Abstract
Sgt1 was described previously in yeast and humans to be a Hsp90 co-chaperone and required for kinetochore assembly. We have identified a mutant allele of Sgt1 in Drosophila and characterized its function. Mutations in sgt1 do not affect overall kinetochore assembly or spindle assembly checkpoint. sgt1 mutant cells enter less frequently into mitosis and arrest in a prometaphase-like state. Mutations in sgt1 severely compromise the organization and function of the mitotic apparatus. In these cells, centrioles replicate but centrosomes fail to mature, and pericentriolar material components do not localize normally resulting in highly abnormal spindles. Interestingly, a similar phenotype was described previously in Hsp90 mutant cells and correlated with a decrease in Polo protein levels. In sgt1 mutant neuroblasts, we also observe a decrease in overall levels of Polo. Overexpression of the kinase results in a substantial rescue of the centrosome defects; most cells form normal bipolar spindles and progress through mitosis normally. Taken together, these findings suggest that Sgt1 is involved in the stabilization of Polo allowing normal centrosome maturation, entry and progression though mitosis.
Keywords: mitosis, Polo, Sgt1
Introduction
Accurate chromosome segregation requires organization and proper function of the mitotic apparatus, including correct attachment of sister chromatids to spindle microtubules. Microtubule–kinetochore interaction is highly complex and involves many factors. It is now clear that kinetochores are highly conserved and contain a large number of proteins, including internal structural components that ensure DNA binding, external components for microtubule binding and proteins involved in signalling processes, such as those required for the activation of the spindle assembly checkpoint (SAC) (Musacchio and Salmon, 2007). Previously, it was suggested that in yeast and human cultured cells, assembly or at least proper localization of some kinetochore components requires the activity of the Hsp90 co-chaperone Sgt1 (Kitagawa et al, 1999; Steensgaard et al, 2004). Sgt1 was initially identified in a screen aimed at isolating suppressors of a temperature-sensitive allele of skp1 and characterized as a subunit of both the core kinetochore and SCF (Skp1-Cul1-F-Box) ubiquitin ligase complexes (Kitagawa et al, 1999). Sgt1 contains protein interaction motifs, such as the tetracopeptide repeat domain (TPR domain) and p23-like CHORD domain (also called CS domain), both found in proteins that interact with chaperones, as well as an Sgt1-specific domain (SGS domain) required for its interaction with adenyl cyclase. As expected, human Sgt1 was shown to bind chaperone Hsp90 by its CHORD domain that is very similar to the well-known Hsp90 co-chaperone p23 (Lee et al, 2004). The interaction between Sgt1 and Hsp90 was also described both in genetic and biochemical studies in yeast, where the complex was described as being important for assembly and turnover of the essential centromere binding factor 3 (Bansal et al, 2004; Lingelbach and Kaplan, 2004; Rodrigo-Brenni et al, 2004; Catlett and Kaplan, 2006).
In human cells, a severe reduction of Sgt1 levels was shown to compromise kinetochore assembly and specifically localization of a number of SAC proteins, including Mad1, Mad2 and BubR1 (Steensgaard et al, 2004). This resulted in a weaker SAC response when cells were challenged with spindle poisons that depolymerize microtubules. However, not only SAC proteins failed to accumulate, but also other constituents of the human kinetochore, including Hec1, CENP-E, CENP-F and CENP-I were absent after depletion of Sgt1 (Steensgaard et al, 2004). Interestingly, studies in human cells with an inhibitor of Hsp90 (17-AAG) suggested that this chaperone is important for kinetochore assembly, as it causes delocalization of various centromeric proteins, mitotic arrest, failure of chromosome congression and aneuploidy (Niikura et al, 2006). Moreover, it was shown that depletion of Sgt1 causes cells to become sensitized to 17-AAG treatment, suggesting that Sgt1–Hsp90 co-chaperone activity is important for kinetochore assembly and function. Hsp90 has received much attention as a target for cancer therapy, given that this protein is involved in multiple pathways and often overexpressed in a variety of tumours (Kamal et al, 2003).
Although these data clearly indicate that Sgt1 and Hsp90 interact and that this interaction is associated with kinetochore assembly, other functional studies of Hsp90 have failed to substantiate these observations. In Drosophila, genetic studies showed that Hsp90 is required for the successful completion of mitosis because, in its absence, centrosomes fail to maintain their integrity (Lange et al, 2000). This centrosome phenotype was directly linked with an overall decrease in the levels of Polo, an essential mitotic kinase (de Carcer et al, 2001). Polo is involved in centrosome maturation during G2 and is known to be involved in many other aspects of mitotic progression (Donaldson et al, 2001).
To address the function of Sgt1 further, we have carried out a genetic analysis of Sgt1 in Drosophila. We identified the Sgt1 homologue and described its subcellular localization during mitosis. Analysis of mutant cells shows that most fail to enter mitosis, but if they do, they arrest in a prometaphases-like state with hypercondensed chromosomes. The mitotic arrest is SAC dependent, and unlike previous studies in yeast and human cells in culture, we observe proper localization of SAC proteins at kinetochores. Overall kinetochore structure is not affected. Interestingly, low levels of kinetochore-associated Polo are consistently observed. We also find that Sgt1 mutant cells fail to organize bipolar spindles and spindle poles do not have normal centrosomes even though centriole duplication is not affected. In Sgt1 mutant cells, most centrosomes fail to accumulate Polo and consequently fail to mature. Importantly, overexpression of Polo is able to significantly rescue all mitotic phenotypes resulting from the mutation in sgt1. Our results suggest that, in Drosophila, Sgt1 is not required for normal kinetochore structure or SAC activity, but it is essential for the functional organization of centrosomes through stabilization of the Polo protein.
Results
Identification and subcellular localization of Sgt1 in Drosophila
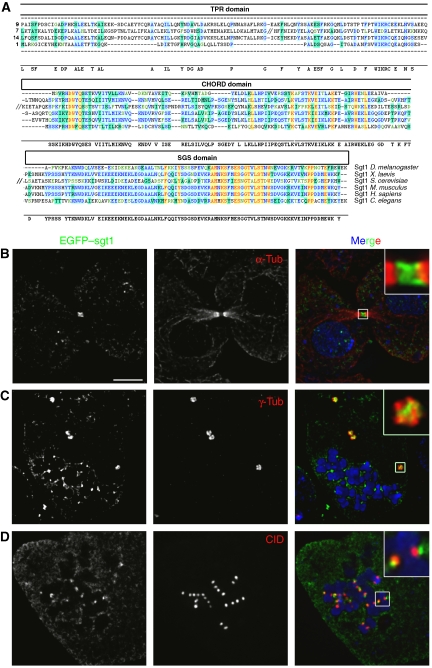
The Drosophila orthologue of Sgt1 was identified through blast searches using the human Sgt1 protein sequence. A single highly conserved putative protein encoded by the gene CG9617 was identified with 41% amino-acid identity (Figure 1A). Sequence analysis shows that most members of the Sgt1 family are characterized by having three distinct domains: a TPR motif, a p23-like CHORD domain (also called CS domain) and the Sgt1-specific domain (SGS). Although the Drosophila orthologue lacks the TPR domain, the function of Sgt1 as a co-chaperone (Bansal et al, 2004) is unlikely to be affected, as the TPR and CS domain are both chaperone-interacting domains that may be redundant. Sgt1 has been reported in human cells to be a soluble protein without any particular localization during mitosis (Steensgaard et al, 2004). To determine if Drosophila Sgt1 localizes in a similar manner, the coding sequence was tagged with EGFP, transfected into S2 Drosophila cells and expressed under the control of an inducible promoter (Figure 1B–D). In asynchronous cultures, no specific accumulation of EGFP–Sgt1 is observed besides a clear localization at the mid-body during very late mitosis (Figure 1B). However, if cells are arrested in mitosis with colchicine, EGFP–Sgt1 shows not only diffuse cytoplasmic staining but also strong accumulation at centrosomes and kinetochores (Figure 1C and D). Expression of EGFP–Sgt1 in interphase or of EGFP alone in either interphase or mitosis gave no specific localization (data not shown). These results indicate that unlike yeast and human cells, Sgt1 in Drosophila shows a specific subcellular localization at different stages of mitosis when spindle microtubules are depolymerized.
