Abstract
Pdcd4 is a tumour suppressor protein. It inhibits translation through interaction with translation initiator eIF4A, resulting in the suppression of neoplastic transformation and tumour invasion. Here, we present the crystal structures of an N-terminal-truncated Pdcd4 in free form and in complex with eIF4A. Upon binding to eIF4A, Pdcd4 undergoes a marked conformational change to form a heterotrimeric complex with eIF4A, with one Pdcd4 binding to two eIF4A molecules in two different modes. The binding of Pdcd4 to eIF4A is required to inhibit the enzymatic activity of eIF4A, translation initiation, and AP-1-dependent transcription. Both MA3 domains are required to efficiently compete with the C-terminal domain of eIF4G (eIF4Gc) for binding to eIF4A whereas a single MA3 is sufficient to inhibit translation. Our structural and mutational analyses reveal that Pdcd4 inhibits translation initiation by trapping eIF4A in an inactive conformation, and blocking its incorporation into the eIF4F complex.
Keywords: Pdcd4, RNA helicase, translation initiation, translational control, tumour suppressor
Introduction
The PDCD4 gene was first isolated as an MA-3 gene (also known as TIS/H731/DUG) in a differential display search for genes upregulated in apoptosis-induced mouse cells (Shibahara et al, 1995). Subsequently, the Pdcd4 protein was identified as a tumour suppressor in a mouse model of tumour promotion, in which high level of Pdcd4 was shown to suppress neoplastic transformation and tumour phenotype (Yang et al, 2001, 2003). Pdcd4 has been shown to suppress skin tumorigenesis and tumour progression in transgenic mice (Jansen et al, 2005). Loss of Pdcd4 expression has been strongly implicated in the development and progression of lung, colon, and breast cancer (Chen et al, 2003; Afonja et al, 2004). Pdcd4 protein levels have been shown to be reduced in human lung-, renal- and glia-derived tumours (Jansen et al, 2004). More recently, downregulation of Pdcd4 has been shown to promote tumour invasion and activate both β-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells (Wang et al, 2008).
Pdcd4 has two potential phosphorylation sites at Ser67 and Ser457 and has been shown to be a regulatory target of the protein kinases Akt (protein kinase B) (Palamarchuk et al, 2005) and S6K1 (Dorrello et al, 2006). Phosphorylation of Pdcd4 by these kinases leads to its rapid degradation mediated by the E3 ubiquitin ligase complex SCFβTrCP, thereby promoting protein translation and cell growth (Dorrello et al, 2006; Schmid et al, 2008). Recently, oncogenic micro-RNA miR-21 was shown to target the PDCD4 gene and to downregulate its expression in cancer cells (Asangani et al, 2008; Frankel et al, 2008; Lu et al, 2008; Zhu et al, 2008).
Pdcd4 inhibits tumour promoter-induced transformation by inhibiting transactivation of the activator protein 1 (AP-1) transcription factor (Yang et al, 2001) and this inhibitory effect is achieved through interaction with translation initiation factor eIF4A (Goke et al, 2002; Yang et al, 2003), the key component of the eIF4F complex, which includes two other translation initiation factors eIF4E and eIF4G. eIF4A is a member of the DEAD-box protein family, and functions as an ATP-dependent RNA helicase to catalyze the unwinding of mRNA secondary structure at the 5′UTR (Rogers Jr et al, 2002). eIF4G is a scaffold protein containing two eIF4A-binding sites: one located within the middle one-third (eIF4Gm) and the other located within the C-terminal one-third (eIF4Gc) (Morino et al, 2000). The binding of eIF4A to eIF4Gm increases helicase activity and is sufficient for cap-dependent translation (De Gregorio et al, 1998; Korneeva et al, 2001, 2005) whereas the interaction of eIF4A with eIF4Gc is thought to provide a modulatory function (Korneeva et al, 2001).
Pdcd4 contains two tandem MA3 domains, both of which are protein-interaction modules that belong to the functionally diverse HEAT-repeat family of proteins. Pdcd4 interacts with eIF4A and eIF4G through its highly conserved MA3 domains (Yang et al, 2004). Both MA3 domains are required for efficient binding to eIF4A whereas the C-terminal MA3 domain competes with eIF4Gc for binding to eIF4A (Yang et al, 2004). Through its association with eIF4A, Pdcd4 inactivates the helicase function of eIF4A and suppresses cap-dependent translation (Yang et al, 2003), and consequently inhibits AP-1 transactivation (Yang et al, 2001, 2003). The observation that Pdcd4 inhibits both tumour formation and tumour progression by targeting translation is consistent with the evidence that translation initiation factors are linked to carcinogenesis (Zimmer et al, 2000; Holland et al, 2004).
The structure of the C-terminal MA3 domain of mouse Pdcd4 has been determined by both crystallography and NMR (LaRonde-LeBlanc et al, 2007; Waters et al, 2007). The C-terminal MA3 domain is structurally very similar to the MA3 domain of eIF4Gc (eIF4G-MA3), competes with eIF4Gc for binding to eIF4A and is sufficient to inhibit cap-dependent translation (LaRonde-LeBlanc et al, 2007). The most recently reported structure of the N-MA3 domain of human Pdcd4 (Suzuki et al, 2008) showed that the two MA3 domains are structurally very similar and have similar eIF4A-binding surfaces. Moreover, like the C-terminal MA3, the N-terminal MA3 competes efficiently with eIF4G-MA3 for binding to eIF4A (Suzuki et al, 2008), suggesting both MA3 domains are structurally and functionally similar. However, how two MA3 domains act cooperatively to bind eIF4A and inhibit translation remains elusive although the interaction of both MA3 domains with eIF4A has been shown to form a more stable complex (Suzuki et al, 2008).
Here, we report the crystal structures of an N-terminal-truncated human Pdcd4 containing both MA3 domains (Pdcd4ΔN) in free form and in complex with full-length eIF4AI. The structure revealed how two tandem MA3 domains of Pdcd4 act synergistically to bind eIF4A specifically and inhibit translation. Structure-based mutagenesis confirmed that the interaction between Pdcd4 and eIF4A is required for Pdcd4 to exert its inhibitory effects on translation and AP-1-dependent transcription, as well as the helicase activity of eIF4A. Our results have unraveled a mechanism underlying the translational inhibition by Pdcd4.
