Abstract
The Tetrahymena thermophila origin recognition complex (ORC) contains an integral RNA subunit, 26T RNA, which confers specificity to the amplified ribosomal DNA (rDNA) origin by base pairing with an essential cis-acting replication determinant—the type I element. Using a plasmid maintenance assay, we identified a 6.7 kb non-rDNA fragment containing two closely associated replicators, ARS1-A (0.8 kb) and ARS1-B (1.2 kb). Both replicators lack type I elements and hence complementarity to 26T RNA, suggesting that ORC is recruited to these sites by an RNA-independent mechanism. Consistent with this prediction, although ORC associated exclusively with origin sequences in the 21 kb rDNA minichromosome, the interaction between ORC and the non-rDNA ARS1 chromosome changed across the cell cycle. In G2 phase, ORC bound to all tested sequences in a 60 kb interval spanning ARS1-A/B. Remarkably, ORC and Mcm6 associated with just the ARS1-A replicator in G1 phase when pre-replicative complexes assemble. We propose that ORC is stochastically deposited onto newly replicated non-rDNA chromosomes and subsequently targeted to preferred initiation sites prior to the next S phase.
Keywords: cell cycle, origin recognition complex, replicator, ribonucleoprotein complex, Tetrahymena
Introduction
The conserved Origin Recognition Complex (ORC) determines the sites for replication initiation in eukaryotic chromosomes and serves as a scaffold for pre-replicative complex (pre-RC) assembly. Although ORC subunits are conserved in eukaryotes, the cis-acting DNA sequence requirements for replicator function are not. Saccharomyces cerevisiae ORC binds in a sequence-specific manner to a short motif present at all origins. ORC binding to autonomously replicating sequence (ARS) elements is required for origin activation, whereas other protein–DNA interactions serve lesser, auxiliary roles (Marahrens and Stillman, 1992; Bolon and Bielinsky, 2006). In contrast, Schizosaccharomyces pombe, Drosophila melanogaster and human ORC bind non-specifically to AT-rich DNA sequences (Kim and Huberman, 1998; Vashee et al, 2003; Remus et al, 2004). S. pombe replicators consist of blocks of degenerate sequence that create multiple ORC-binding sites (Segurado et al, 2003; Dai et al, 2005).
In Drosophila and the rat, ORC is tethered to the respective chorion gene (DAFC-66D) and aldolase origins by associating with unrelated sequence-specific DNA-binding proteins (Beall et al, 2002; Minami et al, 2006). The interaction of human ORC with HMGA1a similarly creates functional origins in heterochromatic regions (Thomae et al, 2008). Whether tethering is commonly used to recruit ORC to metazoan origins is unclear. To add to the complexity, metazoan replicators vary in size and local density of initiation sites. Although the 1.2 kb human lamin B2 replicator initiates at a single discrete site (Abdurashidova et al, 1998), many origins fire within the 20 kb segment downstream of the hamster DHFR gene (Hamlin and Dijkwel, 1995).
Tetrahymena thermophila (Tt) ORC is unusual in that it contains an integral RNA subunit that uses Watson-Crick base pairing to bind to its cognate DNA target in the ribosomal DNA (rDNA) replication origin (Mohammad et al, 2007). This DNA sequence, the type I element, is required for developmentally programmed amplification and cell cycle-controlled vegetative replication of rDNA minichromosomes (Figure 1A) (reviewed in Tower, 2004). Remarkably, the ORC RNA subunit, 26T RNA, corresponds to the terminal 282 nucleotides (nt) of 26S rRNA (Mohammad et al, 2007). Mutations that perturb RNA pairing with the type I element T-rich strand disrupt rDNA origin recognition and activation. Type I elements are recognized by additional single-stranded binding factors, including TIF1p, which binds to the A-rich strand at the origin and controls the timing of rDNA origin activation (Saha et al, 2001; Morrison et al, 2005).
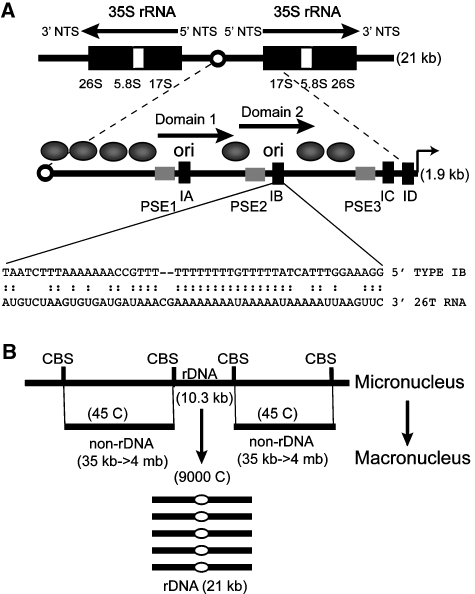
Figure 1.
Organization of rDNA and non-rDNA macronuclear chromosomes. (A) Schematic of the palindromic rDNA minichromosome. Expanded view of the 5′ NTS includes positioned nucleosomes (ovals) type I elements (black rectangles), pause site elements (grey rectangles), rRNA promoter (thin arrow) and replication origins (ori) which reside in the 430 bp imperfect duplicated sequences, domains 1 and 2 (thick arrows). The sequence of the type IB element T-rich strand and flanking DNA are shown, including predicted base pair interactions with 26T RNA. (B) Chromosome reorganization and replication during macronuclear development. A portion of the micronuclear chromosome encoding the single copy rRNA gene is shown (CBS: chromosome-breakage sequences). See text for additional details.
rDNA and non-rDNA chromosomes are differentially replicated during Tetrahymena development. This property stems from the partitioning of chromosome functions into two distinct nuclei within each cell: the ‘germline' micronucleus and the ‘somatic' macronucleus (reviewed in Karrer, 2000). The non-transcribed micronucleus contains the chromosomes that are transmitted to progeny during conjugation. As such, it undergoes conventional mitosis and meiosis. The transcribed, amitotic macronucleus confers the phenotype of the cell. During conjugation pronuclei are exchanged and fuse to generate a new diploid micronucleus. Following two rounds of DNA replication and nuclear division, two of the four micronuclei differentiate into macronuclei. At this time, the five monocentric chromosomes are fragmented at chromosome-breakage sequence (CBS) elements and further rearranged, generating ∼280 macronuclear chromosomes (size range: 21–>4000 kb) (Figure 1B). Non-rDNA chromosomes re-replicate to a final copy number of ∼45 C, whereas the single copy rDNA locus is rearranged into a 21 kb minichromosome and amplified ∼5000-fold. Once development is complete, micro- and macronuclear chromosomes replicate once per vegetative cell division. Despite the absence of centromeres, macronuclear chromosomes are maintained at a relatively constant copy number.
