function in Sall2 localization
NGF activates TrkA and p75NTR in PC12 cells, whereas BDNF, NT-3 and NT-4 activate p75NTR, but not TrkA. This selective receptor activation, and the inability of ligands that do not activate TrkA to induce nuclear translocation of Sall2, led us to evaluate whether TrkA has a role in the re-localization of Sall2. First, we evaluated whether K252a, an inhibitor of TrkA (Berg et al, 1992), but not p75NTR (von Bartheld et al, 1996), would impair NGF-induced movement of Sall2 into the nucleus. PC12 cells were incubated in the absence or presence of 10 nM K252a. This concentration of K252a is the lowest that effects near-complete inhibition of the capacity of NGF to induce the tyrosine phosphorylation of TrkA in PC12 cells (Berg et al, 1992). Confocal microscopy (Figure 6A), and quantification of nuclear staining (Figure 6B), shows that NGF fails to alter Sall2 localization in cells treated with K252a.
To still more definitively establish the importance of TrkA to Sall2 localization, we used PC12nnr5 cells, which express p75NTR, but not TrkA (Loeb et al, 1991). Control experiments showed that both PC12 and PC12nnr5 cells express Sall2 (Supplementary Figure 2A), but confirmed that only PC12 cells express TrkA (Supplementary Figure 2B). We found that NGF did not induce Sall2 nuclear localization in PC12nnr5 cells (Figure 6C and D), as it had in PC12 cells (Figure 5). Because re-expression of TrkA in PC12nnr cells rescues the NGF responsiveness of the mutant cell line (Loeb et al, 1991), our experiment establishes the causal necessity of TrkA to the nuclear movement of Sall2.
We examined the amino-acid sequence of Sall2 to gain insight into how TrkA might exert an effect on this transcription factor. This led us to identify a conserved MAPK phosphorylation motif in the N-terminal region of Sall2, which is in the p75NTR interaction domain (Supplementary Figure 4A). On the basis of this discovery, we assayed NGF-induced activation of members of the MAPK family in PC12 cells and PC12nnr5 cells. NGF activated ERK1/2 and p38 MAPK in PC12 cells, but not PC12nnr5 cells (Supplementary Figure 4B). Activated ERK1/2, but not p38MAPK, was implicated in the nuclear translocation of Sall2, as confocal microscopy showed that U0126, an inhibitor of ERK1/2, but not SB203580, an inhibitor of p38MAPK, impaired NGF-induced nuclear localization of Sall2 (Supplementary Figure 4C and D).
The observations described above show that TrkA has an obligate function in the nuclear movement of Sall2 and indicate that ERK activation is necessary for Sall2 to mobilize to the nucleus.
Sall2 is induced during neuronal differentiation
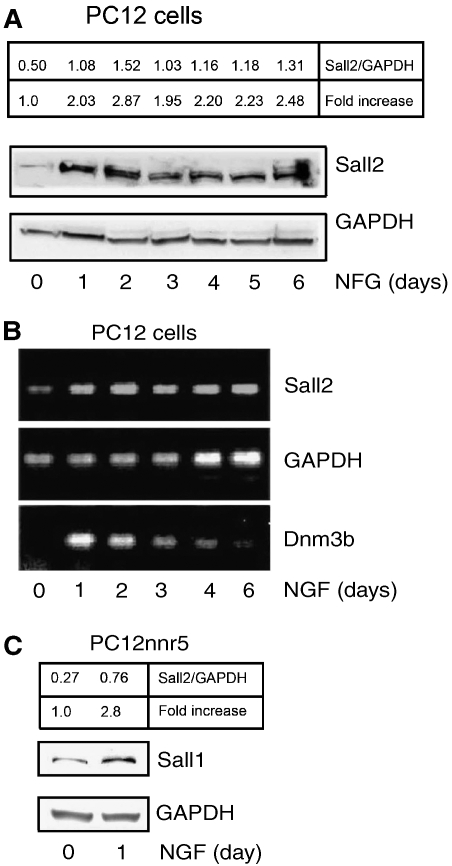
We next investigated whether Sall2 expression is regulated during PC12 cell differentiation, because if true, this would support the conclusion that Sall2 has a function in differentiation. Differentiation medium, which contains NGF, increased expression of the Sall2 protein within a day; the elevated expression of Sall2 then remained high throughout the differentiation process (Figure 7A). Analysis of Sall2 mRNA by RT–PCR showed that Sall2 is transcriptionally upregulated within 24 h (Figure 7B). In this experiment, DNA methyl transferase 3b (Dnmt3b), a gene induced during NGF-dependent differentiation (Bai et al, 2005), served as a positive control. The increase of Sall2 mRNA detected by RT–PCR (Figure 7B) correlated with the elevation of the Sall2 protein detected by western blotting (Figure 7A) during the differentiation of PC12 cells. Finally, we tested whether TrkA participates in the induction of Sall2 mRNA by using TrkA-deficient PC12nnr5 cells. We observed that NGF induced Sall2 in PC12nnr5 cells (Figure 7C), an observation consistent with augmented Sall2 staining in these cells after NGF treatment (Figure 6C). These results suggest that p75NTR, but not TrkA, induces Sall2 expression.
Figure 7.
Regulation of Sall2 expression during neuronal differentiation. (A) PC12 cells were serum-starved for 24 h and then incubated with NGF in 1.5% serum for up to 6 days. Expression of Sall2 was evaluated by immunoblotting. GAPDH expression served as a loading control. The ratio of Sall2 to GAPDH and the fold increase of Sall2 over that at day 0 were calculated after densitometry and are in the upper box. (B) RT–PCR on total RNA from cells treated as described in (A) was assayed for the expression of Sall2, GAPDH and Dnm3b RNA. (C) PC12nnr5 cells were serum-starved for 24 h and then treated with NGF for 1 day. Expression of Sall2 and GAPDH was assayed by immunoblotting. The ratio of Sall2 to GAPDH and the fold increase of Sall2 expression were calculated as in (A) and is in the upper box.