Figure 1.
Identification of Sgt1 in Drosophila and cellular localization in S2 cells. (A) Sequence homology between Sgt1 proteins from different species. The conserved domains are indicated in boxes; tetracopeptide domain (TPR domain), p23-like CHORD domain and Sgt1-specific domain (SGS domain). (B–D) Localization of EGFP–Sgt1 in S2 cells. (B) In asynchronous cells, EGFP–Sgt1 (green) can be detected only at the cleavage furrow during late mitosis as shown by the co-staining with α-tubulin (red). When cells are treated with colchicine, we can observe (C) accumulation of EGFP–Sgt1 (green) at the centrosomes identified by γ-tubulin (red) and (D) at the outer region of the kinetochores as shown by co-staining with the centromeric marker CID (red). For all immunolocalization, DNA was stained with DAPI (blue). Scale bar, 5 μm.
Identification of an sgt1 mutant allele
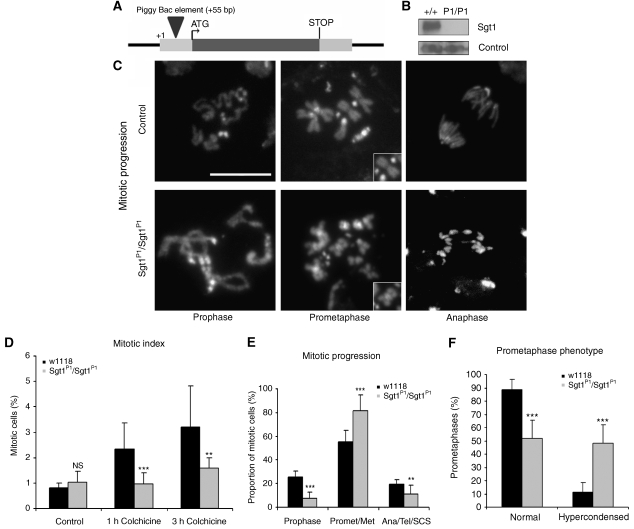
To study the function of Sgt1 in Drosophila, we identified two putative mutant strains (C01268 and C01428) (see Materials and methods) (Figure 2A). Analysis of the DNA sequence indicates that the transposable-elements are inserted at the same site in both strains (+55 bp). Both strains are lethal when homozygous for the insertion or as trans-heterozygotes or hemizygotes over the deficiency Df(3R)CA3, which uncovers this region (data not shown), suggesting that the insertions are associated with the lethality. For all other experiments reported here, we used the C01268 strain referred as sgt1P1. To confirm that the lethality is due to the insertion, we generated precise excisions of the transposable element. In all excisions obtained, we observed a full restoration of viability. Western blot analysis of total protein extracts from either wild-type or sgt1P1/sgt1P1 third instar larvae brains show that the homozygote mutant cells have highly reduced levels of Sgt1 protein, suggesting that sgt1P1 is either a severe hypomorph or a null mutant (Figure 2B).
Figure 2.
Identification and characterization of sgt1P1 alleles. (A) Diagram showing the TE element insertion in gene CG9617 of the mutant strain sgt1P1. (B) Western blot of total protein extracts from control and sgt1P1 neuroblasts probed for Sgt1 and α-tubulin, as a loading control. (C) Cytological analysis of mitotic progression of control (w1118) and mutant (sgt1P1/sgt1P1) third instar larval neuroblasts. Insets in the central panels highlight the chromosome hypercondensation phenotype. (D) Quantification of mitotic parameters in control and mutant neuroblasts in the absence and presence of colchicine (n=10 brains for each condition, and 2000 cells scored). (E) Quantification of mitotic progression in wild-type and sgt1P1 neuroblasts (n=10 brains for each condition, and 100 mitotic figures scored). (F) Quantification of prometaphase phenotype in neuroblasts (n=12 brains for each condition, and 100 prometaphases scored). Not statistically significant (NS); or significantly different: P<0.01 (**) and P<0.001 (***); scale bar, 5 μm.
Sgt1 localizes to specific structures during mitosis, suggesting that the protein might have relevant functions during cell division. To test this hypothesis, we carried out a classical cytological analysis of third instar larval sgt1P1 neuroblasts (Figure 2C). We observe abnormal mitotic phenotypes, including prometaphases with hypercondensed chromosomes (see insets in Figure 2C) and anaphases with lagging chromatids. Quantitative analysis of mitotic progression indicates that mutation in sgt1 does not have an overall effect upon mitotic index (Figure 2D). However, treatment of sgt1P1 neuroblasts with colchicine does not lead to the accumulation of cells in mitosis, suggesting that the SAC might be compromised (Figure 2D). sgt1P1 mutant tissue shows a significant reduction in the frequency of prophases, a severe increase in the frequency of prometaphases and a reduction of cells exiting mitosis, when compared with controls (Figure 2E). The most striking phenotype observed by the loss of Sgt1 function is prometaphase cells with hypercondensed chromosomes similarly to control cells arrested with colchicine (Figure 2F). Interestingly, incubation in colchicine does not result in accumulation of Sgt1 mutant cells in mitosis but sister chromatid separation was not observed, indicating that mutant cells have an active SAC. One plausible explanation for this observation is that mutant cells are delayed before entering mitosis, and when they eventually are able to enter mitosis they arrest in prometaphase, resulting in chromosome hypercondensation. Thus, we suggest that sgt1P1 brains have a mitotic index that is not different from controls mostly because of slow mitotic progression and failure to exit mitosis.