Here, we describe a plasmid shuttle assay that was used to isolate the first non-rDNA Tetarhymena replicator, and show that it is comprised of discrete cis-acting determinants. We provide the first evidence for cell cycle-regulated changes in the specificity of ORC for chromosomal DNA in any eukaryote, and show that these changes do not occur in the rDNA minichromosome. We propose a model for ORC binding to non-rDNA chromosomes, in which ORC associates non-specifically with newly replicated daughter chromosomes and then re-localizes to preferred initiation sites prior to the next S phase.
Results
Isolation of non-rDNA Tetrahymena replicators
Previous transformation studies showed that the 1.9 kb rDNA 5′ NTS supports autonomous DNA replication in Tetrahymena (Pan et al, 1995; Reischmann et al, 1999). Here, we developed a shuttle assay to isolate functional replicators that are not amplified (Figure 2A). To assure that plasmids replicated in Tetrahymena, DNA was isolated from Tetrahymena transformants and digested with the methylation-sensitive restriction endonuclease, DpnI, prior to re-transforming E. coli. Although plasmid DNA prepared from dam+ E coli is sensitive to DpnI, inefficient DNA methylation in Tetrahymena renders replicated molecules resistant to cleavage.
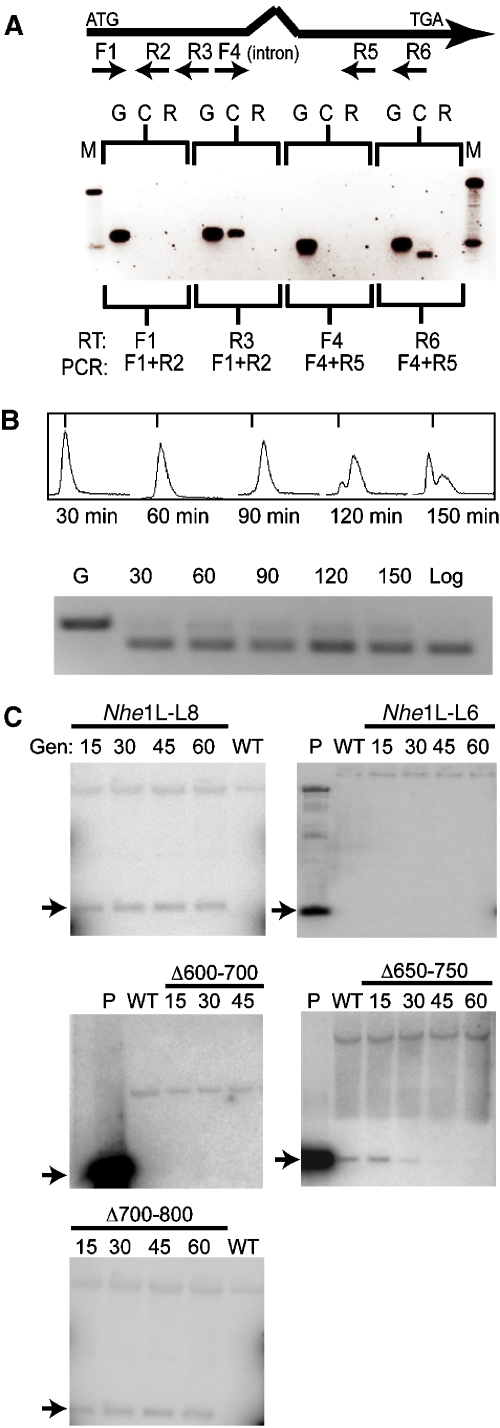
Figure 2.
Isolation of non-rDNA replicators. (A) Schematic of the plasmid shuttle assay used to isolate Tetrahymena ARS1. DpnI-sensitive (DpnIs) plasmids containing T. thermophila genomic DNA were co-transformed into Tetrahymena. Plasmid DNA from Tetrahymena transformants was re-introduced into E. coli. ARS-containing plasmids should be DpnI-resistant (DpnIr) following replication in Tetrahymena, and can be recovered by re-transforming E. coli. (B) Ethidium bromide negative stain image of BamHI-digested plasmid DNA from two clones obtained from the plasmid shuttle assay (Vector: plasmid backbone; Insert: Tetrahymena sequences). (C) Southern blot of undigested Tetrahymena genomic DNA from ‘en masse' transformations using intact ARS1 or a derivative containing just the internal NheI fragment (see Figure 3A for details on the NheI2 derivative). DNA was isolated from pmr co-transformants propagated for 25 generations. ARS1input plasmid DNA (+) was used as a Southern blotting control. Probe: radiolabelled pCC1FOS (no insert). (D) Southern blot of BamHI-digested DNA from wild type Tetrahymena (WT, CU428) and ‘en masse' ARS1 co-transformants propagated for up to 60 generations (gen). Probe: radiolabelled ARS1 insert.
An E. coli plasmid library was created from T. thermophila DNA fragments in the 5–7 kb size range. Pools of 10 colonies were screened by PCR to eliminate plasmids that contained the rDNA origin. DNA from 100 rDNA-negative colonies was co-transformed into the developing macronucleus with plasmid AN101, which rearranges into a linear rDNA minichromosome and confers paromomycin resistance. Circular plasmid DNA was isolated from co-transformants grown en masse for ∼25 generations, digested with DpnI, and electroporated into E. coli. Twelve kanamycin-resistant colonies were obtained. Two plasmids contained inserts in the expected size range (Figure 2B), whereas the remaining plasmids had small inserts and/or lacked adjoining vector sequences (data not shown). This was not unexpected, as AT-rich Tetrahymena sequences frequently rearrange in E. coli. The DNA sequences of the two large insert clones were identical, corresponding to a 6674 bp segment within a 407 kb macronuclear chromosome (locus identifier: CH445556.2, DNA sequence identifier: AAGF01003027; http://www.ciliate.org).
This plasmid, designated ARS1, was reintroduced into Tetrahymena and autonomous replication was assessed by PCR or Southern blotting. In addition to en masse propagation of co-transformants, clonal lines were grown for 60 generations and assayed for retention of plasmid sequences. Approximately half (9/16) of the pmr clones stably propagated the vector backbone (PCR assay, data not shown), and Southern blot analysis of undigested DNA confirmed that the DNA was extrachromosomal (Figure 2C; ARS1). Further analysis of restriction digested DNA with an ARS1-specific probe revealed that plasmid copy number was relatively constant throughout the duration of the experiment (60 generations), approximating that of the endogenous (45 C) macronuclear chromosome (Figure 2D).
