Abstract
Cloning by somatic cell nuclear transfer can result in the birth of animals with phenotypic and gene expression abnormalities. We compared adult cloned pigs and adult pigs from naturally bred control females using a series of physiological and genetic parameters, including detailed methylation profiles of selected genomic regions. Phenotypic and genetic analyses indicated that there are two classes of traits, one in which the cloned pigs have less variation than controls and another characterized by variation that is equally high in cloned and control pigs. Although cloning creates animals within the normal phenotypic range, it increases the variability associated with some traits. This finding is contrary to the expectation that cloning can be used to reduce the size of groups involved in animal experimentation and to reproduce an animal, including a pet, with a homogenous set of desired traits.
Keywords: assisted reproductive technology, developmental biology, embryo, gene regulation
INTRODUCTION
Cloning by somatic cell nuclear transfer can result in the birth of normal cattle offspring [1]; however, many pregnancies are lost throughout gestation and many calves die soon after birth [2]. Although the rates of pregnancy loss and fetal and placental abnormalities vary among species [3], available evidence suggests that the cloning process introduces some epigenetic defects that can lead to developmental abnormalities [4]. Germline epigenetic modifications characterized by the addition of a methyl group to a cytosine nucleotide adjacent 5′ to a guanine nucleotide are initiated during differentiation of germ cells into either spermatozoa or oogonia and are stably maintained throughout development of the offspring [5]. Establishment of imprinted genes [6], X-chromosome inactivation in females [7], suppression of retrotransposons [8], chromatin stability [9], and inactivation of tissue-specific promoters [10] are examples of epigenetic modifications imparted to the genome during gametic and embryonic development. Introduction of the differentiated somatic nucleus in nuclear transfer, however, results in a genomic template different from that characteristic of the sperm and oocyte. Therefore, following nuclear transfer, the reprogramming machinery of the cytoplast must attempt to reorganize this new nucleus into a functional, pluripotent genome capable of carrying out stage and developmentally specific events required for proper differentiation of placental and fetal cell lineages. Characterization of both genome-wide and locus-specific methylation patterns in cloned animals indicates improper or incomplete reprogramming at some loci, demonstrating that either the reprogramming process or the epigenetic state of the donor cell influences the patterns of methylation [11–13].
Although in some previous studies, adult cloned mice have shown abnormal gene expression profiles and global methylation differences [4, 14], in other studies adult cloned animals have been healthy and normal [1, 15]. To clarify some of these issues, we compared cloned pigs with naturally bred control pigs on a series of physiological and genetic parameters, including a detailed methylation profile of selected genomic regions. To properly account for environmental effects versus epigeneic effects, clones were compared with age-, breed-, and sex-matched naturally bred controls. For physiological measurements, multiple blood parameters were selected because they provide an over-view of the physiological status of the animal, are widely used in diagnostic settings, and allow comparison of normal and abnormal values. The genomic regions selected, PRE-1 SINE and centromeric satellites, provide an excellent overview of methylation status in euchromatic or heterochromatic regions of the genome. A hierarchical set of phenotypic and epigenetic effects were found in cloned swine, with some phenotypes and genomic regions being differentially affected in cloned versus noncloned pigs.
MATERIALS AND METHODS
Animals
Two litters of cloned female Duroc swine (clone 1 and clone 2), consisting of five and four pigs, respectively, and two control litters of four purebred female Duroc pigs each (control 1 and control 2) were used in this study. Clone 1 and clone 2 were born 6 wk apart. The cloned litters of piglets were from the same fetal cell line and were age, breed, and sex matched to litters of piglets from their naturally bred Duroc counterparts. The clones were produced as described previously and were confirmed to be genetically identical using microsatellite DNA analysis [16]. The pure-bred control swine obtained from a local breeder consisted of a litter of full sibs (control 1) and a litter of half-sibs from three sows mated to the same boar (control 2). All swine were farrowed in conventional farrowing crates and weaned at 5–6 wk of age. At weaning, cloned and control pigs were placed in adjacent identical pens (1.8 × 6 m) containing a bedded sleeping area and were given continuous access (ad libidum) to a standard commercial ration and water.