Sgt1 and the SAC
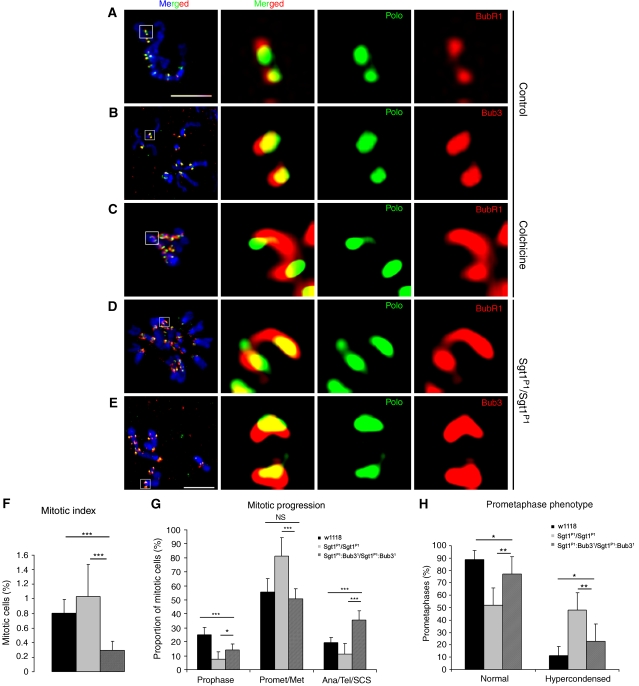
Previous reports have shown that Sgt1 is required for full kinetochore assembly and localization of checkpoint proteins, such as BubR1, Mad1 and Mad2 (Steensgaard et al, 2004). Consequently, it was suggested that the absence of Sgt1 reduces the potency of the SAC (Steensgaard et al, 2004). Therefore, we analysed whether sgt1P1 mutation affects kinetochore localization of BubR1 and Bub3, two well-characterized SAC components (Logarinho et al, 2004; Lopes et al, 2005). We find that both proteins localize to kinetochores of sgt1P1 early prometaphase cells similarly to control cells (Figure 3A and B). However, we consistently observe that in mutant cells BubR1 and Bub3 kinetochore distribution are significantly enlarged (Figure 3D and E), as described for wild-type cells treated with colchicine (Figure 3C). Enlargement of the kinetochore is well documented in cells treated with colchicine and might be an adaptive response to increase the chance of kinetochore–spindle microtubule interaction (Logarinho et al, 2004). Taken together, these results suggest that sgt1P1 mitotic cells might not be able to establish normal microtubule–kinetochore attachment.
Figure 3.
The SAC in sgt1P1 neuroblasts. Immunofluorescence localization of SAC proteins BubR1 and Bub3 in (A–C) control or (D, E) sgt1P1 neuroblasts. In the merged image, DNA (blue) is shown, and in the panels on the right, the kinetochores (white box in the merged image) are shown at a higher magnification. Anti-Polo antibody was used as control, but the signal had to be intensified in sgt1P1 due to the low level of this protein at the kinetochore. During prometaphase (A, B, D, E) control cells show clearly defined localization of both BubR1 and Bub3, but in mutant cells, both proteins show a broad distribution resembling the distribution of BubR1 in (C) control cells when incubated with colchicine. (F) Quantification of the mitotic index in control, sgt1P1 and sgt1P1;bub31 double-mutant neuroblasts. Note that in the double mutant cells, the mitotic index is significantly reduced, indicative of the loss of SAC activity (n=10 brains for each condition, and 2000 cells scored) (G) Quantification of mitotic progression in control, sgt1P1 and sgt1P1;bub31 double-mutant neuroblasts. (n=10 brains for each condition, and 100 mitotic figures scored) (H) Quantification prometaphase with hypercondensed chromosomes in control, sgt1P1 and sgt1P1;bub31 double-mutant neuroblasts (n=12 brains for each condition, where 100 prometaphases were quantified). Not statistically significant (NS); or significantly different: P<0.05 (*), P<0.01 (**) and P<0.001 (***); Scale bar, 5 μm.
Although these observations suggest that SAC proteins can indeed localize properly in sgt1P1 cells, they do not confirm that they are functional. To address this, we constructed strains carrying the sgt1P1 mutation and bub31, an allele of Bub3 that renders the SAC inactive (Lopes et al, 2005). Quantitative analysis of this strain (sgt1P1;bub31) shows a very low mitotic index (Figure 3F). Furthermore, analysis of mitotic progression shows that when Bub3 expression is impaired in the sgt1P1 strain, these cells enter mitosis more readily, do not accumulate in prometaphase and exit mitosis more frequently as shown by the increase in anaphase and telophases (Figure 3G). Also, sgt1P1;bub31 mutant cells do not show chromosome hypercondensation, suggesting that mitotic cells do not arrest at any stage of mitosis (Figure 3H). Taken together, these observations clearly show that in sgt1P1 cells the SAC is active and is responsible for the mitotic delay observed in prometaphase.
sgt1P1 cells fail to progress normally through the cell cycle
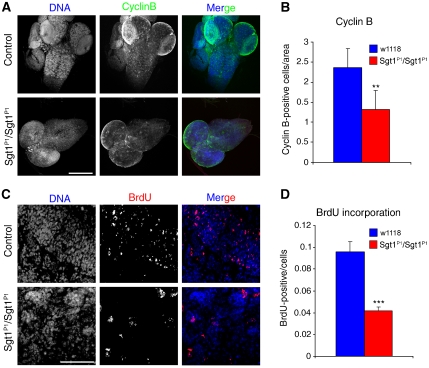
Our results show that sgt1P1 cells can activate the SAC; however, they do not explain why these cells fail to accumulate in mitosis in response to spindle damage. As we do not observe neuroblasts exiting mitosis, a possible explanation is that, in the absence of Sgt1, cells fail to enter mitosis readily and are delayed at other stages of the cell cycle. To test this hypothesis, we analysed the overall ability of third instar larval neuroblasts to express Cyclin B, an essential protein required for mitotic entry (Lehner and O'Farrell, 1990) (Figure 4A). Quantitative immunofluorescence analysis shows that sgt1P1 brains have significantly lower number of cyclin B-positive neuroblasts when compared with controls (Figure 4B). This suggests that most mutant cells fail to reach G2 and therefore are most likely to be delayed in S or G1 phase. Accordingly, we incubated sgt1P1 neuroblasts with BrdU to ascertain whether they enter S phase (Figure 4C). Quantification of BrdU shows that when compared with control, much fewer sgt1P1 cells incorporate BrdU (Figure 4D). Both findings strongly suggest that loss of Sgt1 imposes a delay in cell cycle progression, most likely during G1.
Figure 4.
Analysis of cell cycle progression in control and sgt1P1 neuroblasts. (A) Brains were dissected from control or sgt1P1 mutant larvae and immunostained to reveal cyclin B and counterstained for DNA. (B) Quantification of the number of cyclin B-positive cells in either control or mutant brains. (C) High magnification of a random area from either control or mutant larvae showing BrdU and DNA. (D) Quantification of BrdU-positive cells in relation to the total number of cells. Difference not statistically significant (NS); or significantly different: P<0.01 (**) and P<0.001 (***); scale bar, 50 μm.
Kinetochore structure in sgt1P1 mutant cells
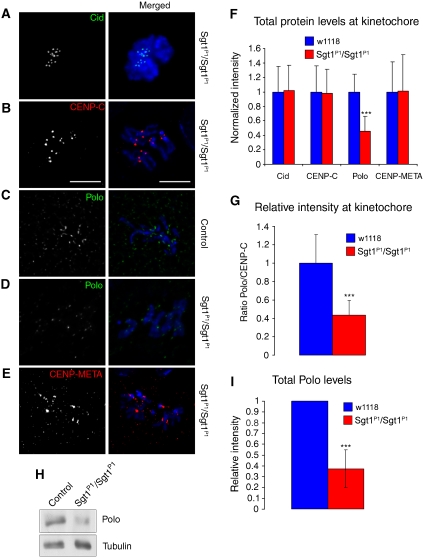
Previous work has suggested that Sgt1 is not only required for kinetochore localization of checkpoint proteins but also the overall structure of the kinetochore. Accordingly, we analysed the localization of several kinetochore proteins in sgt1P1 neuroblasts (Figure 5). The kinetochore is a highly complex multi-layered structure built over CENP-A-containing centromeric heterochromatin (Musacchio and Salmon, 2007). We find that Cid, the Drosophila homologue of CENP-A, the inner kinetochore protein CENP-C, Polo and CENP-META, the Drosophila homologue of CENP-E, all show proper kinetochore localization (Figure 5). However, detailed quantitative analysis of signal intensity for the various kinetochore components shows that whereas Cid, CENP-C and CENP-META remain mostly unaltered in the absence of Sgt1 (Figure 5F), the level of Polo protein is significantly reduced at kinetochores when compared with wild-type control cells (Figure 5F and G). This reduction in kinetochore localization can be due to either mislocalization of the protein or to a reduction in overall protein levels. To address this, we performed western blot of total protein extracts from wild-type and sgt1P1 brains (Figure 5H). We find that mutant cells have a severe reduction (70%) in the total amount of Polo protein levels (Figure 5I), strongly suggesting that Sgt1 is required for its stabilization, in agreement with previous results showing that HSP90, a known interactor of Sgt1, is required to stabilize the levels of Polo protein (de Carcer et al, 2001).