Similar to the initial library screen, plasmid DNA from ARS1 Tetrahymena transformants was reintroduced into E. coli. A mixture of full-length clones and deletion derivatives was obtained. As a control, the empty pCC1FOS vector and AN101 were co-transformed. Southern blot analysis failed to detect vector sequences in pmr progeny and kanamycin-resistant E. coli clones were not recovered. We conclude that ARS1 confers autonomous DNA replication in Tetrahymena.
ARS1 contains two autonomous replicators
Eukaryotic replication origins generally reside in intergenic (IG), AT-rich DNA segments and rarely encroach into genes (Paixão et al, 2004; Wang et al, 2004). Computer annotation predicts that ∼80% of the cloned ARS1 interval encodes two proteins of unknown function (Figure 3A). Less than half of the 600 bp intergenic segment, IG-1, resides in the clone, whereas the entire 1.2 kb IG-2 segment was present. To assess the functional organization of ARS1, restriction enzymes were used to generate deletion derivatives or subclone internal fragments into the smaller pSMART vector (Figure 3A). Plasmids were co-transformed into Tetrahymena. En masse cultured co-transformants were propagated and assayed for the presence of plasmid DNA by Southern blotting.
Figure 3.
ARS1 deletion mapping. (A) Upper schematic: predicted genes (arrows) and intergenic (IG-1, IG-2) segments in the chromosomal ARS1 interval. The end points of deleted (Δ) or retained sequences in ARS1 plasmid derivatives are indicated below. Asterisks denote fragments that were subcloned into pSMART. The remaining inserts are in the original pCC1FOS vector backbone. (B) Southern blot analysis of ARS1 transformants and five of the seven depicted deletion derivatives. DNA was prepared from ‘en masse' co-transformants at defined intervals (generations: gen). BamHI-digested samples were probed with ARS1 fragments containing just those sequences present in the plasmid under examination. C: endogenous chromosomal DNA BamHI fragment; P: plasmid-derived BamHI fragment.
Deletion of the internal NsiI or NheI fragments had no effect on plasmid DNA replication or long-term maintenance (Figure 3A and B; ΔNsiI; ΔNheI). To our surprise, plasmids containing either the left (NheIL, 1–966) or right (NheIR, 3798–6774) ends of ARS1 were stably propagated in Tetrahymena. Conversely, internal fragments subcloned into the pSMART vector backbone failed to support autonomous DNA replication (Figure 2C, NheI2; Figure 3B, NsiI2), suggesting that replicator function is sequence-dependent. A pSMART derivative containing just the IG-2 segment (NheIR2) was stably maintained, indicating that this vector backbone does not inhibit DNA replication in Tetrahymena. We conclude that ARS1 contains two autonomous replicators rather than one, hereafter designated ARS1-A and ARS1-B (Supplementary Figure S1). An analogous situation has been documented in the human β-globin locus (Wang et al, 2004).
Deletion mapping of ARS1-A
Approximately 70% ARS1-A (Figure 3A, nt 300–966) is predicted to encode protein (Tetrahymena Genome Database gene prediction Therm 00579140). Reverse transcription (RT)–PCR verified that this segment is actively transcribed. Two DNA intervals were examined, one of which spans a predicted 40 nt intron (Figure 4A). PCR products of the expected size were detected with cDNA (C) and genomic DNA (G) prepared from a log phase vegetative culture, whereas no product was generated from RNA (R) that was not reverse transcribed. The steady state level of this transcript was relatively constant across the cell cycle (Figure 4B), suggesting that transcription factors are constitutively bound to cis-acting regulatory sequences.
Figure 4.
Deletion mapping of the ARS1-A replicator. (A) RT–PCR analysis of the ARS1-A interval. The diagram shows the relative positions of forward (F1, F4) and reverse complementary (R2–R6) primers, intron 1, and start (ATG) and stop (TGA) codons in the predicted gene. PCRs were performed on genomic DNA (G), reverse transcribed RNA (cDNA, C) and total RNA (R). A negative image of the ethidium bromide-stained gel is shown. (B) Lower panel: RT–PCR analysis of RNA from cells synchronized by centrifugal elutriation (RT primer R6, PCR primers F4 and R5). Log: RNA from an asynchronous cell culture. Upper panel: flow cytometry profile of elutriated cells at defined culturing intervals (min). (C) Southern blot analysis of NheIL-L8 and NheIL-L6 transformants, and three of the six tested NheIL-L8 deletion derivatives (see Table I). DNA was prepared from pmr ‘en masse' ARS1 co-transformants at defined intervals (generations: gen). BamHI-digested DNA was probed with ARS1 fragments containing just those sequences present in the plasmid under examination. Arrow: plasmid-derived ARS1A fragment.
To assess whether cis-acting replication determinants reside in protein-coding sequence and examine the overall organization of the ARS1-A replicator, thirteen terminal or internal deletion derivatives were tested for the ability to support autonomous DNA replication (Table I). Deleting the last 166 bp (Nhe1L-L8; nt 1–800) had no effect on plasmid propagation; however, removal of an additional 200 bp led to the failure to support autonomous replication (Nhe1L-L6; nt 1–600) (Figure 4C). The internal deletion mutant Nhe1L Δ600–800 also failed to replicate (data not shown), indicating that the protein-coding interval contains at least one functional DNA replication determinant. Internal deletion derivatives, NheIL Δ200–400 and NheIL Δ400–600, were competent for plasmid DNA replication and long-term plasmid maintenance; however, the Nhe1L-R8 derivative (Δ1–200) was not (data not shown). We conclude that the right and left portions of ARS1-A contain essential replication determinants. To test whether essential intergenic (nt 1–200) and coding region (nt 600–800) sequences were interchangeable, a second copy of the first 200 bp was introduced into NheI-L6. The resulting plasmid, NheIL-L6/+1–200, failed to replicate (data not shown), indicating that the respective sequences serve distinct functions.
Table 1.
Deletion mapping of the ARS1-A replicator
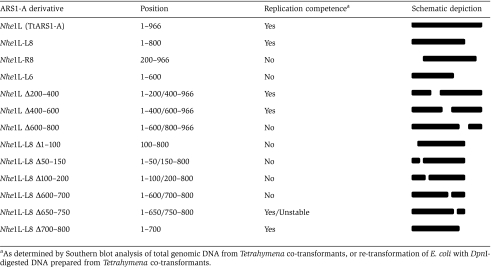 |
Overlapping 100 bp deletion derivatives of NheIL-L8 were examined to further localize replication determinants (Table I, last six constructs). All three deletions in the intergenic interval (NheIL-L8 Δ1–100, NheIL-L8 Δ50–150, NheIL-L8 Δ100–200) failed to support autonomous replication (data not shown). However, the three deletions in the protein-coding segment (NheIL-L8 Δ600–700, NheIL-L8 Δ650–750, NheIL-L8 Δ700–800) behaved differently (Figure 4C). NheIL-L8 Δ700–800 was maintained at a constant level, whereas NheIL-L8 Δ650–750 initially replicated, but was lost during vegetative propagation. NheIL-L8 Δ600–700 failed to support autonomous DNA replication, indicating that it contains an essential cis-acting replication determinant. We conclude that ARS1-A contains dispersed essential and non-essential replication determinants.