PCR Amplification of Mitochondrial Hypervariable Region
For amplification of the swine D-loop, polymerase chain reaction (PCR) assays were performed on total DNA extracted from control and cloned pig tissue samples. Primer sets used to amplify the swine hyper-variable region were 5′-TCCCCAAGACTCAAGGAAGGAG-3′ and 5′-GGCTGGCACGAGATTTACCAAC-3′. Ten microliters of the PCR product were electrophoresed on a 1.5% ethidium bromide-agarose gel to confirm sizes, and amplicons were cloned into the TOPO4 Sequencing Vector (Invitrogen, Carlsbad, CA). A minimum of three colonies/pig were isolated, sequenced on an ABI Prism 3700 (Applied Biosystems, Foster City, CA), and analyzed using a MacVector Clustal arrangement (Accelrys, San Diego, CA). All sequences were compared with the haplotype of the control 1 animals, and similarity over a 520-base pair (bp) area was expressed as a percentage.
Physical Measurements
All piglets were weighed at 27 wk of age. Matched litters were weighed and measured on the same day.
Blood Parameters
Blood (15 ml) was collected via jugular venipuncture at 15 and 27 wk of age. A serum chemistry profile was generated by the Veterinary Diagnostic Laboratory (Texas A&M University, College Station, TX) that included creatinine, alkaline phosphatase (ALP), blood urinary nitrogen (BUN), serum glutamate pyruvate transaminase (SGPT), albumin, phosphorus, calcium, serum proteins, glucose, globulins, and albumin:globulin (A:G) ratio. Basal cortisol and total triiodothyronine (T3) concentrations were determined using commercially available RIA kits (Diagnostic Procedure Company, Costa Mesa, CA). Duplicates that differed by >7% were reassayed. For the cortisol assay, the intraassay coefficient of variation (CV) averaged 0.96% and the interassay CV was 3.4%. The T3 determinations were from a single assay with an intraassay CV of 4%. Matched litters had blood taken on the same day within a 1-h period. For statistical analysis, the degree of variation between clones and controls was compared using F-values. Differences were considered significant at P < 0.05.
Additional Phenotypic Analyses
The number of teats for all control and cloned pigs were recorded. For skin morphology, skin punches obtained approximately 5 cm lateral to the tail were fixed in 10% formalin and processed for histological analysis, and serial sections were stained with hematoxylin and eosin. If needed, a second sample was obtained from an adjacent region of skin.
Genomic DNA Isolation and Bisulfite Treatment
Tissue samples were taken from skin punches of cloned and control pigs. Genomic DNA was isolated using a Wizard Genomic DNA Purification kit (Promega, Madison, WI). Bisulfite conversion of unprotected cytosine nucleotides to uracil (thymine) nucleotides was performed using a CpG Bisulfite Conversion Kit (Serologicals Corporation/Intergen, Nor-cross, GA). Bisulfite-treated DNA was resuspended in double-distilled water and stored at −20°C for later use.
PCR Amplification, Cloning, and Sequencing of Bisulfite-Converted DNA
For amplification of the PRE-1 SINE region, the sequence was obtained from GenBank (X64127, Y00104, and AJ251914). Primer sets used to amplify PRE-1 SINE were 5′-TTAACRAATCCRACTAAAAACCATA-3′ and 5′-GTTGGTTTATMTTAGAGTTATAGTAA-3′ The amplification parameters were 45 cycles at 94°C for 60 sec, 55°C for 60 sec, and 69°C for 1 min, finishing with an extension of 72°C for 10 min [13]. The centromeric satellite sequence was obtained from GenBank (Z75640). Primers used to amplify centromeric satellite DNA were 5′TTTGTAGAATGTAGTTTTTAGAAG-3′ and 5′-AAAATCTAAACTACCTCTAACTC-3′. The amplification parameters were 45 cycles at 94°C for 60 sec, 55°C for 60 sec, and 69°C for 1 min, with a final extension of 72°C for 10 min [13]. The PCR products (10 µl) were electrophoresed on a 2% ethidium bromide gel to confirm sizes and then cloned into the TOPO4 PCR Sequencing Vector (Invitrogen).