Figure 5.
Organization of the kinetochore in the absence of Sgt1. Immunolocalization of different kinetochore proteins in (A, B, D, E) sgt1P1 or (C) control neuroblasts. In the merged images CID and Polo (green), CENP-E and CENP-meta (red) and DNA (blue) are shown. Note reduced Polo signal in sgt1P1 when compared with control neuroblasts. (F) Quantification of the signal intensity of Cid, CENP-C, Polo and CENP-META at kinetochores in both control and sgt1P1 neuroblasts. The prometaphase cells were randomly selected and the intensities of kinetochore proteins in the sgt1 mutant cells were normalized to the average in control cells. All images were acquired using identical parameters. (G) Quantification of the ratio between kinetochore levels of Polo and CENP-C in control and sgt1P1 neuroblasts. (H) Western blot of total protein extracts from control or sgt1P1 neuroblasts and (I) quantification of the signal showing a significant reduction in Polo protein levels in mutant brains. α-tubulin was used as a loading control. (For all the quantifications, n=200 kinetochores from five different preparations in each condition and 20 cells scored). Significantly different: P<0.001 (***); scale bar, 5 μm.
Analysis of spindle organization in sgt1P1 neuroblasts
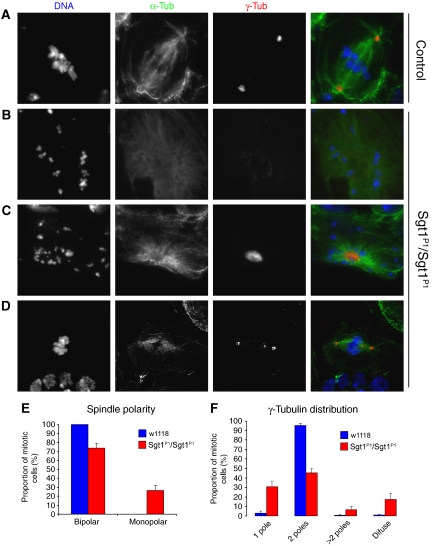
The mitotic phenotype observed in sgt1P1 cells is similar to the phenotype observed in Polo mutants, such as the prometaphase delay and chromosome overcondensation (Llamazares et al, 1991). Polo is known to be responsible for centrosome maturation and spindle bipolarity (Sunkel and Glover, 1988; Llamazares et al, 1991), therefore reduced levels of Polo should lead to centrosome dysfunction and abnormal spindle formation. Therefore, we isolated wild-type and sgt1P1 neuroblasts and studied their mitotic apparatus (Figure 6). We find that although most control cells assemble a bipolar spindle with two well-defined centrosomes, one at each pole (Figure 6A), a significant proportion (22%) of sgt1P1 neuroblasts have monopolar spindles (Figure 6B and C) with either a dispersed (Figure 6B) or well-focused (Figure 6C) centrosome at the centre. The majority (78%) of sgt1P1 cells are able to assemble a bipolar spindle (Figure 6D and E), but more than half (55%) have an abnormal number of centrosomes (30% have just one) or centrosome structure (20% with a diffuse centrosome) (Figure 6F). These results indicate that loss of Sgt1 protein leads to centrosome and spindle abnormalities that are highly reminiscent of loss of Polo function.
Figure 6.
Organization of the mitotic apparatus in the absence of Sgt1. (A) Control and (B–D) sgt1P1 neuroblasts were immunostained to show spindle microtubules (α-tubulin in green), centrosomes (γ-tubulin in red) and DNA (blue). (A) Control cells showing a normal mitotic spindle with γ-tubulin staining at both poles. Mutant cells show many spindle abnormalities including (B) monopolar spindles with a diffuse staining of γ-tubulin, (C) monopolar spindles with only one centrosome or (D) bipolar spindles with more than two defined spots of γ-tubulin staining. (E) Quantification of spindle organization. (F) Quantification of MTOCs with centrosomes as ascertained by γ-tubulin staining (in all the conditions, n=8 brains); scale bar, 5 μm.
In vivo analysis of mitotic progression in sgt1P1 neuroblasts
To further analyse progression through mitosis and spindle formation in the absence of Sgt1, we generated a strain carrying the sgt1P1 mutation and fluorescently tagged centromeres (Cid–mRFP) (Schuh et al, 2007) and microtubules (GFP–tubulin) (Rebollo et al, 2004).
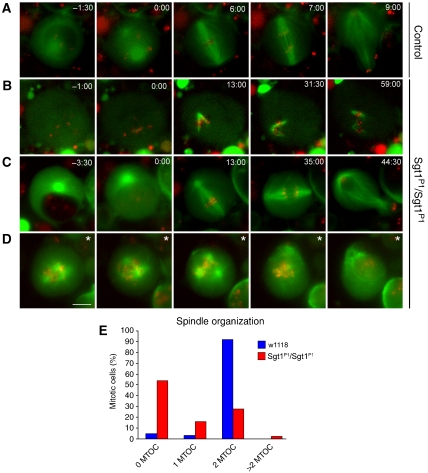
Neuroblasts were observed as primary cultures (Figure 7). Wild-type neuroblasts enter mitosis with two well-defined microtubule-organizing centres (MTOCs), and the bipolar spindle forms on average 2:30±0:30 min after nuclear envelop breakdown (NEBD). Anaphase initiates 7±1:30 min after NEBD and the cell divides asymmetrically (Figure 7A and Supplementary movie 1). In contrast, sgt1P1 neuroblasts show different behaviours. More than half of the cells (55%) enter mitosis with no apparent MTOC, but after NEBD are able to form a microtubule array that grows out from the kinetochores, they are unable to focus at the spindle poles and mitotic progression is delayed in prometaphase for more than 1 h (Figure 7B and E, see also Supplementary movie 2). We also find some (15%) mutant cells that enter mitosis with a single MTOC, form a bipolar spindle and reach metaphase with a short delay but eventually exit mitosis with a significant delay (35 min after NEBD) (Figure 7C and E, see also Supplementary movie 3). We also find few cells (5%) that enter mitosis with more than two MTOCs, fail to organize a proper bipolar spindle and consequently fail to congress the chromosomes and arrest for long periods (Figure 7D and E, see also Supplementary movie 4). Finally, we observe sgt1P1 neuroblasts (25%) that enter mitosis with two MTOCs (Figure 7E). These observations indicate that, in the absence of Sgt1, most cells have severe difficulties in organizing a proper bipolar spindle, do not show normal MTOCs and are mostly arrested in prometaphase.