Bioinformatic analysis of ARS1-A and ARS1-B replicators
A distinguishing feature of the rDNA replicator is the presence of two reiterated regulatory sequences that participate in origin activation—the type I and PSE element (Figure 1A) (Gallagher and Blackburn, 1998; Reischmann et al, 1999; Saha et al, 2001; TL Morrison and GM Kapler, unpublished results). Type I elements form Watson–Crick base pair interactions with the ORC RNA subunit (26T RNA) which targets ORC to this origin (Mohammad et al, 2007). Biochemical data indicate that 26T RNA is an integral component of all ORC complexes. Significant base pairing potential between 26T RNA and DNA sequences in the ARS1-A or ARS1-B intervals was not detected.
rDNA origin- and promoter-proximal type I and PSE elements function as in vivo binding sites for the non-ORC protein, TIF1p (Saha et al, 2001). As both sequences are absent from ARS1-A and ARS1-B, we predicted that TIF1p would not associate with these DNA segments. Chromatin immunoprecipitation (ChIP) analysis with a TIF1 peptide antibody supported this prediction (Supplementary Figure S2). These data and results from experiments described below suggest that 26T RNA does not target ORC to the ARS1-A and ARS1-B origins.
To look for shared motifs in ARS1-A and ARS1-B, or similarity with other sequences in the rDNA replication origin, the three replicators were subjected to pairwise analysis using BLAST, CLUSTALW and LALIGN. Short dispersed blocks of sequence identity were detected in each analysis, consisting of AT-rich sequences 7–10 bp in length (Supplementary Figure S3). No DNA motifs were present in all three replicators. Pairwise analysis of ARS1-A or ARS1-B with two arbitrarily chosen intergenic sequences produced matches of similar length. Thus, compelling evidence for sequence conservation was lacking.
ORC targeting to rDNA and non-rDNA macronuclear chromosomes
The ability of ARS1-A and ARS1-B to support episomal DNA replication suggests that ORC is recruited to these sites in endogenous macronuclear chromosomes. To address this prediction, chromatin ChIP was performed with epitope-tagged Orc1p across a 60 kb segment spanning the endogenous ARS1 chromosomal locus (Figure 5B, schematic). Control reactions on rDNA origin (O), promoter (P) and coding (C) sequences produced the expected results: Orc1p bound the rDNA origin and did not associate with other segments in the rDNA minichromosome (Figure 5A). Identical results were obtained for chromatin from an asynchronous logarithmic vegetative culture or G0/G1 cells synchronized by starvation.
Figure 5.
ORC targeting to rDNA and non-rDNA replicators. (A) Orc1p ChIP analysis of the rDNA domain 1 origin (O), promoter (P) and coding (C) regions. ChIP was performed on T. thermophila strain TD102 using an antibody directed against the protein A IgG-binding epitope-tag in Orc1p. I: total input DNA; (−): no antibody ChIP control; (+): Orc1p ChIP pellet. (B) Orc1p ChIP analysis of the 60 kb segment spanning ARS1 in the endogenous macronuclear chromosome (strain TD102). PCR products derived from primer sets A1 to A12 are spaced at ∼5 kb intervals. (C, D) Streptavidin (SA) chromatin pull-down analysis of rDNA and ARS1 chromosome intervals. Strain MM201 produces a 26T RNA variant that bears a sequence tag extension (Ext), whereas MM202 expresses an aptamer-tagged (Apt) 26T RNA derivative that binds to streptavidin. I: total input DNA; (−) uncoupled sepharose chromatin pull-down pellet, (+): SA-sepharose chromatin pull-down pellet. (E) ChIP analysis of the ARS1 interval with Tetrahymena Mcm6p antibodies.
ChIP analysis of the ARS1-containing chromosome produced very different results (Figure 5B). All examined intervals were enriched in the Orc1p immunoprecipitate for chromatin prepared from the asynchronous vegetative culture. The tested segments consisted of ARS1-A, ARS1-B and seven randomly chosen coding and non-coding sequences spanning 60 kb. In contrast, only the ARS1-A A6 segment was enriched in starvation-induced G0/G1 chromatin immunoprecipitates (Figure 5B).
To verify that we were studying in vivo binding of the ORC holocomplex and not just Orc1p, chromatin pull-down assays were performed with 26T RNA derivatives that contained a 5′ sequence extension (26T-Ext) or an aptamer sequence tag (26T-Apt), the later of which confers binding to streptavidin (SA) (Srisawat and Engelke, 2002). As reported earlier (Mohammad et al, 2007), the rDNA origin region was selectively enriched in the SA-sepharose pull-down fraction in cells expressing the aptamer-tagged 26T RNA (Figure 5C). rRNA promoter and coding sequences were not enriched in chromatin pull-downs from log phase and starved cultures (compare 26T-Apt to 26T-Ext controls).
In contrast, all nine tested segments in the 60 kb ARS1 interval were enriched in chromatin pull downs in asynchronous log phase cells expressing aptamer-tagged 26T RNA, but not the 26T-Ext variant (Figure 5D and data not shown). Moreover, the aptamer-tagged RNA associated with just the ARS1-A A6 fragment in chromatin prepared from G0/G1 synchronized cells. We conclude that 26T RNA is a component of non-rDNA origin binding ORC complexes. The differential binding of ORC to non-rDNA chromosomes in log phase and starved cells cannot be attributed to the presence or absence of 26T RNA.
Starvation not only synchronizes the vegetative cell cycle, it prepares Tetrahymena for conjugation and the associated DNA replication programme. To ask whether Orc1p binding to non-rDNA chromosomes is cell cycle regulated, centrifugal elutriation was used to obtain a highly enriched population of G1 phase cells to examine ORC regulation in an unperturbed cell cycle (Figure 6A; see Supplementary Figures S4 and S6 for 0 min time points). Cell cycle-dependent changes in the abundance of Orc1p were observed, including a precipitous drop in Orc1p levels during S phase (Supplementary Figure S4), analogous to mammalian and Drosophila Orc1p (Mendez et al, 2002; Araki et al, 2003).
Figure 6.