Results
Structure determination
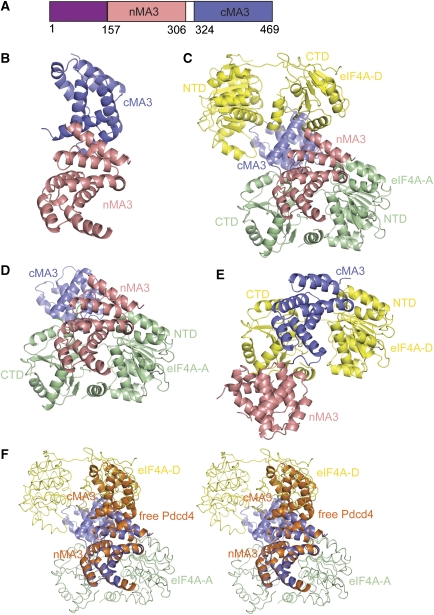
Full length human Pdcd4 (hPdcd4FL) contains an N-terminal putative RNA-binding domain (hPdcd4-NTD, residues 1–140) and two conserved MA3 domains, designated as nMA3 and cMA3, respectively (Figure 1A). As purified full-length protein failed to crystallize, we set out to identify a truncated protein which was amenable for crystallization. Limited proteolysis yielded a fragment from residue 153 onwards as the minimum portion that supports binding to eIF4AI. This data combined with sequence alignments and secondary structure predictions led to the identification of a compact fragment of hPdcd4 containing both the nMA3 and cMA3 domains (hPdcd4ΔN; residues 157 to 469). Purified hPdcd4ΔN yielded diffraction-quality crystals, which belonged to space group P3121 and contained two molecules in the asymmetric unit (AU). The structure was solved by the single-wavelength anomalous dispersion phasing method and refined at a resolution of 2.87 Å to working and free R factors of 23.1 and 27.3%, respectively with no outliers in the Ramachandran plot. The final refined model lacks the linker region (residues 307–323) between the two MA3 domains and the C-terminal tail (residues 451–469) as no interpretable electron density was observed in these regions, which are assumed to be disordered.
Figure 1.
The structures of hPdcd4ΔN and the mPdcd4ΔN-eIF4A complex. (A) Domain architecture of human/mouse Pdcd4 showing the putative N-terminal RNA-binding domain (purple), the N-terminal MA3 domain (nMA3; salmon), and the C-terminal MA3 domain (cMA3; blue). Numbers below the schematic protein outline represents the amino acid position for the domain boundaries. (B) Overall structure of hPdcd4ΔN. The ribbon diagrams of nMA3 and cMA3 are shown in salmon and blue, respectively. (C) Overall structure of the mPdcd4ΔN–eIF4A complex. The colouring scheme for nMA3 and cMA3 of Pdcd4, and the orientation of nMA3 are as in (B). The two copies of eIF4A, eIF4A-A and eIF4A-D are shown in green and yellow, respectively. Individual domains are labelled. (D) The subcomplex comprising of mPdcd4ΔN and one copy of eIF4A (eIF4A-A). In this binding mode, the nMA3 of Pdcd4 interacts with both the NTD and CTD of eIF4A with additional interactions mediated by the cMA3 domain and the CTD of eIF4A. The view and colouring scheme are as in (C). (E) The subcomplex consisting of mPdcd4ΔN and another copy of eIF4A (eIF4A-D). In this binding mode, only the cMA3 domain interacts with both the NTD and CTD of eIF4A-D. The ribbon diagram is re-oriented by superposition of eIF4A-D and eIF4A-A at NTD. The colour coding is as in (C). (F) Conformational change of Pdcd4 upon binding to eIF4A. Stereo views of the nMA3 of hPdcd4ΔN superimposed with that of mPdcd4ΔN in the mPdcd4ΔN-eIF4A complex. Pdcd4 in free form and in complex with eIF4A are shown in orange and blue, respectively, and two copies of eIF4A are shown in worm with the same colours as in (C).
The structure of mouse Pdcd4 (mPdcd4ΔN, residues 120–469) in complex with full-length mouse eIF4AI (designated as mPdcd4ΔN–eIF4A) was solved by the method of molecular replacement using the MA3 domains of hPdcd4ΔN, and the two RecA-like domains of Saccharomyces cerevisiae eIF4AI as search models, and refined at a resolution of 3.5 Å to working and free R factors of 25.0 and 29.0%, respectively. The final model of the mPdcd4ΔN–eIF4A complex contains one mPdcd4ΔN and two eIF4A molecules (designated as eIF4A-A and eIF4A-D) in the AU, with the model of eIF4A-D being more complete than that of eIF4A-A. In mPdcd4ΔN, the N- and C termini and a portion of the linker region are disordered, whereas the disordered regions in eIF4A are located mostly in the loop regions. Statistics of structural determination and refinement for hPdcd4ΔN and mPdcd4ΔN-eIF4A are summarized in Supplementary Table S1 (see Materials and methods).
Structural overview
As shown in Figure 1B, the structure of hPdcd4ΔN contains two α-helical MA3 domains of the HEAT-repeat family that resemble a short solenoid. The two MA3 domains interact with each other to create an inter-domain interface. This observation is consistent with the finding by Suzuki et al. (2008), which showed that two MA3 domains interact in cis through electrostatic interactions.
The structure of the mPdcd4ΔN–eIF4A complex (Figure 1C) shows that Pdcd4 is sandwiched between two eIF4A molecules to create a compact global architecture, with each MA3 domain of Pdcd4 binding to one eIF4A molecule respectively, giving rise to a 1:2 binding stoichiometry between Pdcd4 and eIF4A in the AU. The nMA3 and cMA3 domains of Pdcd4 bind to eIF4A in a nearly perpendicular manner with each MA3 domain contacting both the NTD and CTD of eIF4A, which fixes their relative orientation with respect to each other.
In the mPdcd4ΔN–eIF4A complex, one Pdcd4 binds to the two eIF4A molecules in two different modes. In one mode, the two MA3 domains of Pdcd4 interact with both the NTD and CTD of eIF4A (Figure 1D) to create three interfaces, nMA3–NTD-A, nMA3–CTD-A, and cMA3–CTD-A with a total buried surface area of 3445 Å2. In the other mode, only the cMA3 domain interacts with eIF4A in two interfaces cMA3–NTD-D and cMA3–CTD-D (Figure 1E) with the buried surface area of 1974 Å2. The observation that the CTD of eIF4A is involved in more interactions with Pdcd4 contradicts the finding that Pdcd4 binds to eIF4A-NTD but not to eIF4A-CTD (Suzuki et al. 2008).
The overall structure of the individual MA3 domain is largely unchanged upon complex formation. Superposition of the isolated domains of free Pdcd4 with those in the complexed form gives an r.m.s.d. of 0.60 Å for equivalent Cα atoms. However, the orientation of cMA3 relative to nMA3 differs by 65° when the nMA3 in the complex is superimposed with that in the free form. Such a marked conformational change relieves the steric hindrance between one of the MA3 domains and eIF4A and allows both MA3 domains to interact with eIF4A to form a stable complex (Figure 1F).
eIF4A is comprised of two RecA-like domains (NTD and CTD) joined by a flexible linker. The individual domains of two eIF4A molecules are very similar with an r.m.s.d. of 0.60–1.0 Å for all Cα atoms. Despite these high structural similarities, the orientation of CTD relative to NTD in two eIF4A molecules differs by 15° when the NTD is superimposed (Supplementary Figure S1A). Each eIF4A molecule in the complex adopts an open dumbbell-like conformation with no interactions between its NTD and CTD.