Sall2 promotes p21WAF1/CIP1 expression and activity to inhibit cell proliferation
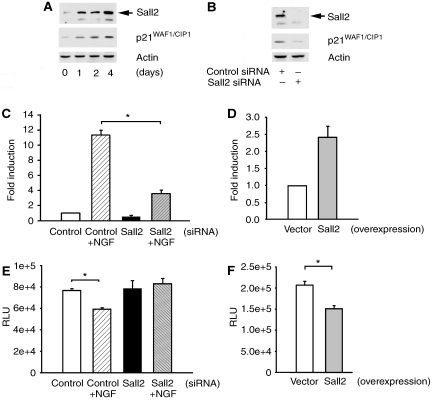
Sall2 mediates the cessation of DNA synthesis in serum-deprived cells (Liu et al, 2007) and inhibits the proliferation of ovarian cancer cells by inducing p21WAF1/CIP1 (Li et al, 2004). We observed that Sall2 and p21WAF1/CIP1 expression increased in parallel in PC12 cells treated with differentiation medium, which contains NGF (Figure 8A). This led us to test whether Sall2 affects NGF-dependent p21WAF1/CIP1 expression in PC12 cells. Sall2 siRNA induced a parallel decrease of Sall2 and p21WAF1/CIP1 protein expression in lysates of cells treated with differentiation medium for 4 days (Figure 8B), suggesting that Sall2 is necessary for p21WAF1/CIP1 expression. To test whether Sall2 regulates p21WAF1/CIP1 transcription, PC12 cells were co-transfected with a p21WAF1/CIP1 reporter that contains the p21WAF1/CIP1 promoter region, and control or Sall2 siRNA. Cells were grown in differentiation medium for 2 days and then reporter activity was evaluated. Differentiation medium increased p21WAF1/CIP1 reporter activity 12-fold in cells treated with control siRNA, but only increased activity 4-fold in cells treated with Sall2 siRNA (Figure 8C). We next determined whether elevated expression of Sall2 would bypass the necessity of stimulating cells with NGF to activate the p21WAF1/CIP1 reporter. To accomplish this, we overexpressed HA–Sall2 in PC12 cells, as shown in Supplementary Figure 5A and B, and found that the activity of the gene reporter was 2.5-fold greater in cells in which Sall2 was overexpressed than in control cells (Figure 8D). Our observations showing that Sall2 increases the expression and activity of p21WAF1/CIP1 led us to surmise that Sall2 might inhibit cell growth. To test this hypothesis, we assayed BrdU incorporation into DNA in PC12 cells depleted of Sall2, or in which Sall2 was overexpressed. Depletion of Sall2 impaired the capacity of NGF to inhibit BrdU incorporation (Figure 8E), whereas overexpression of Sall2 was sufficient to inhibit BrdU incorporation (Figure 8F). Our results show that Sall2 mediates the NGF-induced increase of p21WAF1/CIP1 expression and activity in PC12 cells and promotes the cessation of cell proliferation. Our results also show that Sall2 by itself has the capacity to activate p21WAF1/CIP1 and to induce cell growth arrest.
Figure 8.
Sall2 is required for NGF-dependent p21WAF1/CIP1 expression and cell cycle arrest. (A) PC12 cells were cultured in differentiation medium (DM) containing low serum for several days. Immunoblotting assayed expression of Sall2, p21WAF1/CIP1 and actin. (B) PC12 cells were transfected with control or Sall2-specific siRNA and then incubated in differentiation medium for 4 days. Immunoblotting assayed expression of Sall2, p21WAF1/CIP1 and actin. (C) Sall2 regulates p21WAF1/CIP1 promoter activity. The effect of Sall2 on the p21 promoter reporter was assayed using a firefly luciferase assay in which PC12 cells were transfected with control or Sall2-specific siRNAs (50 nM). After 48 h in the absence or presence of differentiation medium, luciferase activities were determined and normalized to β-gal. Results are the average fold induction±s.e.m. for three independent experiments performed in triplicate, *Student's t-test (P<0.005). (D) Cells were co-transfected with empty vector or HA–Sall2 together with a p21 gene reporter and β-gal and then cultured in same medium as in (C), but without NGF. After 2 days, p21 reporter activity was assayed and is presented as a bar graph as described under (C). Results are the average of fold induction calculated from three independent experiments, each performed in triplicate. (E) PC12 cells were transfected with control or Sall2-specific siRNA and then grown in the absence or presence of differentiation medium for 48 h. BrdU incorporation (in RLUs, relative luciferase units) into DNA was then assayed using a kit from Roche Inc. Results are the average±s.e.m. from three independent experiments performed in sextuplicate. *Student's t-test (P<0.001). (F) PC12 cells transfected with empty vector or HA–Sall2 were grown for 2 days in the same medium as in (E) but without NGF. BrdU incorporation was assayed and is presented as described in (D). Results are the average±s.e.m. from three independent experiments performed in sextuplicate. *Student's t-test (P<0.001).