Cell cycle-regulated ORC binding to the ARS1-A chromosomal locus. A log phase Tetrahymena culture was subjected to centrifugal elutriation. The G1 fraction was further cultured and samples harvested at defined intervals (min) for western blotting of Orc1p (Supplementary Figure S4), flow cytometry and ChIP. (A) Flow cytometry profiles of re-fed cultures (the left line demarcates the G1 propidium iodide (PI) peak, and the right line marks the G2 peak). (B) Orc1p cell cycle ChIP analysis (see Figure 5B for PCR primers locations). I: input; (−): no antibody ChIP pellet; (+): Orc1p ChIP pellet. (C) Orc1p ChIP analysis of the rDNA origin (O), promoter (P) and rRNA coding (C) regions (see Figure 5A schematic).
Similar to starvation-synchronized chromatin, Orc1p selectively associated with the ARS1-A origin in elutriated G1/early S phase cells (Figure 6B, 30 min, A6 region; Supplementary Figure S6B, 0 and 30 min). ARS1-A enrichment was lost as cells progressed through S phase, when Orc1p levels decline (Figure 6B, 60 and 90 min). Newly synthesized Orc1p was detected for all nine ARS1 region probes in G2 phase cells, showing no preference for the A6 fragment (Figure 6B, 120 and 150 min). We conclude that ORC binding to the non-rDNA ARS1 chromosomal interval is dynamically regulated across the cell cycle. ORC appears to be stochastically deposited onto the newly replicated chromosome, and re-localize to the ARS1-A region prior to the next S phase.
As ARS1-B supported episomal DNA replication, but was not bound by Orc1p when pre-RCs assemble on the endogenous chromosome, this segment does not appear to function as a replicator in its normal chromosomal context. Alternatively, the site for ORC binding might have been distal to the PCR primers used in the initial analysis. To address this concern, immunoprecipitated chromatin from the G1 and G2 phase cells was subjected to PCR with five ARS1-B primer sets spanning a 2.1 kb interval (Supplementary Figure S5A). Products were detected for all primer sets in G2 phase chromatin; however, none of these segments was enriched in G1 preparations (Supplementary Figure S5B). In a comparable analysis of the ARS1-A interval, three consecutive primer sets were amplified in immunoprecipitated G1 phase chromatin. Thus, the ARS1-A locus is the preferred site for ORC binding in the endogenous macronuclear chromosome.
Cell cycle ChIP analysis of the rDNA minichromosome revealed origin-specific Orc1p binding only, analogous to log phase and starve vegetative cultures (Figure 6C). Origin binding was detected in G1 and early S phase cells (T=30 min) and disappeared later in S phase (T=60 min), concurrent with the decline in Orc1p protein levels. In contrast to the ARS1 chromosome, subsequent rebinding of Orc1p to rDNA chromatin was restricted to the origin region (T=150 min). The collective results indicate that ORC targeting to rDNA and non-rDNA origins occurs by different mechanisms.
The MCM complex is selectively targeted to the ARS1-A region
To address whether ARS1-A functions as an initiation site in the endogenous chromosome, we examined the association of Mcm6p, a component of the pre-RC that is recruited by ORC and subsequently moves with the replication fork (reviewed in Bell and Dutta, 2002). ChIP analysis was performed for the entire 60 kb ARS1 interval and at multiple sites proximal to the ARS1-A replicator. Similar to Orc1p, Mcm6p immunoprecipitates were enriched for just the ARS1-A region in G1 phase cells synchronized by starvation (Figure 5E, Supplementary Figure S5C) or centrifugal elutriation (Supplementary Figure S6A). Like Orc1p, no DNA binding was detected in log phase cultures or synchronized S phase cells. However, although Orc1p was distributed throughout the 60 kb ARS1 interval in G2 phase cells, no Mcm6 binding was observed at this time. We conclude that ARS1-A is the preferred site for ordered assembly of pre-RC components in the endogenous macronuclear chromosome.
Discussion
Biochemical and genetic studies have documented several distinct mechanisms for targeting ORC to replication initiation sites. They include sequence-specific DNA recognition (S. cerevisiae) (Marahrens and Stillman, 1992), non-specific binding of ORC to degenerate AT-rich sequences (S. pombe, Drosophila, humans) (Kong and DePamphilis, 2001; Vashee et al, 2003; Remus et al, 2004), and tethering of ORC to sequence-specific DNA-binding proteins (Drosophila, rat and humans) (Beall et al, 2002; Minami et al, 2006; Thomae et al, 2008). In the case of Epstein–Barr virus, G-rich RNAs have been recently shown to form a bridge between ORC and EBNA1, the later of which recognizes reiterated sequences at the viral origin (Norseen et al, 2008).
We previously discovered a direct role for RNA in origin recognition. Tetrahymena ORC contains a novel, integral RNA subunit, 26T RNA, that forms Watson–Crick base pairs with complementary sequences at the ribosomal DNA replication origin (Mohammad et al, 2007). Just as type I elements mutations in the rDNA diminish origin utilization (Larson et al, 1986; Yaeger et al, 1989; Gallagher and Blackburn, 1998), so do mutations in 26T RNA that disrupt base pairing interactions (Mohammad et al, 2007). While this RNA may play a role in the selective amplification of rDNA molecules during development, it is also present in ORC complexes during the vegetative cell cycle, when rDNA and non-rDNA origins are coordinately regulated.
These findings prompted us to ask whether ORC is recruited to rDNA and non-rDNA origins by a common mechanism, and whether Tetrahymena replicators contain conserved cis-acting determinants, analogous to the S. cerervisiae ARS consensus sequence (Lee and Bell, 1997; Wyrick et al, 2001; Nieduszynski et al, 2006). To this end, we isolated and characterized two ARS elements derived from chromosomes that are not amplified during development. We found no evidence for sequence conservation between rDNA, ARS1-A and ARS1-B replicators. More importantly, we discovered intrinsic differences in the association of ORC with rDNA and non-rDNA chromosomes as cells progressed through the cell cycle. Whereas ORC binds exclusively to the rDNA origin, it associates in an apparently random, non-specific manner to newly replicated non-rDNA chromosomes, and is subsequently re-localized to a preferred site prior to the next S phase. To the best of our knowledge, this is the first demonstration that the specificity of ORC for DNA changes across the cell cycle. We speculate that intrinsic differences in ORC binding to rDNA and non-rDNA chromosomes contribute to the differential regulation of replication origins during development.