Figure 7.
Live imaging of control and mutant neuroblasts. Primary cultures of neuroblasts were imaged by time-lapse spinning confocal microscopy to visualize spindle microtubules (GFP–α-tubulin in green) and the centromeres (Cid–mRFP in red). (A) Wild-type cell showing two microtubule asters at opposite poles of a well-organized bipolar spindle with centromeres congressing and then separating during anaphase. (B) sgt1P1 cell showing a spindle without asters and microtubules that appears to be nucleated from the chromosomes that remains in mitosis for at least 1 h after NEB. (C) sgt1P1 cell with a spindle organized from a single MTOC that is able to enter anaphase after a delay of 35 min in metaphase. (D) sgt1P1 cell showing a multipolar spindle that remained in prometaphase for at least 60 min. (*) This cell was filmed in prometaphase therefore the time of NEB could not be determined. (E) Distribution of the spindle organization observed after in vivo analysis. The number of MTOCs was determined by the presence of asters at the poles of the mitotic spindle (control n=65 cells; sgt1P1 n=192). Scale bar, 5 μm.
Analysis of centrosome structure in sgt1P1 cells
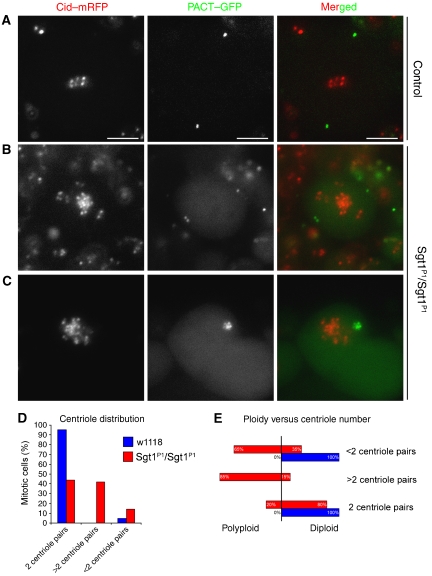
To determine whether abnormal MTOC phenotype is associated with abnormal centriole duplication, we constructed a strain carrying the sgt1P1 allele, Cid–mRFP to label the centromeres and EGFP–PACT (Martinez-Campos et al, 2004) to label the centrioles, and neuroblasts were then analysed as described previously (Figure 8). In wild-type mitotic neuroblasts, most (90%) centrioles are present as pairs of two distinct dots, at each side of the metaphase plate (Figure 8A). However, in sgt1P1 cells, we frequently find an irregular number of centrioles at the poles (Figure 8B) or a single cluster of several PACT–GFP-labelled centrioles (Figure 8C) or even cells with less than two dots of PACT–GFP (data not shown). Quantification shows that most (90%) wild-type cells show the expected two centriole pairs and just a small proportion present less than two centriole pairs (Figure 8D). However, only 40% of sgt1P1 mutant cells show a normal set of centriole pairs, approximately 40% of mutant cells have more than two centriole pairs and some cells have less than two centriole pairs (Figure 8D). Moreover, when we compare the ploidy of sgt1P1 mutant cells with the number of centrioles, we observe a clear correlation between increase in centriole number and increase in ploidy (85% of the cells) (Figure 8E). This correlation suggests that the appearance of additional centrioles is most likely to be a consequence of a failure in a previous division and a subsequent replication of the centrioles in the following cell cycle. Therefore, these observations suggest that although loss of Sgt1 affects centrosome maturation, centrioles replicate normally.
Figure 8.
Determination of centriole numbers in sgt1P1 mutant cells. In vivo analysis of the centrioles was carried out using PACT–GFP and the centromeric marker Cid–mRFP. (A) Wild-type neuroblast showing two well-defined centriolar spots (arrows) at opposite sides of the metaphase plate. (B) sgtP1 mutant neuroblast showing irregular number of centrioles (arrows), two on one side and a single pair on the other side of the cluster of centromeres. (C) Polyploid cell from an sgtP1 mutant brain showing a cluster of centrioles (arrow) in one side of the large centromere cluster. (D) Quantification of the number of centrioles in control and mutant neuroblasts. (E) Quantification of cellular ploidy and the number of centrioles in control and mutant neuroblasts (control n=40; sgt1P1 n=121 cells). Scale bar, 5 μm.
Overexpression of Polo rescues Sgt1 centrosome phenotype and mitotic progression
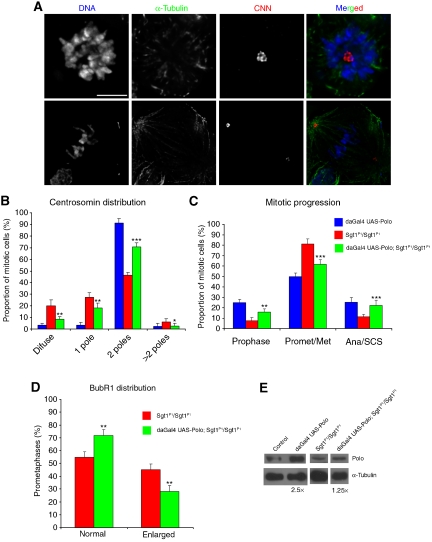
To test if Sgt1 centrosome maturation function is indeed a consequence of failure in Polo stabilization or an indirect consequence of the absence of Sgt1, we overexpressed Polo protein in sgt1P1 or control flies and analysed centrosome, spindle organization and mitotic progression (Figure 9). In control cells, we observed that Polo overexpression has no effect in centrosome numbers (Figure 9B) or mitotic progression (Figure 9C); however, Polo levels are increased 2.5 times as shown by western blot (Figure 9E). However, when Polo was overexpressed in an sgt1P1 background, we observed significant rescue of the mutant phenotype, although Polo levels only increase 1.25 times (Figure 9E). The number of cells that do not show CNN staining is significantly reduced, as are the cells with just one centrosome, whereas cells with normal number of centrosomes and a normal bipolar spindle increase significantly (Figure 9A and B). Moreover, overexpression of Polo in Sgt1 mutant background restores mostly normal mitotic progression, showing that they do not arrest in prometaphase (Figure 9C). We also analysed kinetochore organization in sgt1P1 cells after Polo overexpression. For this, we quantified the number of cells showing at least 50% enlarged kinetochores when stained for the SAC protein BubR1 (Figure 9D). We find that after Polo overexpression in sgt1P1 cells exhibiting BubR1, enlarged kinetochores decreases significantly (Figure 9D). These observations suggest that Polo overexpression results in a significant recovery of kinetochore microtubule attachment in accordance with the improved frequency of normal mitotic exit. Overall, these data indicate that the mitotic phenotypes caused by mutation in sgt1 appears to result from destabilization and consequent reduction in Polo protein levels.
Figure 9.