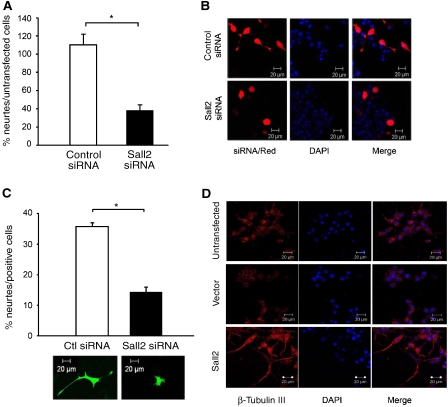
Sall2 silencing inhibits neurite outgrowth from PC12 cells
When PC12 cells are incubated with NGF they stop dividing, extend neurites and differentiate. As p21WAF1/CIP1 is induced by Sall2 and has an important function in mediating neuronal differentiation, we investigated the effect of Sall2 on neurite outgrowth by using two different assays. PC12 cells transfected with control or Sall2-specific siRNA were cultured for 2 days in collagen-coated cell culture inserts containing differentiation medium. Neurite formation was then quantified by a colorimetric assay (Smit et al, 2003) (also see Materials and methods). We found that control siRNA did not affect neurite outgrowth, whereas Sall2 siRNA diminished outgrowth by more than 50% (Figure 9A). Next, we co-transfected control or Sall2 siRNAs together with pEGFP or dsRed transfection markers. Neurite outgrowth was evaluated by confocal microscopy after 2 days in differentiation medium. Sall2 siRNA significantly decreased neurite outgrowth of PC12 cells (Figure 9B and C). As Sall2 silencing did not affect NGF-dependent survival (Supplementary Figure 6), our results show that Sall2 contributes to neurite outgrowth from PC12 cells.
Figure 9.
Sall2 regulates neurite outgrowth from PC12 cells. (A) Equal numbers of PC12 cells transfected with control or Sall2-specific siRNA were seeded into cell culture inserts coated with collagen and then cultured in differentiation medium (DM) consisting of 1.5% serum and 100 ng/ml NGF for 2 days. Neurite outgrowth was quantified by measuring cresyl violet (A562 nm) extracted from stained neurites. Data are the mean±s.e.m. of results from three independent experiments performed in triplicate. *Student's t-test (P<0.001). The percentage of neurite outgrowth is obtained by dividing neurite formation from cells transfected with control siRNA, or Sall2 siRNA, by that from untransfected cells, and multiplying by 100. (B) PC12 cells co-transfected with control or Sall2-specific siRNA and the transfection marker dsRed were serum-starved for 24 h and then cultured with differentiation medium for 2 days. Confocal microscopy shows the phenotype of the transfected cells after 2 days in differentiation medium. (C) The bar graph shows the percentage of cells in panel B extending neurites at least one and a half cell bodies in length. About 100 cells were counted for each condition. Data are the mean±s.e.m. of results from three independent experiments, *Student's t-test (P<0.001). A representative picture of a cell transfected with control or Sall2-specific siRNA and pEGFP is shown under the bar graph. (D) PC12 cells transfected with empty vector or HA–Sall2 were cultured in the medium described under (A) but without NGF. The cells were stained with an antibody to β-tubulin III, and with DAPI, and then examined by fluorescence microscopy at × 40 magnification.
As overexpression of Sall2 increases p21WAF1/CIP1 promoter activity (Figure 8D) and induces the cessation of cell proliferation (Figure 8E), we determined whether overexpression of Sall2 also induces neurite outgrowth from PC12 cells. Cells transfected with an empty vector or Sall2 were grown in NGF-free medium for 2 days. At that time, the morphology of cells that stained positively for β-tubulin III was characterized by fluorescence microscopy. We found that none of the vector-transfected cells extended neurites, whereas numerous Sall2 transfectants did so (Figure 9D). Furthermore, β-tubulin III staining increased in cells transfected with Sall2, indicating that these cells differentiated spontaneously. Thus, Sall2 has the capacity to induce neurite extension.
Sall2 is necessary for the neurite outgrowth from primary hippocampal cells
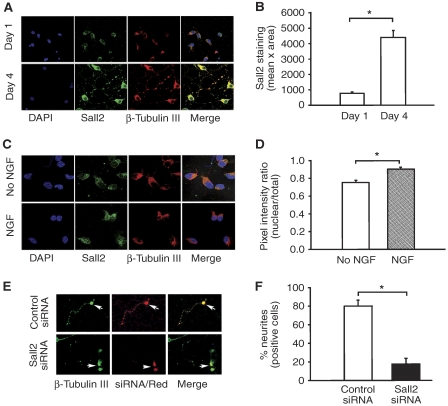
To reinforce the importance of Sall2 for neurite outgrowth, we studied hippocampal cells, which express p75NTR and TrkA (Culmsee et al, 2002), and from which NGF accelerates neurite outgrowth (Brann et al, 1999). By using confocal microscopy to examine Sall2 expression, we specifically focused on neuronal cells that stained positively for β-tubulin III. We found that Sall2 and β-tubulin III expression increased between 1 and 4 days after plating (Figure 10A and B). The relationship of Sall2 expression to neuronal differentiation (Figure 10A and B) was consistent with the likelihood that Sall2 would participate in neurite outgrowth from primary hippocampal cells. Also consistent with a likely role for Sall2 in neurite outgrowth is our observation that NFG induced a significant (20%) increase in the nuclear translocation of Sall2 in primary hippocampal cells (Figure 10C and D). To test the hypothesis that Sall2 is important for neurite outgrowth, primary hippocampal cells were co-transfected with siRNA and the dsRed transfection marker. After 24 h, NGF was added to the medium and 16 h later, neurite outgrowth was evaluated by confocal microscopy (Figure 10E). Quantification of dsRed-stained neurons (identified by expression of β-tubulin III/green) showed that 80% of cells transfected with control siRNA extended neurites, whereas only 15% of cells transfected with Sall2 siRNA did so (Figure 10F). Thus, Sall2 facilitates neurite outgrowth from primary hippocampal cells.
Figure 10.