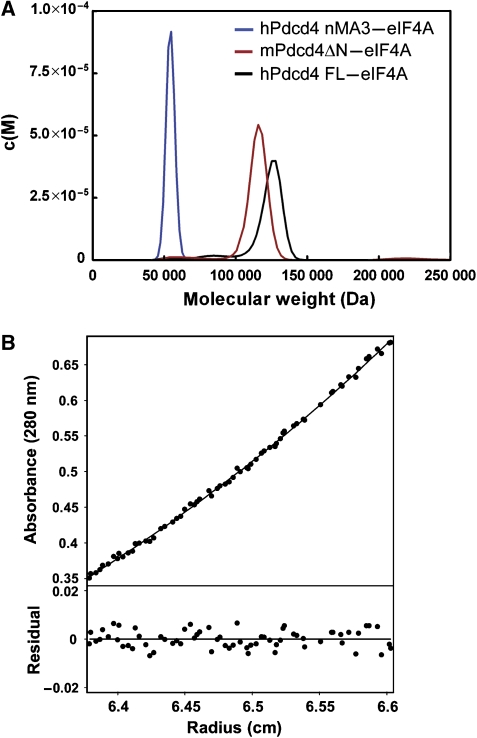
Pdcd4 and eIF4A bind in a 1:2 stoichiometry in solution
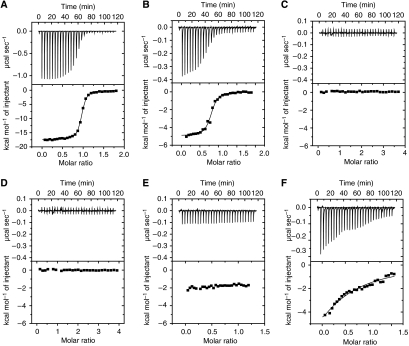
Consistent with a 1:2 stoichiometry between Pdcd4 and eIF4A observed in the crystal, size-exclusion chromatography analyses (data not shown) indicated the presence of a complex with a higher molecular weight than would be expected for a heterodimer, that is, 1:1 stoichiometric ratio (85 kDa). To validate the 1:2 stoichiometry in the crystal, analytical ultracentrifugation was employed to examine the association state between Pdcd4 and eIF4A. We first analyzed the purified mPdcd4ΔN–eIF4A complex by sedimentation velocity. The data fitted well to the c(S) and c(M) distributions, giving a peak that corresponds to a molecular mass of 117 kDa (Figure 2A). As the calculated molecular weights of monomeric mPdcd4ΔN and eIF4A are 39 kDa and 46 kDa, respectively, the calculated molecular mass of a 1:2 mPdcd4ΔN-eIF4A heterotrimeric complex is 131 kDa, which agrees with the molecular mass measured by sedimentation velocity. The complexes of hPdcd4FL with eIF4A and the nMA3 domain of hPdcd4 with eIF4A were also analyzed by sedimentation velocity. The data of hPdcd4FL-eIF4A or nMA3-eIF4A fitted well to the c(S) and c(M) distributions and generated a single peak corresponding to a molecular weight of 120 kDa or 55 kDa, respectively (Figure 2A). These derived molecular weights are close to the calculated values for a 1:2 hPdcd4FL–eIF4A complex (144 kDa) and a 1:1 nMA3–eIF4A complex (62 kDa).
Figure 2.
Pdcd4 binds to eIF4A with a 1:2 stoichiometry in solution. (A) The complexes of hPdcd4 nMA3–eIF4A, mPdcd4ΔN–eIF4A, and hPdcd4FL-eIF4A were analyzed by sedimentation velocity and fitted to the c(S) and c(M) size-distribution functions. The molar mass distribution, c(M) is shown for all three samples. (B) The mPdcd4ΔN–eIF4A complex was also analyzed by sedimentation equilibrium and fitted to an ideal single-species model. A representative fit is shown.
The mPdcd4ΔN–eIF4A complex was further analyzed by sedimentation equilibrium over a wide concentration range. After correcting for buoyancy and viscosity, the data fitted well to an ideal single-species model, giving an apparent weight—average molecular weight of 123 kDa (Figure 2B), very close to that of the heterotrimer. Taken together, these results demonstrated that one Pdcd4 molecule binds to two eIF4A molecules in solution and indicated that the 1:2 stoichiometry between Pdcd4 and eIF4A observed in the crystal is functionally relevant.
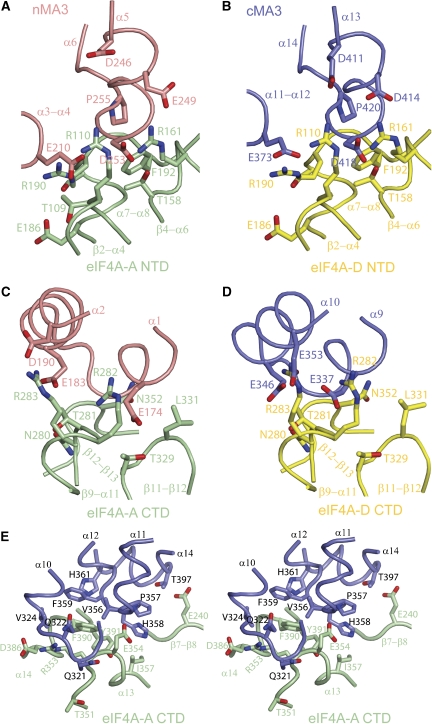
The Pdcd4–eIF4A interface
Except for the cMA3–CTD-A interface, both MA3 domains make strikingly similar contacts with their respective eIF4A partner, with the interfaces of nMA3–NTD-A and nMA3–CTD-A equivalent to cMA3–NTD-D and cMA3–CTD-D, respectively (Figures 3A to D). Specifically, the α3-α4 and α5-α6 loops, and helix α5 in the nMA3 domain, which are equivalent to the α11-α12 and α13-α14 loops, and helix α13 in the cMA3 domains fit into the shallow groove formed by the β2-α4, α7-α8, and β4-α6 loops in the NTD of eIF4A whereas the helices α1 and α2 in the nMA3, which correspond to the helices α9 and α10 in the cMA3 domain binds to the groove formed by the β9-α11, β11-β12, and β12-α13 loops in the CTD of eIF4A (Figures 3A to D). This observation is consistent with the findings that both MA3 domains are structurally similar and bind to eIF4A using similar binding interfaces (Suzuki et al. 2008).
Figure 3.
The Pdcd4–eIF4A interface. Colour codings are as in Figure 1C with the view rotated 45° around x axis relative to the orientation in Figure 1C. Secondary structures and residues involved in the interface are labelled. Superposition of eIF4A-A and eIF4A-D at the NTD showing that nMA3 and cMA3 of Pdcd4 make similar contacts with their respective eIF4A molecules to generate two pairwise equivalent interfaces, nMA3–NTD-A (A) and cMA3–NTD-D (B). Superposition of eIF4A-A and eIF4A-D at the CTD showing that nMA3 and cMA3 of Pdcd4 make similar contacts with their respective eIF4A molecules to generate two more pairwise equivalent interfaces, nMA3–CTD-A (C) and cMA3–CTD-D (D). (E) Stereo view showing the additional interface formed between cMA3 and the CTD of eIF4A-A (cMA3-CTD-A).