Organization of ARS1 replicators
ARS assays have been used to study the cis- and trans-acting requirements for origin activation in S. cerevisiae and S. pombe (reviewed in Cvetic and Walter, 2005). Although this approach has been used to enrich for metazoan replicators (Gerhardt et al, 2006), further genetic dissection has not been possible due to high plasmid loss rates. Stable ARS activity was previously reported for Tetrahymena rDNA plasmids that contain two tandem copies of the 1.9 kb 5′ NTS (Pan et al, 1995). However, this configuration is poorly suited for functional studies of cis-acting replication determinants, as these plasmids recombine freely, generating molecules with up to 20 tandem 5′ NTS copies. In contrast, the non-rDNA ARS1 episome described here does not oligomerize and is stably maintained at a copy number equivalent to the macronuclear chromosome of origin (∼45 C). These attributes allowed us to examine the cis-acting requirements for origin function.
Our initial experiments uncovered the presence of two independent replicators rather than one. Although this situation is thought to be uncommon, it is not without precedent. Using site-specific recombination to integrate human β-globin locus derivatives into the same ectopic site, Aladjem and co-workers identified two closely spaced replicators (Wang et al, 2004). Nascent strand PCR analysis of the wild type β-globin locus revealed a relatively broad (>4 kb) initiation zone, suggesting that both replicators may function in the endogenous human chromosome.
By analogy, the Tetrahymena ARS1-A and ARS1-B segments contain sufficient sequence information to support autonomous DNA replication. However, ChIP analysis with Orc1p and Mcm6p antibodies indicate that ARS1-A is the preferred site for pre-RC assembly in the endogenous macronuclear chromosome. ORC binding was restricted to the ARS1-A region in G1 and early S phase cells. Thus the ARS1-B replicator appears to be dormant under normal physiological conditions. This result came as a surprise since ARS1-B is entirely intergenic and ∼70% of ARS1-A codes for protein.
Analysis of the Drosophila chorion gene amplicon (DAF-66D) revealed that dispersed cis-acting replication determinants can span several genes. Like most characterized replicons, cis-acting regulatory sequences and replication initiation sites map to intergenic DNA segments (reviewed in Tower, 2004). Whereas transcription has also been shown to inhibit replication initiation in protein-coding regions (Saha et al, 2004), this does not appear to be the case in Tetrahymena, as the ARS1-A gene is constitutively transcribed. Intragenic replication initiation was first described for the Syrian hamster CAD locus (Kelly et al, 1995), however, the cis-acting requirements for activation of this origin are not known. Essential replication determinants in the human β-globin and lamin B2 genes map to an intron and the 3′ untranslated region, respectively (Paixão et al, 2004; Wang et al, 2004). By comparison, two of the three essential replication determinants in Tetrahymena ARS1-A reside in protein-coding sequence. To our knowledge, this constitutes the first example in which protein-coding DNA has been shown to further function as a cis-acting DNA replication determinant.
The organization of ARS1-A resembles metazoan replicators (reviewed in Aladjem and Fanning, 2004), in that it is comprised of unique, dispersed replication determinants. Two of the three regulatory sequences are absolutely required for episomal DNA replication, and might serve as binding sites for ORC or other factors that recruit ORC to the DNA. We speculate that the non-essential determinant is dispensable for plasmid re-replication during macronuclear development, as the initial copy number of the Nhe1L8 Δ650–750 mutant derivative was comparable to the endogenous macronuclear chromosome (∼45 C). The gradual loss of this plasmid is reminiscent of B3 element deletion derivatives of S. cerevisiae ARS1 (Marahrens and Stillman, 1992). In this example, ABF1 binding to the B3 element prevents nucleosomes from encroaching on the ORC-binding site (Lipford and Bell, 2001).
Although it seems unlikely that transcription factors associate with ARS1-A protein-coding sequence, non-coding replication determinants in ARS1-A could function in this way. By analogy, the 5′ border of the nucleosome-free rDNA domains 1 and 2 contains a reiterated DNA sequence, the pause site element (PSE), which facilitates origin activation (Figure 1A) (Saha et al, 2001; TL Morrison and GM Kapler, unpublished results). Furthermore, the promoter-proximal, rmm8 mutation diminishes activation of origins up to ∼1 kb upstream, and induces a change in chromatin structure at the distal Domain 1 and Domain 2 origins (Gallagher and Blackburn, 1998).
ORC targeting to rDNA and non-rDNA replication origins
The most intriguing and unprecedented finding from this study is the dynamic cell cycle-regulated change in ORC binding to non-rDNA chromosomes. The rDNA minichromosome behaved as expected: ChIP analysis with epitope-tagged Orc1p and chromatin pull downs with aptamer-tagged 26T RNA revealed that ORC associates with origin sequences and is excluded from the remainder of the 21 kb rDNA minichromosome (Mohammad et al, 2007; this study). Although the mechanism for sequence-specific recognition differs from S. cerevisiae, the TtORC–rDNA interaction is functionally equivalent to ScORC binding to the yeast ARS1 origin (Diffley and Cocker, 1992; Lee and Bell, 1997).
A similar analysis of the Tetrahymena ARS1 chromosome generated completely different profiles when an asynchronous vegetative (log phase) population or synchronized G2 phase cells were examined. ORC bound to all 17 tested DNA segments in a 60 kb interval, only three of which span the ARS1-A replicator (Figures 5 and 6, Supplementary Figures S4, S5 and S6). This apparent lack of specificity cannot be attributed to the absence of 26T RNA, as Orc1p ChIP and 26T RNA pull down assays generated the same results (Figure 5). The stochastic distribution of Tetrahymena ORC is reminiscent of ORC binding to S. pombe chromosomes (Dai et al, 2005). However, specificity is ultimately achieved in Tetrahymena at the time of pre-RC assembly, when ORC binding to the non-rDNA ARS1 chromosome is restricted to the ARS1-A locus. This newly gained specificity cannot be attributed to sequence-specific RNA–DNA interactions with 26T RNA, as ARS1-A lacks complementarity to this species. The collective data support a model in which rDNA and non-rDNA replicators are recognized by different mechanisms.
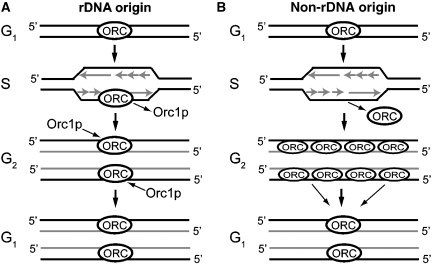
We first consider the rDNA minichromosome (Figure 7A). ORC binding to this origin is mediated in part by 26T RNA–rDNA interactions. Origin activation during S phase results in the turnover of Orc1p (Supplementary Figure S4), analogous to mammals and Drosophila (Mendez et al, 2002; Araki et al, 2003). As the distribution of Orc1p on the rDNA does not change following de novo synthesis of Orc1p in late G2 and G1, we speculate that other ORC subunits may not be displaced from the rDNA origin during S phase. If true, they could mark the site for new ORC-DNA complex assembly. Re-formation of holocomplexes in late G2/G1 would then allow for the recruitment of other pre-RC components to rDNA replication origins.