Overexpression of Polo in sgt1P1 and wild-type neuroblasts. (A) CNN and α-tubulin staining of neuroblasts in which Polo was overexpressed, using the daGAL4 driver (daGAL4:UAS-Polo; sgt1P1/sgt1P1). The top panel shows a cell with a monopolar spindle similar to those observed in sgtP1/sgtP1 homozygote, with one well-defined pole with strong staining of CNN. In the bottom panel, a typical Polo-overexpressing cell showing a bipolar spindle similar to controls with one defined spot of CNN at each pole. (B) Quantification of the CNN distribution in both control (daGAL4:UAS-Polo; +/+), sgt1P1 mutant (sgt1P1/sgt1P1) and Sgt1 mutant overexpressing Polo (daGAL4:UAS-Polo; sgt1P1/sgt1P1) (n=10 brains). (C) Quantification of mitotic progression in control (daGAL4:UAS-Polo; +/+), sgt1P1/sgt1P1 and Sgt1 mutant overexpressing Polo (daGAL4:UAS-Polo; sgt1P1/sgt1P1) (n=10 brains). (D) Quantification of the number of cells showing enlarged BubR1 kinetochore accumulation in either Sgt1 mutant (sgt1P1/sgt1P1) and Sgt1 mutant overexpressing Polo (daGAL4:UAS-Polo; sgt1P1/sgt1P1) neuroblasts. Normal staining indicates a dot-like BubR1 accumulation, whereas enlarged staining indicates cells that have more than 50% of their kinetochores with BubR1 staining similar to kinetochores of wild-type cells treated with colchicine (n=5 different brains and 200 cells scored). (E) Western blot and quantification of Polo protein levels in the different genotypes. Not statistically significant (NS); or significantly different: P<0.05 (*), P<0.01 (**) and P<0.001 (***); all statistically significant differences correspond to comparisons between sgt1P1/sgt1P1 and daGAL4:UAS-Polo; sgt1P1/sgt1P1 strains. Scale bar, 5 μm.
Discussion
Drosophila Sgt1 protein is essential for cell proliferation
We identified the Drosophila melanogaster homologue of the human Sgt1 protein encoded by the gene CG9617. This protein is partially conserved from yeast to humans, suggesting that it might have key functions in cellular processes. Members of the Sgt1 protein family contain three highly conserved domains in many species: TPR, p23-like CHORD and SGS domains. The TPR and p23-like CHORD domains are characteristic of proteins that interact with chaperones directing them to a specific group of proteins named ‘client proteins'. Therefore, Sgt1 was classified as a co-chaperone, as it interacts with the chaperone HSP90 through the p23-like domain. Moreover, Sgt1 is thought to form a complex that, through the TPR domain, activates Skp1, a protein involved in kinetochore assembly (Kitagawa et al, 1999; Bansal et al, 2004; Rodrigo-Brenni et al, 2004; Steensgaard et al, 2004). Although, Drosophila Sgt1 lacks the TPR domain, it is unlikely that its co-chaperone function is affected, as in human cells the interaction with HSP90 is made through the p23-like CHORD domain (Lee et al, 2004), which is also conserved in the Drosophila protein. However, in Drosophila, Sgt1 protein is unlikely to interact with Skp1 and therefore might not be required for kinetochore assembly (Bansal et al, 2004; Rodrigo-Brenni et al, 2004). Our genetic analysis indicates that sgt1 is an essential gene in Drosophila. Immunofluorescence studies show that Sgt1 localizes to the mid-body in late mitosis and in the absence of microtubule to the outer kinetochore domain and also to the centrosomes. This localization pattern suggests that Sgt1 might have a function in kinetochore–microtubule interaction and in centrosome function. While our results show that Sgt1 is required for somatic cell proliferation, meiosis might also require Sgt1, as ovaries and testis are highly abundant in Sgt1 mRNA (Chintapalli et al, 2007). Cytological analysis of the mutant neuroblasts showed strong mitotic phenotypes, including disorganized metaphase plates and hypercondensed chromosomes. A low frequency of cells with lagging chromosomes during anaphase, aneuploidy and polyploidy were also observed. The strong mitotic phenotypes are consistent with the hypothesis that Sgt1 is most likely to have essential functions during cell division. Similarly, human cells depleted of Sgt1 by RNAi show similar mitotic phenotypes, including chromosome hypercondensation, failure in chromosome alignment and mitotic delay (Steensgaard et al, 2004).
Sgt1 is not required for activation or maintenance of the SAC
Studies in yeast (Bansal et al, 2004; Rodrigo-Brenni et al, 2004) and RNAi experiments in human cells (Steensgaard et al, 2004) have suggested that Sgt1 might be involved in kinetochore assembly. Furthermore, it was shown that reduction in Sgt1 levels leads to a weak SAC response and to the mislocalization of checkpoint proteins. However, this weak checkpoint is able to delay mitotic cells for several hours, and only when Mad2 was co-depleted, cells no longer arrested in mitosis, suggesting that even with reduced levels of kinetochore proteins, these cells have an active SAC (Steensgaard et al, 2004). These observations lead to the proposal that Sgt1 is essential for kinetochore assembly but not for the normal activation of SAC. Unlike human cells, we find that in Drosophila sgt1 mutant neuroblasts, spindle checkpoint proteins, such as BubR1 and Bub3, do accumulate normally at kinetochores. However, similar to HeLa cells (Steensgaard et al, 2004), when mutant Drosophila tissue is incubated in colchicine, the mitotic index does not increase as in controls. We propose that the mitotic index does not increase in response to microtubule depolymerization mostly because Sgt1 mutant cells progress through the cell cycle more slowly and therefore enter mitosis less frequently than control cells. In support of this explanation, our analysis of mitotic progression showed that mutant cells have a significant reduction in the number of prophases when compared with control cells. Moreover, sgt1 mutant cells do not progress through the cell cycle normally, as fewer cells incorporate BrdU or accumulate cyclin B. Accordingly, genetic analysis in yeast showed that mutant cells show a delay at the G1/S transition (Kitagawa et al, 1999). These observations strongly suggest that in the absence of Sgt1, cells are stalled early in interphase, and when they eventually enter mitosis, they become arrested in prometaphase for long periods, similarly to what was observed in human cells (Steensgaard et al, 2004). Also, we find that in Drosophila, this transient arrest is clearly SAC dependent, as sgt1PI:bub3 double mutants do not arrest in prometaphase nor show hypercondensed chromosomes, but show premature sister chromatid separation and accelerated mitotic exit. Accordingly, we conclude that Sgt1 must have an essential function during interphase to promote cell cycle progression and is also required during mitosis to promote normal mitotic progression.
Sgt1 is not required for kinetochore assembly
In yeast and human cells, Sgt1 has been shown to be required for kinetochore assembly, a requirement we did not observe in Drosophila. Interestingly, treatment of HeLa cells with 17-AAG, an HSP90 inhibitor, resulted in failure to localize several kinetochore proteins (Niikura et al, 2006). Nonetheless, when these cells are analysed by electron microscopy, the structure of the kinetochore is not affected and kinetochores bind microtubules normally (Niikura et al, 2006). Therefore, if Sgt1 has an important function as co-chaperone of Hsp90, this is unlikely to include important kinetochore components. Accordingly, in Drosophila, sgt1P1 cells arrest in prometaphase with an active SAC response and accumulate normal levels of SAC proteins at the kinetochore. Moreover, all the structural components of the kinetochore that we analysed localized normally and at similar levels to controls, with the exception of Polo kinase (see later on this discussion). We consider two possible explanations for these observations. It is possible that in Drosophila the kinetochore assembly pathway is very different from the one described in yeast and humans, being Sgt1 independent, which is very unlikely, given the high conservation of kinetochore proteins across these organisms (Przewloka et al, 2007; Schittenhelm et al, 2007). Alternatively, given that the Drosophila Sgt1 lacks the TPR domain, it is possible that other TPR-containing proteins interact with HSP90, thus fulfilling the requirements for kinetochore assembly. Drosophila has several proteins containing the TPR domain but none have the SGS domain characteristic of Sgt1-like proteins. Therefore, as in Drosophila, many proteins have been shown to diverge significantly from their yeast or human homologues or even divided the function into two separate proteins, as in the case of separase (Jager et al, 2001) it is possible that another protein performs this kinetochore-specific function.