Sall2 regulates neurite outgrowth from primary hippocampal cells. (A) Expression of the Sall2 increases during culture of primary hippocampal cells. Sall2 expression was evaluated by confocal microscopy 1 and 4 days after plating. Cells were fixed and stained with anti-Sall2 (green) and anti-β-tubulin III (red). DAPI-stained nuclei are in blue. β-Tubulin was a neuronal marker. (B) Fluorescence intensity (mean pixel × total area) of Sall2 staining was measured by digital image analysis. Data are the mean±s.e.m. (n=30 cells) from three independent experiments, *Student's t-test (P<0.001). (C) NGF induces nuclear translocation of Sall2 in primary hippocampal cells. At 1 day after plating, cells were cultured for 16 h in the absence or presence of NGF, fixed, probed with antibodies to Sall2 and β-tubulin III, and then evaluated by confocal microscopy. DAPI-stained nuclei are in blue. (D) Fluorescence intensity of nuclear and total Sall2 staining was measured by digital image analysis. Fluorescence intensity was quantified by calculating the ratio of nuclear pixel intensity × nuclear area/total pixel intensity × total area. Data are the mean±s.e.m. (n=25 cells) from three independent experiments, *Student's t-test (P<0.005). (E) Silencing of Sall2 inhibits neurite outgrowth. At 1 day after plating, primary hippocampal cells were co-transfected with control or Sall2-specific siRNA and dsRed. After 2 days, cells were cultured with NGF for 16 h, fixed and evaluated by confocal microscopy. (F) A bar graph shows the percentage of cells that stained positive for β-tubulin III (green) and dsRed positive extending neurites in cells transfected with control or Sall2 siRNA. Data are the mean±s.e.m. (n=20–25 cells) of results from two independent experiments performed in triplicate, *Student's t-test (P<0.001).
Discussion
In the present study, we used the death domain of p75NTR to screen a fetal brain two-hybrid library for interacting proteins. This led us to identify Sall2 as a new protein that interacts directly with p75NTR. Multiple clones that encode overlapping regions of an interacting protein may sometimes be identified by screening two-hybrid libraries of activation domain hybrids, permitting partial characterization of amino-acid sequences required for an interaction to occur (Phizicky and Fields, 1995). Here, the capacity of clones encompassing amino acids 119–144 and 237–383 to independently associate with the death domain of p75NTR shows that Sall2 interacts with p75NTR through multiple, non-contiguous amino-acid sequences. Sall2 is unique among the Sall family proteins in containing a p75NTR interaction motif that is conserved among species, suggesting that the Sall2 interaction with p75NTR is functionally important.
Sall proteins are C2H2 zinc-finger transcription factors in which the zinc-finger motifs are distributed over the entire protein. They contain glutamine-, proline- and alanine-rich sequences typical of transcriptional activators or repressors (Kuhnlein et al, 1994). Four members of the human family have been identified, Sall 1–4. Although human Sall-1, -3 and -4 are associated with malformation syndromes (Kohlhase et al, 1998, 1999, 2002; Kohlhase, 2000), Sall2 is not. Rather, Sall2 is a potential tumour suppressor. The gene maps to 14q11.1–13, a region associated with loss of heterozygosity in half of human ovarian cancers (Bandera et al, 1997). Sall2 is well expressed in ovarian epithelial cells but depleted in ovarian carcinoma cells (Li et al, 2004). Expression of Sall2 in ovarian carcinoma cells inhibits DNA synthesis and colony formation, increases expression of p21WAF1/CIP1 and reduces tumorigenicity. Also consistent with a role for Sall2 as a tumour suppressor is its significant role in the induction of a transcriptional programme that mediates cellular quiescence (Liu et al, 2007).
Human and mouse Sall2 are highly homologous; they are expressed in the kidney, and lung, and highly expressed in brain (Kohlhase et al, 2000). The carboxy-terminal zinc-finger of Sall2 only distantly resembles zinc-fingers in other Sall proteins; however, the remaining zinc-fingers are very similar to those in other Sall proteins, indicating that Sall2 is a bona fide member of the Sall gene family (Kohlhase et al, 2000). Here, we show that the Sall2 protein is expressed by neuronal, but not astroglial, cells in mouse brain regions that also express p75NTR. Sall2 and p75NTR colocalized outside the nucleus and the endogenous proteins co-immunoprecipitate from mouse brain as well as from cells. Our observations of the brain and our identification of discrete, evolutionarily conserved p75NTR interaction regions unique to Sall2, establish Sall2 and p75NTR as interacting proteins in a tissue in which their association must be physiologically relevant.
Sall2 and p75NTR constitutively interact. NGF dissociates p75NTR/Sall2 complexes and induces the translocation of Sall2 into the nucleus. Two other transcription factors, SC-1, which inhibits cell cycle progression (Chittka and Chao, 1999; Chittka et al, 2004), and pro-apoptotic NRIF (Casademunt et al, 1999), also associate with p75NTR when NTs are absent. As with Sall2, NGF, but not BDNF, NT-3, or NT-4, induces nuclear redistribution of SC-1 (Chittka and Chao, 1999), whereas BDNF, but not NGF, dissociates p75NTR/NRIF complexes (Geetha et al, 2005). The basis for the selectivity with which NTs act on and through p75NTR deserves further study. However, it is now apparent that p75NTR has a general function in the latency of a group of transcription factors and that NTs selectively induce the release, nuclear movement and activities of these factors.
The redistribution of Sall2 induced by NGF requires TrkA, as shown by the inability of NGF to induce nuclear localization of Sall2 in PC12 cells in which TrkA was inhibited or absent. Thus, p75NTR sequesters and TrkA activates Sall2 by inducing its redistribution into the nucleus, where it activates target genes. The involvement of TrkA in the activation of a p75NTR-interacting transcription factor is not unique to Sall2. SC-1 transcriptional activity is increased by overexpression of TrkA and diminished by the TrkA inhibitor, K252a. However, the mechanism through which TrkA affects SC-1 localization is not known (Chittka et al, 2004). The specific role played by TrkA in the nuclear translocation of Sall2 has not yet been characterized and is outside the scope of this inaugural article on Sall2, but it could involve post-transcriptional modifications. Modification could be mediated by a direct effect of TrkA on Sall2, although we have not yet been able to detect interaction of the endogenous proteins. Alternatively, TrkA could exert an effect on Sall2 indirectly by activating downstream kinases. Indeed, our observations suggest that the activation of Erk1/2 downstream of TrkA is involved in the nuclear translocation of Sall2 (Supplementary Figure 4). It is also possible that coordinate actions of p75NTR and TrkA are required for nuclear entry of Sall2. That p75NTR can have a function in inducing a post-translational modification and the nuclear translocation of an associated factor is illustrated by the observation that the p75NTR-interacting transcription factor NRIF requires polyubiquitination by TRAF6, another p75NTR-associated factor, to move into the nucleus (Geetha et al, 2005).