Figure 7.
Model for ORC binding to rDNA and non-rDNA chromosomes. (A) ORC binding to the rDNA is restricted to the origin region, and occurs throughout the cell cycle due to RNA–DNA base pairing interactions between 26T RNA and rDNA origin type I element (T-rich strand). Degradation of Orc1p generates a sub-complex that remains bound to the origin. (B) ORC binding to non-rDNA replicators is independent 26T-RNA/DNA base pairing and is cell cycle regulated. In this case, the entire ORC complex dissociates from the origin upon Orc1p degradation. The holocomplex randomly binds newly synthesized daughter chromosomes and re-localizes to the preferred initiation site prior to the next S phase.
The association of ORC with non-rDNA chromosomes is much more dynamic, oscillating between sequence-specific and non-specific interactions (Figure 7B). Sequence-specificity is restricted to G1 and early S phase, when pre-RCs are assembled and activated. As this interaction is not mediated by base pairing with 26T RNA, it may be inherently less stable than the ORC–rDNA interaction. We speculate that the entire complex dissociates from non-rDNA origins once Orc1p is degraded. In support of this, ORC was distributed throughout the 60 kb ARS1 chromosomal interval in G2 phase cells. As cells entered G1 in preparation for DNA replication, ORC was ‘re-focused' to a preferred site (ARS1-A). Re-localization could be mediated by other DNA-binding proteins that interact with ORC, analogous to the Drosophila Myb–MuvB complex and rat AIF-2 (Beall et al, 2002; Minami et al, 2006), post-translational modification of ORC proteins, or epigenetic changes in chromatin structure (reviewed in Antequera, 2004). As 26T RNA is present in all ORC complexes (Mohammad et al, 2007), it could form a bridge between ORC and sequence-specific DNA-binding proteins, analogous to the situation in Epstein–Barr virus (Norseen et al, 2008).
Prior studies in Drosophila and Xenopus document an increase in the density of chromosomal replication origins in early stage embryos compared to later developmental stages or adult tissues (Blumenthal et al, 1974; Hyrien et al, 1996; Lemaitre et al, 2005). This difference is presumed to reflect dynamic changes in the concentration and/or distribution of chromatin-associated replication initiation factors. In the work presented here, we show that the distribution of ORC changes within each cell cycle. We speculate that a similar process might occur in broad replication initiation zones in metazoan chromosomes (reviewed in Aladjem and Fanning, 2004). Accordingly, ORC would be deposited ‘locally' onto newly replicated chromosomes and subsequently targeted to preferred initiation sites. Of relevance to this possibility, a recent study showed that in response to DNA stress, Chinese hamster fibroblasts activate cryptic origins within broad initiation zones. When the source of stress is removed a hierarchical pattern of origin usage is re-established (Courbet et al, 2008). For this change to occur, cells must progress through S phase. Thus, origin choice in distantly related eukaryotes, such as Tetrahymena and mammals, is under cell cycle control.
Materials and methods
Library construction, macronuclear DNA transformation and propagation of Tetrahymena transformants
Fragmented Tetrahymena thermophila genomic DNA (size range 5–7 kb) was cloned into the Eco72I site of pCC1FOS (Lucigen Corporation, Middleton, WI). PCR was used to identify small plasmid DNA pools devoid of rDNA origin sequence. DNA was introduced into the macronucleus of mating Tetrahymena strains (CU427 and CU428) by bioballistic bombardment using a Bio-Rad PDS-1000 apparatus outfitted with a hepta adapter (Bio-Rad Laboratories, Hercules, CA) (Bruns and Cassidy-Hanley, 2000). Circular plasmid DNAs (35 ug of pCC1FOS non-rDNA libraries or ARS1 derivatives) were co-transformed with the pmr rDNA rearrangement vector, AN101 (Saha et al, 2001). Serial dilutions were used to determine transformation efficiency and establish clonal lines. Transformants (with or without cloning) were propagated to assess plasmid stability. Replication in Tetrahymena was initially assessed in a plasmid shuttle assay as follows. Genomic DNA was prepared from a 50 ml ‘en masse' co-transformant Tetrahymena culture. Plasmid DNA was purified on a Qiagen midiprep column, digested with the methylation-sensitive enzyme, DpnI, re-transformed into E. coli EpiMax 300 cells (Epicentre Biotechnologies, Madison, WI), and sequenced. ARS1 deletion derivatives were generated by restriction digestion and re-circularization, or subcloned into the low copy number vector pSMART to minimize DNA rearrangement.
Molecular biology techniques
DNA and RNA isolation, Southern blotting, PCR and RT–PCR were performed as previously described (Mohammad et al, 2007). Chromatin immunoprecipitation and pull-down experiments were performed with strains containing TAP-tagged ORC1 (TD102), sequence-tagged 26T RNA (MM201) or aptamer-tagged 26T RNA (MM202) (Mohammad et al, 2007). The 44 nt S1 sequence on aptamer-tagged 26T RNA confers binding to streptavidin (SA) (Srisawat and Engelke, 2002). For ChIP and chromatin pull-downs, 106 cells were treated with 1% formaldehyde for 10 min prior to addition of sodium dodecylsulfate (SDS) to a final concentration of 1%. ChIP analysis of Orc1p was performed with affinity-purified rabbit peroxidase anti-peroxidase soluble complex antibody, which binds the TAP tag protein A IgG-binding domain (Sigma Chemical, St Louis, MO; product P-1291). Mcm6p ChIP analysis was performed with affinity-purified rabbit antibodies directed against amino acids 34–51 (GKKIKYYREKALLLKIYE) of the T. thermophila MCM6 protein (Tetrahymena Genome Database gene prediction: TTHERM-00448570, e value versus human MCM6: 1.0e-172; http://www.ciliate.org). For 26T RNA chromatin pull-downs, lysates were pre-cleared with sepharose beads (100 μl) and pre-incubated with 15 U avidin (Sigma Chemical) for 30 min at RT to saturate SA-binding sites in biotinylated proteins, prior to incubation with 100 μl of SA sepharose (GE Healthcare, Piscataway, NJ). Eluted DNA was subjected to 30 cycles of PCR. Products ranging in size from ∼130–220 bp were visualized on agarose gels. ChIP and chromatin pull down results were validated in at least three experiments using freshly prepared chromatin.