Absence of Sgt1 results in the failure of centrosome maturation
sgt1 mutant cells show severe mitotic abnormalities in the organization of the mitotic apparatus. We find significant alterations in the spindle organization, namely monopolar spindles, dispersion of the PCM component γ-tubulin and also cells containing only a single centrosome during mitosis. These observations, taken together with its centrosomal localization during mitosis, suggest that Sgt1 might be required for proper organization of the centrosome. Indeed, the abnormal centrosome phenotypes are highly reminiscent to those observed after mutation or inhibition of Hsp90 function in Drosophila and human cells, suggesting that the interaction between Hsp90 and Sgt1 might be impaired (Lange et al, 2000). As Hsp90 has been clearly shown to be involved in the organization and function of centrosomes, we speculate that the interaction between Sgt1 and Hsp90 might underline this process. Likewise, an impairment of the interaction between these two proteins would lead to abnormalities in the establishment of a stable bipolar spindle preventing proper microtubule–kinetochore interaction, which in turn would activate the SAC causing a prometaphase arrest. Moreover, this failure in centrosome organization and function is also most likely to explain the high prevalence of spindle defects observed when Sgt1 protein is absent. However, analysis of centriole components clearly show that loss of Sgt1 function does not result in abnormal centriole duplication. We can conclude that the most significant aspect, mitotic arrest, is most likely to be the result of SAC activation due to abnormal spindles resulting from abnormal centrosomes that fail to mature properly.
Sgt1 stabilizes Polo
Centrosome maturation has been shown to be directly dependent on the function of the Polo kinase (Sunkel and Glover, 1988). Interestingly, we found that in the absence of Sgt1, Polo protein is destabilized and its levels reduce, both at kinetochores and in total protein extracts. As Sgt1 is a co-chaperone of Hsp90 and depletion or inhibition of Hsp90 in Drosophila S2 cells result in low levels of Polo protein with mitotic phenotypes similar to sgt1P1 cells (de Carcer et al, 2001), we speculate that Sgt1 might be important in directing Hsp90 to Polo for its stabilization and proper physiological conformation. Indeed, when Polo is overexpressed in the absence of Sgt1, we observe that its levels do not increase as much as in controls, supporting the idea that Sgt1 is required for its stabilization. However, overexpression of Polo is sufficient for a significant recovery of the Sgt1 mitotic phenotype, suggesting a direct interaction between Sgt1 and Polo. By contrast, in human cells treated with the Hsp90 inhibitor 17-AAG, Plk1 levels remain constant, suggesting that human cells might have backup mechanisms to stabilize this important kinase.
Taken together, our results suggest that in Drosophila, Sgt1 has at least two essential functions, during interphase to promote transit from G1 to S-phase and later in mitosis to promote the stability of Polo kinase.
Materials and methods
Drosophila stocks
W1118 was used as control strain (Bloomington Stock Center, Indiana, USA). Two mutant alleles C01268 and C01428 were obtained from Exelixis Corporation (CA, USA). These lines carry a Piggy Bac transposable element insertion at nucleotide +55 bp upstream of the initiator ATG of sgt1 gene. The mutant allele bub31 (Lopes et al, 2005) was recombined into a chromosome carrying the C01268 mutant allele of Sgt1 to test SAC activity. For in vivo studies, strains were constructed to carry the following transgenes: Cid–mRFP (Schuh et al, 2007), GFP–Tub (Rebollo et al, 2004) and PACT–GFP (Martinez-Campos et al, 2004). To obtain revertants of C01268, this strain was crossed with w1118; CyO, P{Tub-PBacT}2/wgSp-1 (Bloomington Stock Center) and all white-eyed progeny selected. To carry out overexpression of Polo, a UAS-Polo transgene was used (Mirouse et al, 2006) and the expression was induced with the ubiquitous driver daGal4. All stocks were grown at 25°C on standard culture conditions.
Cytological analysis
Third instar larvae brains were dissected in 0.7% NaCl solution and fixed in 45% acetic acid followed by 60% acetic acid. DNA was counterstained with DAPI in Vectashield (Vector, UK). When required, brains were incubated with 10 μM of colchicine (Sigma) before fixation. Mitotic index was determined as the percentage of cells in mitosis over the total cell population. At least 2000 cells were scored from different preparations. All quantifications were performed on a Zeiss Axioskop microscope (Carl Zeiss, Germany) using 1000 × magnification.
Immunostaining of Drosophila neuroblasts
For immunostaining, larval brains were dissected in 0.7% NaCl, fixed in 2% formaldehyde together with 0.1% Triton X-100 in PBS and then transferred to a solution of 45% acetic acid containing 2% formaldehyde. Brains were squashed and immersed in liquid nitrogen followed by permeabilization in PBS with 0.05% Tween 20. Blocking and incubation of primary and secondary antibodies was done in PBS-BT (1% BSA and 0.1% Triton X-100). For immunofluorescence analysis of specific spindle components, brains were dissected, as described previously and fixed in 3.7% formaldehyde in PBS, transferred to 45% acetic acid and then to 60% acetic acid. After squashing and immersion in liquid nitrogen, slides were transferred to absolute ethanol at −20°C. Permeabilization was done in PBS with 0.1% Triton X-100. Blocking and incubation of primary and secondary antibodies was done in PBS with 1% BSA. Images were collected in a Zeiss Axiovert 200M microscope (Carl Zeiss, Germany) using an Axiocam (Carl Zeiss, Germany). Data stacks were deconvolved using the Huygens Essential version 3.0.2p1 (Scientific Volume Imaging BV, The Netherlands). All the images were projected with ImageJ (http://rsb.info.nih.gov) and processed with Photoshop CS (Adobe Microsystems, CA, USA).
Antibodies
The primary antibodies used were anti-α-tubulin mouse B512 (Sigma) used at 1:1000; anti-γ-tubulin mouse GTU88 (Sigma) used at 1:250; anti-Polo mouse monoclonal MA294 (Llamazares et al, 1991) used at 1:10; anti-BubR1 Rb666 rabbit polyclonal (Logarinho et al, 2004) used at 1:2000; anti-CID rat polyclonal (CE Sunkel, unpublished data) used at 1:1500; anti-CENP-meta rabbit used at 1:500; anti-Bub3 (Logarinho et al, 2004) used at 1:500; anti-CENP-C (Heeger et al, 2005) used at 1:4000; anti-CNN (Heuer et al, 1995) used at 1:750; and CP190 (Whitfield et al, 1988) used at 1:500. Antibodies used for immunofluorescence in whole brains were anti-BrdU (Sigma, clone BU-33) used at 1:80 and anti-cyclinB Clone F2F4 (Lehner and O'Farrell, 1990) used at 1:15. Antibodies used for western blot were anti-Polo mouse monoclonal MA294 (Llamazares et al, 1991) used at 1:100, anti-Sgt1 used at 1:1000 and anti-tubulin DM1A (Sigma) used at 1:5000. Antibody against Drosophila Sgt1 was produced by cloning part of the cDNA RH27607 (20–178 amino acids) (Drosophila Genomics Resource Centre) in the pET23a vector. The His-tagged fusion protein was expressed in Escherichia coli BL21 cells, and after purification it was injected in two different rabbits. Antibody specificity for Sgt1 was confirmed by the appearance of the band at the predicted size (23 KDa) that was not observed in the pre-immune serum. The secondary antibodies were obtained from Molecular Probes and Jackson Immunoresearch and used according to the manufacturer's instructions.