Sall2 expression increases during, and correlates with, the differentiation of PC12 and TrkA-deficient PC12nnr5 cells. Our results suggest that NGF mediates its effect on Sall2 expression by acting through p75NTR, and that TrkA is not required for this process. Sall2 expression also increases when primary hippocampal cells are cultured in NGF-free medium (Figure 10A and B), suggesting that autocrine activation of p75NTR, or other receptors, can induce Sall2 expression. Our observations are consistent with a model in which p75NTR sequesters and localizes Sall2 to the cell surface and in proximity to TrkA. NGF dissociates Sall2 from p75NTR and activates TrkA, which has an obligate function in the translocation of Sall2 into the nucleus. NGF not only dissociates p75NTR/Sall2 complexes but also induces Sall2 expression by acting through the p75NTR receptor. These observations suggest that an autoregulatory mechanism may replenish Sall2, such that it may be re-localized to the cell membrane by p75NTR and released as required.
Our results show that Sall2 and p21WAF1/CIP1 expression increase in parallel during NGF-induced differentiation and that depletion of Sall2 diminishes p21WAF1/CIP1 expression in PC12 cells. The effect of Sall2 on p21WAF1/CIP1 expression appears to be direct, because depletion of Sall2 diminishes NGF-induced activation of a p21WAF1/CIP1 promoter that contains previously identified Sall2-binding sites (Li et al, 2004). Sall2 and p21WAF1/CIP1 are also related functionally, because depletion of Sall2 impairs the capacity of NGF to induce growth arrest in PC12 cells. p21WAF1/CIP1 is induced by and mediates the capacity of NGF to induce neuronal cells to exit from the cell cycle, undergo growth arrest and differentiate (Yan and Ziff, 1997; Erhardt and Pittman, 1998; Ito et al, 2002). NGF induces p21WAF1/CIP1 through p53-dependent (Poluha et al, 1997; Hughes et al, 2000) and p53-independent mechanisms (Yan and Ziff, 1997; Billon et al, 1999). In ovarian cancer cells, Sall2 binds the promoter for and induces p21WAF1/CIP1, establishing Sall2 as a transcription factor that can function independently of p53 (Li et al, 2004). The present study extends these cancer-related observations by showing that Sall2 may induce NGF-dependent cell cycle arrest and growth inhibition through the induction of p21WAF1/CIP1.
Depletion of Sall2 also diminishes neurite outgrowth from PC12 cells and primary hippocampal neurons. The mechanisms by which Sall2 regulates neurite extension are at present unclear, but could relate in part to its effect on p21WAF1/CIP1. Cell cycle regulatory proteins, including p21WAF1/CIP1, have been detected in the cytoplasm and nuclei of postnatal and even adult neurons in the Cornu Ammonis subfields and the granule and subgranular layers of the mouse hippocampus (Schmetsdorf et al, 2005; Pechnick et al, 2008). Also, in the developing rat brain, high levels of p21WAF1/CIP1 mRNA and protein are present in postmitotic neurons in the cortex, cerebellum, hippocampus and thalamus. p21WAF1/CIP1 is a well-recognized target of the p53 transcription factor; however, expression of p53 is low or undetectable in neurons of the developing mouse brain that express p21WAF1/CIP1 (van Lookeren Campagne and Gill, 1998). These observations suggest that transcription factors other than p53 could be responsible for the expression of p21WAF1/CIP1 in these brain areas. Interestingly, we found that Sall2 is expressed in postmitotic neurons of the cortex, hippocampus and thalamus in the mouse brain (Figure 3). The presence of negative cell cycle regulatory proteins in postmitotic neurons has been associated with cell cycle withdrawal to support the differentiation and maintenance of different types of neurons. Although the role of nuclear p21WAF1/CIP1 in cell cycle withdrawal is well documented, recent studies have demonstrated that p21WAF1/CIP1 has additional functions outside the nucleus and these are important to the development of mature neurons. Cytoplasmic p21WAF1/CIP1 promotes neurite outgrowth and branching from hippocampal neurons (Bito et al, 2000; Tanaka et al, 2002, 2004). Future work will be necessary to define how p21WAF1/CIP1 regulates neurite outgrowth.
Our observations show that p75NTR impairs and TrkA induces the nuclear localization Sall2. Nuclear Sall2 promotes three NT-induced processes that are hallmarks of the neuronal phenotype: induction of p21WAF1/CIP1, cell cycle arrest and neurite outgrowth. Sall2-deficient mice were originally reported to develop without an obvious abnormal phenotype (Sato et al, 2003). However, results published after submission of the present study show that Sall2-deficient mice display background-specific neural tube defects and suggest that Sall2 is required for embryonic neural tube development (Bohm et al, 2008). Our observations and the phenotype of Sall2-deficient mice reinforce one another in providing a role for Sall2 in the physiology of the nervous system. This compelling conclusion coupled with our demonstration that overexpression of Sall2 induces the neurotropin-independent expression of p21WAF1/CIP1, growth inhibition and neurite outgrowth suggests that a more complete understanding of Sall2 action will provide fundamental insight into the transcriptional regulation of neuronal development.