The interactions of Pdcd4 to eIF4A in the pairwise equivalent interfaces (nMA3–NTD-A versus cMA3–NTD-D and nMA3–CTD-A versus cMA3–CTD-D) are mediated by conserved residues from both proteins (Supplementary Figures S2 and S3) through primarily a combination of salt bridges and hydrogen bonds. For example, in the nMA3–NTD-A interface, Glu249 and Asp253 in nMA3 form salt bridges with Arg161 and Arg110 in eIF4A-A (Figure 3A). Similarly, Asp414 and Asp418 in cMA3 are salt bridged to Arg161 and Arg110 in eIF4A-D (Figure 3B). In support of the importance of these salt bridge interactions, mutations of Glu249, Asp253, Asp414 and Asp418 on Pdcd4 dramatically reduced its binding to eIF4A, and the mutant proteins failed to efficiently inhibit translation (Yang et al, 2003, 2004).
Further recognition specificities between Pdcd4 and eIF4A are provided by the cMA3–CTD-A interface (Figure 1D). In this interface, the linker region between the MA3 domains of Pdcd4, the α10-α11 loop and part of α13 interact with the surface groove formed by the β12-α13, α14-β14 loops and β13 of the CTD of eIF4A-A through predominantly hydrophobic interactions (Figure 3E). The hydrophobic patch comprising of Val356, Pro357, His358 and Phe359 in Pdcd4 interacts with Ile357, Phe390 and Tyr391 in the CTD of eIF4A-A. The extensive interactions of the conserved residues in the cMA3 domain of Pdcd4 with eIF4A suggest that this interface may have an important function in the formation of the mPdcd4ΔN–eIF4A complex.
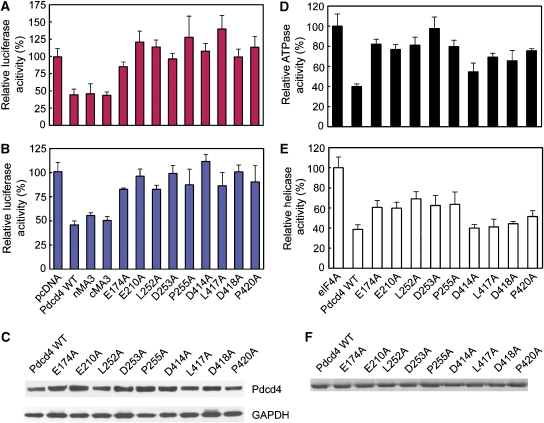
To test the role of the interfaces in the mPdcd4ΔN–eIF4A complex, we selected 10 pairs of residues in Pdcd4, which are conserved between the nMA3 and cMA3 domains and have equivalent interactions with eIF4A molecules, and one additional residue His358 in the cMA3–CTD-A interface (Supplementary Table S2). These include the previously characterized residues Glu249, Asp253, Asp414 and Asp418 (Yang et al, 2003, 2004). We mutated these residues to Ala in the context of hPdcd4FL and examined the effects of these mutations on the interactions of Pdcd4 with eIF4A using isothermal titration calorimetry (ITC). As shown in Supplementary Table S3, wild-type Pdcd4 binds tightly to eIF4A with an equilibrium dissociation constant (Kd) of 110 nM, whereas all of the mutants with the exception of R403A displayed weaker affinities to different extents. Most mutations showed moderately reduced binding affinities (1.5–8.7 fold) with L252A, D253A, H358A, D414A and P420A showing strong defects (13–79 fold). These results indicated that the residues in all five interfaces are required for the binding of Pdcd4 to eIF4A. The observation that single mutations in one MA3 domain failed to disrupt the Pdcd4–eIF4A interaction is consistent with the structural data showing that both domains are required for efficient eIF4A binding.
Mutations that affect mPdcd4DN–eIF4A interaction reduce translation inhibition and AP-1-dependent transcription
To determine whether the Pdcd4 mutants that showed reduced binding to eIF4A lose the ability to inhibit translation and AP-1-dependent transcription in vivo, wild-type Pdcd4 and its mutants E174A, E210A, L252A, D253A, P255A, D414A, L417A, D418A and P420A were overexpressed in JB6 RT101 cells, and luciferase activity was assessed after transient cotransfection with a luciferase reporter construct containing a stem-loop structure in the 5′-untranslated region. Although wild-type Pdcd4 resulted in a 55% inhibition of luciferase mRNA translation, all the mutations lost their abilities to inhibit translation (Figure 4A). An AP-1-dependent transcription assay showed a similar trend of results in which all the mutants tested lost their inhibitory abilities when compared with the wild-type Pdcd4 (Figure 4B). Importantly, the inabilities of these mutants to inhibit translation and AP-1-dependent transcription are correlated strongly with the reduced binding activities to eIF4A (Supplementary Table S3 and Figure S5), strengthening the notion that the binding of Pdcd4 to eIF4A is required for inhibition of translation and AP-1-dependent transcription (Yang et al, 2003, 2004).
Figure 4.
Functional analysis of the Pdcd4–eIF4A interaction. (A) In vivo translational inhibition by wild-type (WT) and mutants Pdcd4 proteins. The luciferase activity in the absence of Pdcd4 is designated as 100%. The mean values and s.d. from three independent experiments are shown. (B) Inhibition of in vivo AP-1 dependent transcription by WT Pdcd4 and its variants measured by the relative luciferase activity. The mean values and s.d. from three independent experiments are shown. (C) Similar expression levels of wild-type and mutant Pdcd4. RT101 cell lysates (10 μg) from transient transfection with wild-type Pdcd4 and Pdcd4 mutant expression plasmids were separated on 4–20% Bis–Tris NuPage gels, transferred to nitrocellulose, and subjected to immunoblotting with Pdcd4 antibody, with visualization by chemiluminescent detection. (D) Defective eIF4A-binding mutants of Pdcd4 are unable to inhibit the ATP hydrolysis activity of eIF4A, depicted as percentage of eIF4A ATPase activity in the absence of Pdcd4 (mean values of three independent experiments±s.d.). (E) Inhibition of eIF4A RNA helicase activity by WT Pdcd4 and its mutants depicted as percentage of eIF4A helicase activity in the absence of Pdcd4. (mean values of three independent experiments±s.d.). (F) Similar protein expression levels of WT and mutant Pdcd4. Purified proteins were separated on 12% SDS–PAGE gels and stained with Coomassie blue.
The binding of Pdcd4 to eIF4A is required to inhibit its enzymatic activities
eIF4A exhibited RNA-dependent ATPase and ATP-dependent RNA helicase activities (Rogers et al, 2002). Previous biochemical data showed that Pdcd4 inhibits translation at least in part by inactivating the helicase activity of eIF4A (Yang et al, 2003). To examine whether the Pdcd4 mutants, which showed correlated defects in eIF4A binding, inhibition of translation and AP-1-dependent transcription, lose the ability to inhibit the ATPase and RNA helicase activities, ATPase and RNA helicase assays of eIF4A were performed in the presence of wild-type and mutant Pdcd4 proteins. As shown in Figure 4D, wild-type Pdcd4 inhibited the RNA-dependent ATPase activity of eIF4A by 60%, whereas all the mutants showed reduced inhibitory effects with D253A restoring ATP hydrolysis to the level comparable to that in the absence of Pdcd4.