Western blot analysis
For western blot analysis, brains were dissected from third instar larvae and collected in sample buffer (10% glycerol, 5% β-mercaptoethanol, 2% SDS, Bromophenol blue in Tris). Samples were separated by SDS–PAGE, transferred to a PVDF membrane using a semi-dry system and blocked overnight at 4°C with 0.5% gelatin from cold water fish skin (FSG) (Sigma), 1% BSA and 8% low-fat milk in TBS 0.05% Tween. Membranes were then incubated with the primary and secondary antibodies diluted in 0.5% FSG and 1% BSA in TBS 0.05% Tween. After antibody incubation, membranes were developed with ECL.
Transfection of S2 cells
Drosophila S2 cells were grown to a yield of 3 × 106 cells in 3 ml of Schneider medium (Sigma) with 10% fetal bovine serum (FBS) at 25°C and then transfected with pMT-EGFP-Sgt1 using calcium phosphate. Expression of the EGFP–Sgt1 construct was induced with CuSO4 (1 mM), and cells were collected after 16 h for immunofluorescence analysis. Where indicated, cells were treated with 10 μM of colchicine for the desired period. The pMT-EGFP-Sgt1 vector was generated by cloning Sgt1 full-length cDNA into the HincII/SacI sites of the MCS of pMT-EGFP-C1 plasmid.
Whole-mount preparations
To detect DNA replication, third instar larvae neuroblasts were dissected in PBS, incubated in 10 μM BrdU (Roche) diluted in PBS, fixed in 5% formaldehyde, then washed in PBST and hydrolysed in 2.2 N HCl. The tissue was then washed with 100 mM sodium tetraborate and PBST. Preparations were blocked in PBST containing 10% FBS and then incubated with a monoclonal BrdU antibody. Anti-mouse Alexafluor 568 was used as secondary antibody. To detect cyclin B, third instar larvae neuroblasts were dissected in 0.7% NaCl, fixed in 3.7% formaldehyde, washed and blocked in 0.7% NaCl with 10% FBS, 0.3% Triton X-100 and RNAse (10 μg/ml). Preparations were incubated with primary antibody and secondary antibodies in the blocking solution and then incubated with topro3 (Molecular Probes) to label the DNA. Imaging was done using a Laser Scanning Confocal Microscope Leica SP2 AOBS SE (Leica Microsystems, Germany). For BrdU quantifications, images of the whole brains were acquired and then used ImageJ to define a threshold for positive cells. Quantification was done in every stack acquired, and then the ratio between BrdU-positive cells and total number of cells was calculated.
In vivo studies
Third instar larvae were dissected in PBS and then transferred to a drop of Schneider medium (Invitrogen) for appropriate incubation. Cells were imaged using a Spinning Disk Confocal System Andor Revolution XD (ANDOR Technology, UK). Frames were collected every 30 s. Time-lapse images were then treated with the microscope software IQ 1.7 (ANDOR Technology, UK).
Supplementary Material
Supplementary movie 1
Supplementary movie 2
Supplementary movie 3
Supplementary movie 4
Supplementary Information
Acknowledgments
We thank M Goldberg, S Heidmann, S Llamazares and J Raff for stocks and all the members of the lab for comments and suggestions during the course of this work. TM and AM hold PhD fellowships from the Fundação para a Ciência e a Tecnologia (FCT) of Portugal. Work in the laboratory of CE Sunkel is financed by project grants from FCT.
References
- Bansal PK, Abdulle R, Kitagawa K (2004) Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol Cell Biol 24: 8069–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett MG, Kaplan KB (2006) Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J Biol Chem 281: 33739–33748 [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- de Carcer G, do Carmo Avides M, Lallena MJ, Glover DM, Gonzalez C (2001) Requirement of Hsp90 for centrosomal function reflects its regulation of Polo kinase stability. EMBO J 20: 2878–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson MM, Tavares AA, Ohkura H, Deak P, Glover DM (2001) Metaphase arrest with centromere separation in polo mutants of Drosophila. J Cell Biol 153: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, Lehner CF (2005) Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev 19: 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer JG, Li K, Kaufman TC (1995) The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development 121: 3861–3876 [DOI] [PubMed] [Google Scholar]
- Jager H, Herzig A, Lehner CF, Heidmann S (2001) Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev 15: 2572–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ (2003) A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 425: 407–410 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Lange BM, Bachi A, Wilm M, Gonzalez C (2000) Hsp90 is a core centrosomal component and is required at different stages of the centrosome cycle in Drosophila and vertebrates. EMBO J 19: 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Jacob J, Michowski W, Nowotny M, Kuznicki J, Chazin WJ (2004) Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J Biol Chem 279: 16511–16517 [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH (1990) The roles of Drosophila cyclins A and B in mitotic control. Cell 61: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach LB, Kaplan KB (2004) The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol Cell Biol 24: 8938–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE (1991) polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev 5: 2153–2165 [DOI] [PubMed] [Google Scholar]
- Logarinho E, Bousbaa H, Dias JM, Lopes C, Amorim I, Antunes-Martins A, Sunkel CE (2004) Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J Cell Sci 117: 1757–1771 [DOI] [PubMed] [Google Scholar]
- Lopes CS, Sampaio P, Williams B, Goldberg M, Sunkel CE (2005) The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J Cell Sci 118: 187–198 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW (2004) The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouse V, Formstecher E, Couderc JL (2006) Interaction between Polo and BicD proteins links oocyte determination and meiosis control in Drosophila. Development 133: 4005–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Niikura Y, Ohta S, Vandenbeldt KJ, Abdulle R, McEwen BF, Kitagawa K (2006) 17-AAG, an Hsp90 inhibitor, causes kinetochore defects: a novel mechanism by which 17-AAG inhibits cell proliferation. Oncogene 25: 4133–4146 [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, Laue ED, McAinsh AD, Glover DM (2007) Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2: e478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Llamazares S, Reina J, Gonzalez C (2004) Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol 2: E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Thomas S, Bouck DC, Kaplan KB (2004) Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol Biol Cell 15: 3366–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF (2007) Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116: 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S (2007) Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17: 237–243 [DOI] [PubMed] [Google Scholar]
- Steensgaard P, Garre M, Muradore I, Transidico P, Nigg EA, Kitagawa K, Earnshaw WC, Faretta M, Musacchio A (2004) Sgt1 is required for human kinetochore assembly. EMBO Rep 5: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM (1988) polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci 89: 25–38 [DOI] [PubMed] [Google Scholar]
- Whitfield WG, Millar SE, Saumweber H, Frasch M, Glover DM (1988) Cloning of a gene encoding an antigen associated with the centrosome in Drosophila. J Cell Sci 89: 467–480 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary movie 1
Supplementary movie 2
Supplementary movie 3
Supplementary movie 4
Supplementary Information