Materials and methods
Sall2 constructs
Full-length human Sall2 was obtained by ligating two cDNA fragments and cloned into a modified eukaryotic expression vector pCMV2-FLAG. Fidelity of the coding sequence was confirmed by dideoxynucleotide sequencing and the sequence of its encoded protein was confirmed by amino-acid sequence analysis.
Two-hybrid screening
The human p75NTR death domain, including seven amino acids upstream and nine amino acids downstream of the motif (amino acids 324–399), was prepared by PCR and ligated into pGBT9 to generate a fusion protein with the GAL4 DNA-binding domain. A human fetal brain two-hybrid library was inserted into pGAD424 to yield fusions with the GAL4 activation domain. The PGBT9-p75NTR/death domain and the pGAD424 human fetal brain two-hybrid library were co-transformed into SFY526 yeast and 75 million double transformants were screened for the lacZ phenotype. To test specificity, four His+, lacZ+ clones were isolated and retransformed into SFY526 yeast, alone, with pGBT9, pGBT9-NTR/death domain, pLAM5′ (the lamin gene) and with the pGBT9-FAS intracellular domain. Clones that activated the lacZ reporter only in the presence of the p75NTR death domain fusion were chosen as true positives.
Cell culture and transfection, isolation of RNA, RT–PCR and BrdU incorporation
Cells were cultured and transfected as described in the Supplementary data. Total RNA was isolated, RT–PCR was conducted and BrdU incorporation was assayed according to our standard methods (see Supplementary data).
Immunofluorescence of Sall2 in mouse brain sections
Frozen whole adult mouse brains were stored at −80°C until sectioned. Coronal sections (10 μm) were made in the UCSF mouse pathology facility using a cryostat and were stored at −80°C. Slides were brought to room temperature, immersed in freshly prepared 1% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.4) for 30 min, washed several times in PBS and blocked by incubation for 1 h in 3% normal goat serum in PBS. Slides were incubated with anti-Sall2, or double stained with anti-Sall2 and anti-β-tubulin III, Sall2 and GFAP, or Sall2 and anti-p75NTR for 90 min and then washed several times with PBS. Slides were then incubated for 30 min with Alexa Fluor-conjugated secondary antibodies and with PBS. Cover slips were sealed with fluorescent Vectashield-mounting medium and the slides were stored flat in the dark at 4°C until evaluation by immunofluorescence microscopy.
Confocal microscopy
Cells were fixed in 4% paraformaldehyde/PBS (pH 7.4) (20 min, room temperature), and washed three times with PBS. Cells were permeabilized with 0.1% Triton X-100/PBS (10 min), washed three times with PBS, blocked with 3% goat serum (1 h at room temperature or overnight at 4°C) and incubated for 1 h with antibody to FLAG, Sall2, β-tubulin III or GFPA. The cells were washed and then incubated with an FITC or Alexa Fluor-conjugated anti-mouse or anti-rabbit secondary antibody. The nucleus was detected by staining with DAPI or Syto 16. Excitation of the stains was performed in a Bio-Rad MRC 1024 Krypton/Argon confocal imaging system under × 60 magnification and analysed using the METAMORPH program. For experiments in PC12 cells, a Zeiss UV Argon LMS 510 confocal microscope was used at × 40 or × 60 magnifications. Data were analysed using the Zeiss LSM510 software. Nuclear Sall2 was assayed by calculating the percentage of total to nuclear cell fluorescence (O'Callaghan et al, 2003).
siRNA
Transfection of a pool of four rat Sall2-specific RNAi duplexes (Dharmacon) into PC12 or hippocampal cells decreased Sall2 expression (Supplementary Figure 3A and B), which remained low for more than 4 days. To estimate the transfection efficiency of cells, siGLO siRNA labelled with a Cy3-fluorescent tag was used. Confocal analysis indicated ∼90% transfection efficiency (Supplementary Figure 3C), a result comparable to that obtained when PC12 cells were transfected with other siRNAs (Thonberg et al, 2004). In some experiments, siRNA was co-transfected with the transfection markers 50 nM dsRed or 50 ng pEGFP. Red- or GFP-positive cells were counted and divided by the total number of cells per × 40 field to calculate the average transfection efficiency (PC12 cells ∼12–14%, hippocampal cells ∼1–2%).
Neurite outgrowth from PC12 cells and primary hippocampal neurons
Neurite outgrowth was assayed by a procedure described by Smit et al (2003) and by counting cells with neurites that were more than 1.5 times the diameter of the cell body (see Supplementary data).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Acknowledgments
The Sall2 antibody was a gift from Dr Thomas Benjamin; Sall2 genomic and cDNA fragments were from Dr Jorgen Kolhase; the p21WAF1/CIP1 luciferase reporter was from Dr Mei-Huey Jeng; cDNA for p75NTR and the p75NTR-9992 antibody were provided by Dr Moses Chao and antiserum against p75NTR (Rex) and TrkA (RTA) by Dr Louis Reichardt. Dr Michael Rauchman provided full-length mouse Sall2 in pcDNAFlu. Dr Ksenya Shchors provided frozen mouse brains. We thank Drs Ariel Castro, Jennifer LaVail, Noelle L'Etoile and Samuel Pleasure for review of the paper and Louis Reichardt for suggestions. Special thanks to Pamela Derish for editing the paper and Prema Idumalla for technical assistance.