A fluorescence-based helicase assay showed wild-type Pdcd4 inhibited the helicase activity of eIF4A by ∼65% whereas all of the Pdcd4 mutants inhibited the helicase activity of eIF4A to a much lesser extent (Figure 4E). In particular, the inhibition was most severely reduced by E174A, E210A, L255A, D253A and P255A, which restored 60% of the helicase activity. These results indicate that the interaction with eIF4A is equally important for Pdcd4 to mediate its effects on the helicase and ATPase activities of eIF4A.
Structural basis for inhibiting the helicase activity of eIF4A by Pdcd4
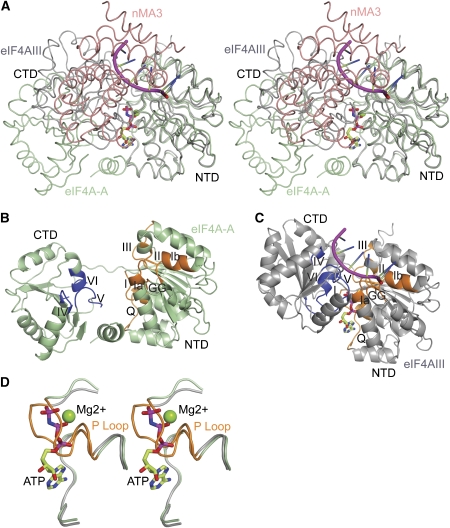
Insights into the structural basis for inhibition of eIF4A by Pdcd4 come from the structural comparison of the eIF4A in our complex structure with eIF4AIII in the exon junction complex (EJC), a close homologue of eIF4AI, which exhibited a closed productive conformation with bound RNA and ATP (Andersen et al, 2006; Bono et al, 2006). Superposition of Pdcd4-bound eIF4A and eIF4AIII in the EJC complex on the NTD of eIF4A shows that the orientations of the CTDs between these two structures differ by 55° (Figure 5A). As a result of this striking conformational difference between eIF4A and eIF4AIII, motifs IV, V, VI in the CTD of the Pdcd4-bound eIF4A face the solvent region rather than facing the NTD (Figures 5B and C), thereby disrupting the RNA-binding site and leaving the ATP-binding site open. Moreover, motif I (Walker A motif or P loop) in the NTD which is involved in binding to the phosphate groups of ATP, moves upwards by ∼5 Å towards the central β-sheet to occupy the position of the phosphate groups of ATP (Figure 5D). Finally, the binding of each MA3 domain of Pdcd4 to both the NTD and CTD of eIF4A has two consequences. First, it sterically clashes with the RNA substrate (Figure 5A), which is consistent with the NMR competition assay showing that Pdcd4 displaced RNA from eIF4A (Suzuki et al. 2008). Second, it blocks the domain closure of eIF4A, thus fixing it in an open nonproductive conformation. Taken together, these results suggest that Pdcd4 inactivates eIF4A by blocking RNA binding directly, disrupting RNA- and ATP-binding sites, and preventing the conformational transition of eIF4A from an open nonproductive state to a closed productive state.
Figure 5.
Structural basis for inhibiting eIF4A by Pdcd4. (A) Stereo view showing the superposition of eIF4AIII (in the EJC; grey) and Pdcd4-bound eIF4A (eIF4A-A; green). RNA is shown in magenta, ATP in ball-and-stick model and nMA3 of Pdcd4 in salmon. The view of eIF4A-A is as in Figure 1C. (B) Ribbon diagram showing the open nonproductive conformation of Pdcd4-bound eIF4A. Motifs Q, I, Ia, Ib, GG, II and III in the NTD are shown in orange whereas motifs IV, V, VI in the CTD are shown in blue. The view is as in (A). (C) Ribbon diagram of the close active conformation of eIF4AIII with bound RNA and ATP in the EJC. The colouring scheme for the motifs is as in (B). and the view is as in (A). (D) A close-up view in stereo showing the conformational difference of P loop in Pdcd4-bound eIF4A and eIF4AIII in EJC.
A single MA3 domain is sufficient to inhibit translation but both MA3 domains are required to compete with eIF4Gc for eIF4A binding
The cMA3 domain alone has been reported to be sufficient to inhibit translation (LaRonde-LeBlanc et al, 2007). Given that nMA3 and cMA3 domains are structurally similar, nMA3 might have the same inhibitory effect on translation as the cMA3. Consistent with this view, our in vivo functional assays showed that nMA3 or cMA3 alone is capable of inhibiting translation and AP-1 transcription with the efficiency comparable to that of the full-length protein (Figures 4A and B), suggesting that both MA3 domains are functionally similar.
Previous data showed that Pdcd4 competes with eIF4Gc for eIF4A binding to inhibit translation (Yang et al, 2003, 2004). Moreover, both the nMA3 domain and cMA3 domain alone have been shown to efficiently compete with eIF4Gc for binding to eIF4A (LaRonde-LeBlanc et al, 2007; Suzuki et al, 2008). However, a surface plasmon resonance (SPR) assay showed that each MA3 domain alone binds to eIF4A at least 8-fold more weakly compared with the MA3-dual that contains both MA3 domains (Suzuki et al, 2008). Deletion mutation studies showed that partial deletion of either the nMA3 or cMA3 domain of Pdcd4 dramatically decreased its binding to eIF4A (Yang et al, 2004). Our ITC data showed that eIF4Gc binds to eIF4A very tightly with a Kd of 60 nM (Figure 6A) whereas a single nMA3 or cMA3 domain of Pdcd4 binds to eIF4A with Kd values of 1.75 μM and 28.25 μM respectively (Supplementary Table S3). Altogether these results indicated that a single MA3 domain may not be sufficient to compete with eIF4Gc for eIF4A binding.
Figure 6.
Both MA3 domains of Pdcd4 are required to compete with eIF4Gc for binding to eIF4A. (A) Measurement of the binding affinity of eIF4Gc to eIF4A using isothermal titration calorimetry (ITC) by titration of eIF4Gc in the syringe into eIF4A in the cell. The upper panel shows the experimental data and the lower panel shows the integrated heat for each injection and fitted to a single-site binding model. See Supplementary Data Table S3 for dissociation constant (Kd) and stoichiometric (N) measurements. (B–F) ITC titrations of Pdcd4 FL WT (B), Pdcd4 nMA3 (C), Pdcd4 cMA3 (D), D253A (E), or P420A (F) in the injection syringe into the eIF4A–eIF4Gc complex in the cell. The data were fitted to a single-site model, where possible.