Cell cycle synchronization
Log phase Tetrahymena cultures were synchronized in G0/G1 by starvation as previously described (Yakisich et al, 2006). G1 phase cells were also obtained by elutriation in a Beckman J6M/E centrifuge. Here, 1.5 l of log phase cells (density of 1 × 105/ml) was loaded into the elutriation chamber (flow rate: 50 ml/min, rotor speed: 850 r.p.m.) and washed with 0.5 l of 2% PPYS (growth media). The pump flow rate was increased to 65 ml/min to harvest G1 cells. Cell density was adjusted to 2 × 105/ml and cells were harvested at 30 min intervals to obtain S and G2 phase populations. Flow cytometry was performed as previously described (Morrison et al, 2005).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Information
Supplementary Figures Legend
Acknowledgments
We thank Ron Godiska (Lucigen Corporation) for advice and construction of the T. thermophila library, and Eileen Hamilton and Ed Orias for the analysis of library complexity. We also thank Sebastian Yakisich and Dorothy Shippen for advice on experiments and writing of the paper. This study was supported by the following grants to GMK: NIH (5R01GM053572) and NSF (MCB-0132675).
References
- Abdurashidova G, Riva S, Biamonti G, Giacca M, Falaschi A (1998) Cell cycle modulation of protein-DNA interactions at a human replication origin. EMBO J 17: 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Fanning E (2004) The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F (2004) Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. EMBO J 23: 4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki M, Wharton RP, Tang Z, Yu H, Asano M (2003) Degradation of origin recognition complex large subunit by the anaphase-promoting complex in Drosophila. EMBO J 22: 6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 42: 833–837 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS (1974) The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223 [DOI] [PubMed] [Google Scholar]
- Bolon YT, Bielinsky AK (2006) The spatial arrangement of ORC binding modules determines the functionality of replication origins in budding yeast. Nucleic Acids Res 34: 5069–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns PJ, Cassidy-Hanley D (2000) Biolistic transformation of macro- and micronuclei. Methods Cell Biol 62: 501–512 [DOI] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M (2008) Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Cvetic S, Walter JC (2005) Eukaryotic origins of DNA replication: could you please be more specific? Sem Cell Devel Biol 16: 343–353 [DOI] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ (2005) DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH (1992) Protein-DNA interactions at a yeast replication origin. Nature 357: 169–172 [DOI] [PubMed] [Google Scholar]
- Gallagher RC, Blackburn EH (1998) A promoter region mutation affecting replication of the Tetrahymena ribosomal DNA minichromosome. Mol Cell Biol 18: 3021–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Jafar S, Spindler MP, Ott E, Schepers A (2006) Identification of new human origins of DNA replication by an origin-trapping assay. Mol Cell Biol 26: 7731–7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JL, Dijkwel PA (1995) On the nature of replication origins in higher eukaryotes. Curr Opin Genet Dev 5: 153–161 [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M (1996) Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997 [DOI] [PubMed] [Google Scholar]
- Karrer KM (2000) Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol 62: 127–186 [DOI] [PubMed] [Google Scholar]
- Kelly RE, DeRose ML, Draper BW, Wahl GM (1995) Identification of an origin of bidirectional DNA replication in the ubiquitously expressed mammalian CAD gene. Mol Cell Biol 8: 4136–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Huberman JA (1998) Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol 18: 7294–7303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong DC, DePamphilis ML (2001) Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol 21: 8095–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DD, Blackburn EH, Yaeger PC, Orias E (1986) Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell 47: 229–240 [DOI] [PubMed] [Google Scholar]
- Lee DG, Bell SP (1997) Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol 17: 7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre J-M, Danis E, Pasero P, Vassetzky Y, Mechali M (2005) Mitotic remodeling of the replicon and chromosome structure. Cell 123: 787–801 [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP (2001) Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7: 21–30 [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823 [DOI] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim S-Y, Tansey WP, Stillman B (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after Initiation of DNA replication. Mol Cell 9: 481–491 [DOI] [PubMed] [Google Scholar]
- Minami H, Takahashi J, Suto A, Saitoh Y, Tsutsumi K (2006) Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin. Mol Cell Biol 26: 8770–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MM, Donti TR, Yakisich JS, Smith AG, Kapler GM (2007) Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J 26: 5048–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TL, Yakisich JS, Cassidy-Hanley D, Kapler GM (2005) TIF1 represses rDNA replication initiation, but promotes normal S phase progression and chromosome transmission in Tetrahymena. Mol Biol Cell 16: 2624–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Knox Y, Donaldson AD (2006) Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 15: 1874–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM (2008) RNA-dependent recruitment of the origin recognition complex. EMBO J 27: 3024–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão S, Colaluca IN, Cubells M, Peverali FA, Destro A, Giadrossi S, Giacca M, Falaschi A, Riva S, Biamonti G (2004) Modular structure of the human lamin B2 replicator. Mol Cell Biol 24: 2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W-J, Gallagher RC, Blackburn EH (1995) Replication of an rDNA gene origin plasmid in the Tetrahymena thermophila macronucleus is prevented by transcription through the origin from an RNA polymerase I promoter. Mol Cell Biol 15: 3372–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischmann KP, Zhang Z, Kapler GM (1999) Long-range cooperative interactions regulate the initiation of replication in the Tetrahymena thermophila rDNA minichromosome. Nucleic Acids Res 27: 3079–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR (2004) DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Nicholson A, Kapler GM (2001) Cloning and biochemical analysis of the Tetrahymena origin binding protein TIF1—Competitive DNA binding in vitro and in vivo to critical rDNA replication determinants. J Biol Chem 276: 45417–45426 [DOI] [PubMed] [Google Scholar]
- Saha S, Shan Y, Mesner LD, Hamlin JL (2004) The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev 18: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication origins at AT-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR (2002) RNA affinity tags for purification of RNAs and ribonucleoprotein complexes. Methods 26: 156–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A (2008) Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci USA 105: 1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J (2004) Developmental gene amplification and origin regulation. Annu Rev Genet 38: 273–304 [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC (2003) Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin CM, Brooks S, Cimbora D, Groudine M, Aladjem MI (2004) The human beta-globin replication initiation region consists of two modular independent replicators. Mol Cell Biol 24: 3373–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Yaeger PC, Orias E, Shaiu W-L, Larson DD, Blackburn EH (1989) The replication advantage of a free linear rDNA gene is restored by somatic recombination in Tetrahymena thermophila. Mol Cell Biol 9: 452–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakisich JS, Sandoval PY, Morrison TL, Kapler GM (2006) TIF1 activates the intra-S-phase checkpoint response in the diploid micronucleus and amitotic polyploid macronucleus of Tetrahymena. Mol Biol Cell 17: 5185–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Information
Supplementary Figures Legend