References
- Bai S, Ghoshal K, Datta J, Majumder S, Yoon SO, Jacob ST (2005) DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol Cell Biol 25: 751–766 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bandera CA, Takahashi H, Behbakht K, Liu PC, LiVolsi VA, Benjamin I, Morgan MA, King SA, Rubin SC, Boyd J (1997) Deletion mapping of two potential chromosome 14 tumor suppressor gene loci in ovarian carcinoma. Cancer Res 57: 513–515 [PubMed] [Google Scholar]
- Berg MM, Sternberg DW, Parada LF, Chao MV (1992) K-252a inhibits nerve growth factor-induced trk proto-oncogene tyrosine phosphorylation and kinase activity. J Biol Chem 267: 13–16 [PubMed] [Google Scholar]
- Billon N, Carlisi D, Datto MB, van Grunsven LA, Watt A, Wang XF, Rudkin BB (1999) Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 18: 2872–2882 [DOI] [PubMed] [Google Scholar]
- Bito H, Furuyashiki T, Ishihara H, Shibasaki Y, Ohashi K, Mizuno K, Maekawa M, Ishizaki T, Narumiya S (2000) A critical role for a Rho-associated kinase, p160ROCK, in determining axon outgrowth in mammalian CNS neurons. Neuron 26: 431–441 [DOI] [PubMed] [Google Scholar]
- Bohm J, Buck A, Borozdin W, Mannan AU, Matysiak-Scholze U, Adham I, Schulz-Schaeffer W, Floss T, Wurst W, Kohlhase J, Barrionuevo F (2008) Sall1, Sall2, and Sall4 are required for neural tube closure in mice. Am J Pathol 173: 1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann AB, Scott R, Neuberger Y, Abulafia D, Boldin S, Fainzilber M, Futerman AH (1999) Ceramide signaling downstream of the p75 neurotrophin receptor mediates the effects of nerve growth factor on outgrowth of cultured hippocampal neurons. J Neurosci 19: 8199–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casademunt E, Carter BD, Benzel I, Frade JM, Dechant G, Barde YA (1999) The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. EMBO J 18: 6050–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV (2003) Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 4: 299–309 [DOI] [PubMed] [Google Scholar]
- Chittka A, Arevalo JC, Rodriguez-Guzman M, Perez P, Chao MV, Sendtner M (2004) The p75NTR-interacting protein SC1 inhibits cell cycle progression by transcriptional repression of cyclin E. J Cell Biol 164: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittka A, Chao MV (1999) Identification of a zinc finger protein whose subcellular distribution is regulated by serum and nerve growth factor. Proc Natl Acad Sci USA 96: 10705–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culmsee C, Gerling N, Lehmann M, Nikolova-Karakashian M, Prehn JH, Mattson MP, Krieglstein J (2002) Nerve growth factor survival signaling in cultured hippocampal neurons is mediated through TrkA and requires the common neurotrophin receptor P75. Neuroscience 115: 1089–1108 [DOI] [PubMed] [Google Scholar]
- Dechant G, Barde YA (2002) The neurotrophin receptor p75(NTR): novel functions and implications for diseases of the nervous system. Nat Neurosci 5: 1131–1136 [DOI] [PubMed] [Google Scholar]
- Erhardt JA, Pittman RN (1998) p21WAF1 induces permanent growth arrest and enhances differentiation, but does not alter apoptosis in PC12 cells. Oncogene 16: 443–451 [DOI] [PubMed] [Google Scholar]
- Geetha T, Kenchappa RS, Wooten MW, Carter BD (2005) TRAF6-mediated ubiquitination regulates nuclear translocation of NRIF, the p75 receptor interactor. EMBO J 24: 3859–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehler S, Gallo G, Veien E, Letourneau PC (2004) p75 neurotrophin receptor signaling regulates growth cone filopodial dynamics through modulating RhoA activity. J Neurosci 24: 4363–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Tischler AS (1976) Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA 73: 2424–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Gollapudi L, Sladek TL, Neet KE (2000) Mediation of nerve growth factor-driven cell cycle arrest in PC12 cells by p53. Simultaneous differentiation and proliferation subsequent to p53 functional inactivation. J Biol Chem 275: 37829–37837 [DOI] [PubMed] [Google Scholar]
- Ito H, Nomoto H, Furukawa S (2002) Role of low-affinity p75 receptor in nerve growth factor-inducible growth arrest of PC12 cells. J Neurosci Res 69: 653–661 [DOI] [PubMed] [Google Scholar]
- Khursigara G, Bertin J, Yano H, Moffett H, DiStefano PS, Chao MV (2001) A prosurvival function for the p75 receptor death domain mediated via the caspase recruitment domain receptor-interacting protein 2. J Neurosci 21: 5854–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara G, Orlinick JR, Chao MV (1999) Association of the p75 neurotrophin receptor with TRAF6. J Biol Chem 274: 2597–2600 [DOI] [PubMed] [Google Scholar]
- Kohlhase J (2000) SALL1 mutations in Townes–Brocks syndrome and related disorders. Hum Mutat 16: 460–466 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Altmann M, Archangelo L, Dixkens C, Engel W (2000) Genomic cloning, chromosomal mapping, and expression analysis of msal-2. Mamm Genome 11: 64–68 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Hausmann S, Stojmenovic G, Dixkens C, Bink K, Schulz-Schaeffer W, Altmann M, Engel W (1999) SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics 62: 216–222 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, Turnpenny P, Winter RM, Reardon W (2002) Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 11: 2979–2987 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W (1998) Mutations in the SALL1 putative transcription factor gene cause Townes–Brocks syndrome. Nat Genet 18: 81–83 [DOI] [PubMed] [Google Scholar]
- Kuhnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, Gehring WJ, Jackle H, Schuh R (1994) spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J 13: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Li E, Huber LJ, Landis SC, Sharpe AH, Chao MV, Jaenisch R (1992) Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 69: 737–749 [DOI] [PubMed] [Google Scholar]