To test whether both MA3 domains of Pdcd4 are required to efficiently compete with eIF4Gc for eIF4A binding, we analyzed the binding of wild-type and mutant Pdcd4 proteins to the purified eIF4A–eIF4Gc complex by ITC. Wild-type Pdcd4 efficiently competed with eIF4Gc for eIF4A binding with an apparent Kd of 0.19 μM (Figure 6B) whereas neither the nMA3 nor cMA3 alone outcompeted eIF4Gc for binding to eIF4A (Figures 6C and D). Furthermore, mutation D253A in the nMA3 domain, which showed the strongest defect for binding to eIF4A, failed to compete with eIF4Gc whereas P420A in the cMA3 domain, which had a 12-fold reduced eIF4A-binding affinity, competed with eIF4Gc for eIF4A binding albeit less efficiently (Kd of 10 μM) (Figures 6E and F). In contrast, D190A, which had a comparable binding affinity to that of WT Pdcd4, competed efficiently with eIF4Gc for binding to eIF4A (Supplementary Table S3). These results demonstrated that both MA3 domain of Pdcd4 are required to compete with eIF4Gc for eIF4A binding, and a single MA3 domain is not sufficient to prevent eIF4Gc binding to eIF4A in contrary to the previous reports (LaRonde-LeBlanc et al, 2007; Suzuki et al, 2008). The observation that a single MA3 domain failed to compete with eIF4Gc for eIF4A binding seems at odds with the finding that either nMA3 or cMA3 alone is sufficient to inhibit translation (Figure 4A). The simplest explanation for this discrepancy is that a single MA3 domain still retains sufficient affinity for binding to eIF4A, thus inactivating its helicase activity and inhibiting translation.
Pdcd4 and eIF4Gm have distinct binding sites on eIF4A
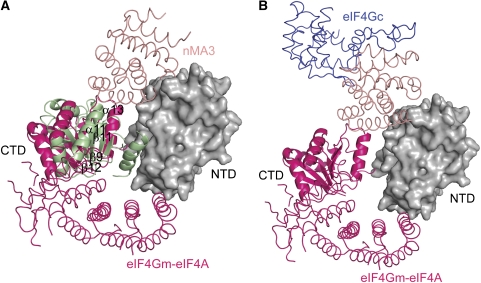
Pdcd4 has been shown to bind to eIF4Gm in the absence or presence of eIF4A (Yang et al. 2003). More recently, the crystal structure of eIF4A in complex with eIF4Gm has been solved (Schutz et al, 2008), therefore allowing us to examine whether Pdcd4 and eIF4Gm can bind simultaneously to eIF4A at the molecular level. Superposition of the mPdcd4ΔN–eIF4A complex and the eIF4Gm–eIF4A complex on the NTD of eIF4A-A showed that Pdcd4 and eIF4Gm occupy diametrically opposite positions on eIF4A (Figure 7A), suggesting that the binding of Pdcd4 and eIF4Gm to eIF4A are not mutually exclusive. Structural superposition also showed that the CTDs in Pdcd4- and eIF4G-bound eIF4A differ in such a way that their central β-sheets lie almost perpendicular to each other (Figure 7A). As a result of such a marked conformational difference, the interaction of the CTD of eIF4A with the MA3 domain of Pdcd4 is weakened due to steric hindrance and the displacement of the regions involved in interactions with Pdcd4 upon eIF4Gm binding. Similarly weakened interactions of the CTD of eIF4A with eIF4Gm would occur upon Pdcd4 binding. These results suggest that Pdcd4 and eIF4Gm could bind to eIF4A simultaneously to form a ternary complex (Supplementary Figure S6), albeit the association of Pdcd4 or eIF4Gm to eIF4A is less tight as compared with their respective binary complexes.
Figure 7.
The putative models of eIF4A-eIF4Gm-MA3 and eIF4A-eIF4Gm-eIF4Gc. (A) Superposition of eIF4A-eIF4Gm (hot pink) and mPdcd4ΔN-eIF4A on the NTD of eIF4A-A (grey surface representation). The CTD of eIF4A and the nMA3 domain of mPdcd4ΔN-eIF4A are coloured in green and salmon respectively. The secondary structures in the regions involved in interactions with nMA3 of Pdcd4 are labelled. The view of eIF4A-A is as in Figure 3A. (B) The putative model of the eIF4A–eIF4Gm–eIF4Gc complex. The model is generated by superposition of eIF4A–eIF4Gm (hot pink) and mPdcd4ΔN–eIF4A on the NTD of eIF4A-A (grey surface representation), and superposition of the MA3 domain (salmon) of eIF4Gc (blue) with the nMA3 domain (not shown for clarity) of Pdcd4 in the mPdcd4ΔN–eIF4A complex. The view of eIF4A-A is as in Figure 3A.
Discussion
Previous reports based on competition experiments suggested that eIF4A interacts with eIF4Gm and eIF4Gc using different binding surfaces (Yang et al, 2003; Korneeva et al, 2005). The high structural similarity between the MA3 domains of eIF4Gc and Pdcd4 allows us to examine the validity of this notion. Superposition of eIF4Gc and eIF4A-bound Pdcd4 on the nMA3 domain suggested that the MA3 domain of eIF4Gc would bind the same site on eIF4A occupied by nMA3 or cMA3 of Pdcd4 (Figure 7B). Given that Pdcd4 and eIF4Gm could bind to the opposite sites of eIF4A, eIF4Gc, and eIF4Gm would bind eIF4A at the same time with eIF4A sandwiched in between (Figure 7B). This view is consistent with the model proposed by Sonenberg and co-workers (Morino et al, 2000) whereby eIF4A has two different binding surfaces for binding to eIF4G, allowing it to be sandwiched between eIF4Gm and eIF4Gc. Similar to the modeled ternary complex of Pdcd4–eIF4Gm–eIF4A wherein Pdcd4 or eIF4Gm binding would weaken the interaction of the other with eIF4A, the binding of eIF4Gm to eIF4A may cause the weakened interaction between eIF4Gc and eIF4A or vice versa. Biochemical data showed that the binding of eIF4Gm to eIF4A increased its helicase activity whereas the binding of eIF4Gc to eIF4A just played a modulatory role (Korneeva et al, 2001, 2005). One possible explanation for the distinct roles of eIF4Gm and eIF4Gc is that the MA3 domain of eIF4Gc first binds to eIF4A to inactivate its helicase activity. Subsequently, the binding of eIF4Gm on the other side of eIF4A changes the relative orientation of the two domains of eIF4A. This change in orientation of eIF4A not only reduces the binding of eIF4Gc to eIF4A but also allows eIF4A to adopt a productive conformation.
Several tumours and tumour cell lines showed elevated levels of translation initiation factors such as eIF4E (De Benedetti and Harris, 1999), eIF4A (Eberle et al, 1997; Shuda et al, 2000) and eIF4G (Bauer et al, 2001) suggesting that these translation factors may function as oncogenic proteins. Therefore, downregulation or inactivation of these translation factors may offer new approaches to develop anticancer drugs that target translation factors including eIF4A. Two natural products pateamine A (PatA) and hippuristanol, and a signalling lipid molecule 15d-PGJ2 have been shown to inhibit translation by regulating the eIF4A activities (Low et al, 2005; Bordeleau et al, 2006; Kim et al, 2007). PatA and 15d-PGJ2 bind to eIF4A to block the interaction between eIF4A and eIF4G whereas hippuristanol specifically binds to the CTD of eIF4A to inhibit its RNA-binding activity. Our structure showed that Pdcd4 inhibits translation in a manner different from that of these compounds by blocking the binding of eIF4Gc to eIF4A and by trapping eIF4A in an inactive conformation. Given that a single MA3 domain of Pdcd4 makes bipartite contacts with both the NTD and CTD of eIF4A to block the domain closure of eIF4A, one strategy for inhibiting eIF4A is to design a peptide molecule that interacts with eIF4A across its domains. This study would pave a way for further structural, chemical, and functional approaches that are required for the development of anticancer agents targeting eIF4A.
Materials and methods
Structure determination of hPdcd4DN
Details of cloning, expression, and purification are described in Supplementary Data. Crystals of SeMet-hPdcd4ΔN were grown at 15°C by hanging-drop vapour diffusion. Equal volumes of protein and crystallization reagent (20% (w/v) PEG MME 2000, 0.1 M Tris–HCl pH 8.5, 0.2 M Trimethylamine N-oxide (TMAO), 10 mM DTT, 10 mM Sarcosine) were mixed. Prior to data collection, the crystals were cryoprotected using mother liquor containing 15% (v/v) ethylene glycol and flash-frozen in liquid nitrogen. Diffraction data were collected at the peak of the selenium K edge (λ=0.9798 Å) on the UK CRG beamline BM14 (ESRF, Grenoble, France) and processed using the CCP4 suite (CCP4, 1994). Here, 17 selenium sites were located using the program SnB (Miller et al, 1994), followed by refinement of the heavy atom sites and phasing with SHARP (De la Fortelle and Bricogne, 1997). Subsequent model building was carried out using the program O (Jones et al, 1991) and the fitted model was refined with CNS (Brunger et al, 1998) and REFMAC5 (Murshudov et al, 1997). The final refinement statistics for hPdcd4ΔN are summarized in Supplementary Table S1.
Structure determination of the mPdcd4DN–eIF4A complex
The crystals of mPdcd4ΔN–eIF4A were obtained at 15°C by hanging-drop vapour diffusion. An equal volume of protein was mixed with crystallization reagent containing 20% (w/v) PEG 3350, 0.1 M Bis-Tris Propane pH 6.5, 0.2 mM Sodium citrate, 0.1 M Taurine. The crystals were transferred to a cryoprotectant buffer containing mother liquor and 20% (v/v) glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected on beamline ID14-4 (ESRF, Grenoble, France) and processed using the CCP4 suite (CCP4, 1994). The structure of the mPdcd4ΔN–eIF4A complex was solved by molecular replacement using PHASER (McCoy et al, 2007). Two MA3 domains of hPdcd4ΔN, and the ATPase domain and C-terminal domain of S. cerevisiae eIF4AI (Benz et al, 1999; Caruthers et al, 2000) were taken as search models. Refinement was performed with CNS (Brunger et al, 1998) and REFMAC5 (Murshudov et al, 1997) and model building was carried out using COOT (Emsley and Cowtan, 2004). The final refinement statistics for the mPdcd4ΔN–eIF4A complex are summarized in Supplementary Table S1.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed at 42 000 r.p.m. and 20°C using a ProteomeLab XL-A analytical ultracentrifuge (Beckman Coulter) in 2-channel centrepieces. Radial scans were collected at 4 min intervals at 280 nm. Prior to centrifugation, proteins were dialyzed against 100 mM NaCl, 20 mM Tris–HCl pH 7.6 and 10 μM Tris[2-carboxyethyphosphine] hydrochloride (TCEP). The solvent density, viscosity, and protein partial specific volumes were calculated using the SEDNTERP program (Sednterp version 1.09, http://www.rasmb.bbri.org). With the program SEDFIT (Schuck, 2000), the data were directly fitted to the boundaries using numerical solutions to the Lamm equation to obtain the sedimentation coefficient distribution c(S) and molar mass distribution c(M).
Sedimentation equilibrium experiments were performed at 20°C in six-channel centrepieces using a ProteomeLab XL-A (Beckman Coulter). Radial scans were taken at 230, 250, and 280 nm. Prior to the run, protein samples were dialyzed into the same buffer as for the velocity run. Samples were centrifuged for 22 h at each speed of 4000, 6000, and 8000 r.p.m. and a further 2 h for data acquisition. After the samples reached equilibrium and no further change could be seen in the distribution, a final 10 h scan at 42 000 r.p.m. was taken to measure the residual absorbance for initial offset values. The data were analyzed using the HETEROANALYSIS program (http://www.biotech.uconn.edu/auf/) and the absorbance gradient was fitted to an ideal single species.
Isothermal titration calorimetry
ITC measurements were performed at 20°C in a VP-ITC microcalorimeter (MicroCal Inc.). Protein samples were dialyzed into a buffer containing 100 mM NaCl, and 20 mM Tris–HCl pH 7.6. For analyzing the binding of Pdcd4 to eIF4A, 100–180 μM wild-type (WT) or mutant Pdcd4 proteins were injected into the calorimetric cell containing eIF4A at 20–30 μM. For the competition-binding assay, the cell was loaded with 20–30 μM of the purified eIF4A–eIF4Gc complex and the injection syringe contained 120–180 μM of hPdcd4 WT or mutants or the nMA3/cMA3 domain at a concentration of 400 μM. Titrations were initiated with one 2 μl injection, followed by twenty-eight 10 μl injections, with 240 s equilibration time between injections. The heat of dilution was measured by additional injections of syringe sample after saturation and subtracted to obtain the effective heat of binding, which was plotted versus the molar ratio of injectant/cell. Data were analyzed using the Origin 7.0 program and fitted to a single-site binding model to obtain the parameters Kd, ΔH, and stoichiometry (N).
ATPase assays
ATPase assays were performed using an enzyme-linked assay that couples the hydrolysis of ATP to the oxidation of NADH using pyruvate kinase and lactate dehydrogenase, resulting in a decrease in NADH absorbance at 340 nm, which is proportional to the rate of steady-state ATP hydrolysis. The raw absorbance was converted into the rate of steady-state ATP hydrolysis (nmol s−1) using the extinction coefficient for NADH of 6300 M−1 cm−1. Reactions (200 μl) containing 50 mM NaCl, 50 mM Tris–HCl pH 7.5, 5 mM DTT, 5 mM MgCl2, 2 mM ATP, 1 mM phosphoenolpyruvate (PEP), 0.4 mM NADH, 1% (v/v) PK/LDH (Sigma), 200 mg ml−1 total yeast RNA (Sigma), and 500 nM eIF4A were assayed at 25°C in a 96-well plate using a Benchmark Plus spectrophotometer (Bio-Rad). For analysis of WT and mutant hPdcd4, 5 μM of heIF4AI and 2.5 μM of hPdcd4 were used. The rates were calculated by linear regression analysis using the GraphPad Prism 4.0 program (GraphPad Software Inc.).