- Li D, Tian Y, Ma Y, Benjamin T (2004) p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP). Mol Cell Biol 24: 3885–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Adler AS, Segal E, Chang HY (2007) A transcriptional program mediating entry into cellular quiescence. PLoS Genet 3: e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb DM, Maragos J, Martin-Zanca D, Chao MV, Parada LF, Greene LA (1991) The trk proto-oncogene rescues NGF responsiveness in mutant NGF-nonresponsive PC12 cell lines. Cell 66: 961–966 [DOI] [PubMed] [Google Scholar]
- Mukai J, Hachiya T, Shoji-Hoshino S, Kimura MT, Nadano D, Suvanto P, Hanaoka T, Li Y, Irie S, Greene LA, Sato TA (2000) NADE, a p75NTR-associated cell death executor, is involved in signal transduction mediated by the common neurotrophin receptor p75NTR. J Biol Chem 275: 17566–17570 [DOI] [PubMed] [Google Scholar]
- Naumann T, Casademunt E, Hollerbach E, Hofmann J, Dechant G, Frotscher M, Barde YA (2002) Complete deletion of the neurotrophin receptor p75NTR leads to long-lasting increases in the number of basal forebrain cholinergic neurons. J Neurosci 22: 2409–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, O'Connell JX, Gilks CB, Sorensen PH, Linn S, West RB, Liu CL, Botstein D, Brown PO, van de Rijn M (2003) Tissue microarray validation of epidermal growth factor receptor and SALL2 in synovial sarcoma with comparison to tumors of similar histology. Am J Pathol 163: 1449–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan DW, Hasdemir B, Leighton M, Burgoyne RD (2003) Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J Cell Sci 116: 4833–4845 [DOI] [PubMed] [Google Scholar]
- Pechnick RN, Zonis S, Wawrowsky K, Pourmorady J, Chesnokova V (2008) p21Cip1 restricts neuronal proliferation in the subgranular zone of the dentate gyrus of the hippocampus. Proc Natl Acad Sci USA 105: 1358–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Fields S (1995) Protein–protein interactions: methods for detection and analysis. Microbiol Rev 59: 94–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluha W, Schonhoff CM, Harrington KS, Lachyankar MB, Crosbie NE, Bulseco DA, Ross AH (1997) A novel, nerve growth factor-activated pathway involving nitric oxide, p53, and p21WAF1 regulates neuronal differentiation of PC12 cells. J Biol Chem 272: 24002–24007 [DOI] [PubMed] [Google Scholar]
- Reichardt LF (2006) Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci 361: 1545–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL, Verdi JM, Barker PA (2000) NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron 27: 279–288 [DOI] [PubMed] [Google Scholar]
- Sato A, Matsumoto Y, Koide U, Kataoka Y, Yoshida N, Yokota T, Asashima M, Nishinakamura R (2003) Zinc finger protein sall2 is not essential for embryonic and kidney development. Mol Cell Biol 23: 62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmetsdorf S, Gartner U, Arendt T (2005) Expression of cell cycle-related proteins in developing and adult mouse hippocampus. Int J Dev Neurosci 23: 101–112 [DOI] [PubMed] [Google Scholar]
- Smit M, Leng J, Klemke RL (2003) Assay for neurite outgrowth quantification. Biotechniques 35: 254–256 [DOI] [PubMed] [Google Scholar]
- Song HY, Dunbar JD, Donner DB (1994) Aggregation of the intracellular domain of the type 1 tumor necrosis factor receptor defined by the two-hybrid system. J Biol Chem 269: 22492–22495 [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Asada M, Mizutani S, Yoshikawa H, Tohyama M (2002) Cytoplasmic p21(Cip1/WAF1) regulates neurite remodeling by inhibiting Rho-kinase activity. J Cell Biol 158: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Yachi K, Fujiwara T, Yoshikawa H, Tohyama M (2004) Cytoplasmic p21(Cip1/WAF1) enhances axonal regeneration and functional recovery after spinal cord injury in rats. Neuroscience 127: 155–164 [DOI] [PubMed] [Google Scholar]
- Teng KK, Hempstead BL (2004) Neurotrophins and their receptors: signaling trios in complex biological systems. Cell Mol Life Sci 61: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonberg H, Scheele CC, Dahlgren C, Wahlestedt C (2004) Characterization of RNA interference in rat PC12 cells: requirement of GERp95. Biochem Biophys Res Commun 318: 927–934 [DOI] [PubMed] [Google Scholar]
- van Lookeren Campagne M, Gill R (1998) Tumor-suppressor p53 is expressed in proliferating and newly formed neurons of the embryonic and postnatal rat brain: comparison with expression of the cell cycle regulators p21Waf1/Cip1, p27Kip1, p57Kip2, p16Ink4a, cyclin G1, and the proto-oncogene Bax. J Comp Neurol 397: 181–198 [DOI] [PubMed] [Google Scholar]
- Vilar M, Murillo-Carretero M, Mira H, Magnusson K, Besset V, Ibanez CF (2006) Bex1, a novel interactor of the p75 neurotrophin receptor, links neurotrophin signaling to the cell cycle. EMBO J 25: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bartheld CS, Williams R, Lefcort F, Clary DO, Reichardt LF, Bothwell M (1996) Retrograde transport of neurotrophins from the eye to the brain in chick embryos: roles of the p75NTR and trkB receptors. J Neurosci 16: 2995–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schack D, Casademunt E, Schweigreiter R, Meyer M, Bibel M, Dechant G (2001) Complete ablation of the neurotrophin receptor p75NTR causes defects both in the nervous and the vascular system. Nat Neurosci 4: 977–978 [DOI] [PubMed] [Google Scholar]
- Yamashita T, Tucker KL, Barde YA (1999) Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron 24: 585–593 [DOI] [PubMed] [Google Scholar]
- Yan GZ, Ziff EB (1997) Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J Neurosci 17: 6122–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Mehlen P, Rabizadeh S, VanArsdale T, Zhang H, Shin H, Wang JJ, Leo E, Zapata J, Hauser CA, Reed JC, Bredesen DE (1999) TRAF family proteins interact with the common neurotrophin receptor and modulate apoptosis induction. J Biol Chem 274: 30202–30208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information