Helicase assays
The substrate used in the fluorescence-based helicase assay was prepared by annealing at a 1:1.2 molar ratio, a 3′ Cy3-labelled 36-mer (GGGGAGA(A4C)3UAGCACCGUAAAGC) (metaBIOn International AG) to a 5′ BHQ-2-labelled 12-mer (GCUUUACGGUGC) (Biosearch Technologies), both RP-HPLC-purified, in 20 mM Tris–HCl pH 7.5, by heating to 90°C for 2 min and cooling slowly to room temperature. Helicase assays (200 μl) containing 1 μM heIF4AI and 1 μM heIF4B were performed in 30 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.075% Triton X-100, 0.05% sodium azide, 25 nM substrate, 312.5 nM unlabelled capture strand (GCACCGUAAAGC) (metaBIOn International AG) and 2.5 mM ATP. Where indicated, WT or mutant Pdcd4 proteins were included at 5 μM. Unwinding was initiated by the addition of ATP and was carried out at 37°C for 90 min. Fluorescence measurements were carried out in a 96-well plate on a Tecan SPECTRAFluor Plus at the excitation and emission wavelengths of 540 and 595 nm respectively, with readings taken every 30 s. Initial rates of unwinding were calculated from the linear region of the assay using the GraphPad Prism 4.0 program (GraphPad Software Inc.).
In vivo translation and AP-1 transcription assays
RT101 cells (1 × 104 cells) were seeded in 24-well plates in EMEM medium containing 4% FBS. For AP-1-dependent transcription assay, cells were transfected with 1 μg of Pdcd4 (or mutant) expressing plasmid, 0.2 μg of AP-1 luciferase reporter plasmid (4 × AP-1-LUC), and 10 ng of Renilla reporter plasmid (pRL-SV40, Promega) using 3.6 μl of TransIT-LT1 reagent (Mirus). After transfection, cells were incubated for 48 h. For translation assay, cells were transfected with 0.5 μg of Pdcd4 (or mutant) expressing plasmid, 0.1 μg of pCMV-SL-LUC (Yang et al, 2004), and 10 ng of pRL-SV40 using 1.8 μl of TransIT-LT1 reagent (Mirus). After 24 h, cells were then serum starved with 0.1% FBS in EMEM medium for 24 h, followed by incubation in EMEM medium containing 4% FBS for additional 24 h. Cells were lysed in 1 × Passive lysis buffer (Promega) and luciferase activity was measured as previously described (Wang et al, 2008).
Accession numbers
The coordinates and structure-factor amplitudes of hPdcd4ΔN and the mPdcd4ΔN–eIF4A complex have been deposited in the Protein Data Bank with accession codes 3EIJ and 3EIQ, respectively.
Supplementary Material
Supplementary Information
Acknowledgments
We thank N Sonenberg for the gift of eIF4B expression plasmid and A McCarthy for access to ID14-4 (ESRF, France). BM14 is funded by the UK Research Councils and the EMBL. This study was financially supported by the Biomedical Research Council of A*STAR (Agency for Science, Technology and Research) (HS). This study was partially supported by CORBE pilot grant at University of Kentucky funded by P20 RR020171 from the National Center for Research Resources to H-SY.
References
- Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH (2004) Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 23: 8135–8145 [DOI] [PubMed] [Google Scholar]
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR (2006) Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27: 2128–2136 [DOI] [PubMed] [Google Scholar]
- Bauer C, Diesinger I, Brass N, Steinhart H, Iro H, Meese EU (2001) Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer 92: 822–829 [DOI] [PubMed] [Google Scholar]
- Benz J, Trachsel H, Baumann U (1999) Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae--the prototype of the DEAD box protein family. Structure Fold Des 7: 671–679 [DOI] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J (2006) Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci USA 97: 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chen Y, Knosel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, Ozaki I, Petersen I (2003) Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol 200: 640–646 [DOI] [PubMed] [Google Scholar]
- De Benedetti A, Harris AL (1999) eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol 31: 59–72 [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Preiss T, Hentze MW (1998) Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA 4: 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Method. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M (2006) S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314: 467–471 [DOI] [PubMed] [Google Scholar]
- Eberle J, Krasagakis K, Orfanos CE (1997) Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro. Int J Cancer 71: 396–401 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283: 1026–1033 [DOI] [PubMed] [Google Scholar]
- Goke A, Goke R, Knolle A, Trusheim H, Schmidt H, Wilmen A, Carmody R, Goke B, Chen YH (2002) DUG is a novel homologue of translation initiation factor 4G that binds eIF4A. Biochem Biophys Res Commun 297: 78–82 [DOI] [PubMed] [Google Scholar]
- Holland EC, Sonenberg N, Pandolfi PP, Thomas G (2004) Signaling control of mRNA translation in cancer pathogenesis. Oncogene 23: 3138–3144 [DOI] [PubMed] [Google Scholar]
- Jansen AP, Camalier CE, Colburn NH (2005) Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 65: 6034–6041 [DOI] [PubMed] [Google Scholar]
- Jansen AP, Camalier CE, Stark C, Colburn NH (2004) Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol Cancer Ther 3: 103–110 [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kim WJ, Kim JH, Jang SK (2007) Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J 26: 5020–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva NL, First EA, Benoit CA, Rhoads RE (2005) Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J Biol Chem 280: 1872–1881 [DOI] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Merrick WC, Rhoads RE (2001) Characterization of the two eIF4A-binding sites on human eIF4G-1. J Biol Chem 276: 2872–2879 [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Santhanam AN, Baker AR, Wlodawer A, Colburn NH (2007) Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol Cell Biol 27: 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO (2005) Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722 [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y (2008) MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27: 4373–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J App Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Gallo SM, Khalak HG, Weeks CM (1994) SnB: crystal structure determination via shake-and-bake. J Appl Cryst 27: 613–621 [Google Scholar]
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 20: 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Cryst D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y (2005) Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res 65: 11282–11286 [DOI] [PubMed] [Google Scholar]
- Rogers GW Jr, Komar AA, Merrick WC (2002) eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72: 307–331 [DOI] [PubMed] [Google Scholar]
- Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH (2008) Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res 68: 1254–1260 [DOI] [PubMed] [Google Scholar]
- Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U (2008) Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA 105: 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Asano M, Ishida Y, Aoki T, Koike T, Honjo T (1995) Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene 166: 297–301 [DOI] [PubMed] [Google Scholar]
- Shuda M, Kondoh N, Tanaka K, Ryo A, Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, Horiuchi S, Shichita M, Yamamoto N, Yamamoto M (2000) Enhanced expression of translation factor mRNAs in hepatocellular carcinoma. Anticancer Res 20: 2489–2494 [PubMed] [Google Scholar]
- Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, Wagner G (2008) PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci USA 105: 3274–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun Z, Yang HS (2008) Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene 27: 1527–1535 [DOI] [PubMed] [Google Scholar]
- Waters LC, Veverka V, Bohm M, Schmedt T, Choong PT, Muskett FW, Klempnauer KH, Carr MD (2007) Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A. Oncogene 26: 4941–4950 [DOI] [PubMed] [Google Scholar]
- Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH (2004) A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 24: 3894–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23: 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH (2001) A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene 20: 669–676 [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18: 350–359 [DOI] [PubMed] [Google Scholar]
- Zimmer SG, DeBenedetti A, Graff JR (2000) Translational control of malignancy: the mRNA cap-binding protein, eIF-4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res 20: 1343–1351 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information