Analysis of CpG Methylation of the PRE-1 SINE and Centromeric Satellite Repeat Regions
The PRE-1 SINE and centromeric satellite were amplified from bisulfite-converted DNA and cloned into the TOPO4 sequencing vector (Invitrogen). Twenty colonies were isolated, and plasmids were purified from the PRE-1 SINE and centromeric satellite transformations for each of the cloned and control pigs. Sequences were analyzed to extrapolate only informative sequences, i.e., those containing complete conversion of non-CpG cytosine to thymine and yielding high-quality sequence. Sequences for the PRE-1 SINE and centromeric satellites for each animal were aligned using MacVector Clustal W (Accelrys). At least 12 fully converted sequences were analyzed per animal. Thirteen and nine CpG pairs were conserved among the sequences of PRE-1 SINE and centromeric satellites, respectively. Presence of CpG and TpG dinucleotides within these conserved regions was used to assess percent methylation at a particular CpG region. The percent methylation at each CpG dinucleotide and total methylation of the repeat element (percent methylation among all CpG dinucleotides within an animal) were determined. Mean comparisons were made with a one-way ANOVA. Comparison of variation between two groups was determined with an F-test. Differences were considered significant at P < 0.05.
RESULTS
Mitochondrial Typing
Sequence analysis revealed that mitochondrial types did not differ in the control 1 group, as expected for full siblings (data not shown). In the control 2 group, three different types of mitochondrial backgrounds were identified, as expected of half-siblings originating from three gilts mated to the same boar. The similarity between the three mitochondrial types was 99.2% across a 520-bp region of the D-loop, suggesting that the mitochondria were highly similar. When this analysis was extended to the cloned pigs, the similarity in identity to the control 1 group across the same 520-bp region ranged from 96.3% to 100%, with two cloned pigs having identical D-loop sequences to the controls, indicating that the mitochondrial backgrounds for both cloned and control pigs was very similar and unlikely to result in major phenotypic differences.
Phenotypic Comparisons
Body weights of the pigs were determined at 27 wk of age to compare variation among cloned and control pigs. When the relative CVs for body weights were compared, no differences between cloned and control pigs were detected. Examination of the range in body weights (Table 1) indicated that with the exception of a single cloned pig, values for cloned and control pigs overlapped, indicating all were in the normal range for swine of this age.
TABLE 1.
Body weights at 27 wk of age for cloned and control pigs.
| Clone 1 | Control 1 | Clone 2 | Control 2 | |||
|---|---|---|---|---|---|---|
| Weight (kg) | (n = 5) | (n = 4) | (n = 4) | (n = 4) | Clone 1 + 2 | Control 1 + 2 |
| Mean ± SD | 95.1 ± 8.8 | 99.8−7.3 | 97.7−7.6 | 108.3 ± 5.1 | 96.2 ± 7.9 | 104.1 ± 7.4 |
| Range | 81.6−102.1 | 90.2−106.6 | 89.8−104.8 | 101.6−113.9 | 81.6−104.8 | 90.2−113.9 |
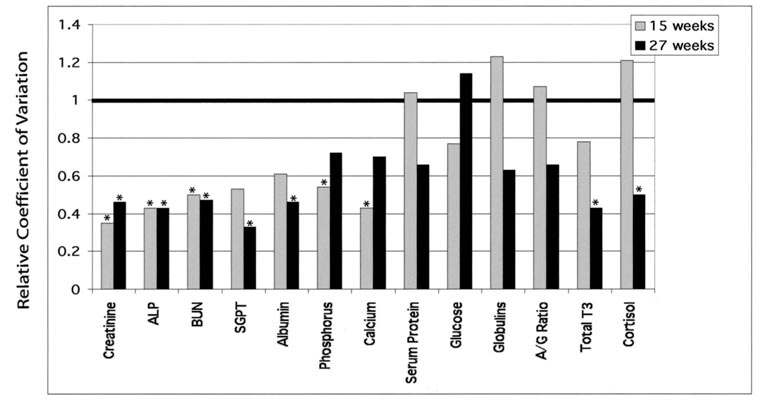
In contrast to results from body weight measurements, variation in blood profiles analyzed at 15 and 27 wk indicated a more complex interaction between cloning effects and phenotype. There were two groups of traits: one in which cloned pigs had less variability than controls, and one in which clones had the same variability as control pigs (Fig. 1). Creatine, ALP, and BUN were less variable in cloned than control pigs at both 15 and 27 wk. However, SGPT, albumin, phosphorus, and calcium were less variable in cloned pigs at one of the two times periods, and there was a nonsignificant trend toward reduced variability at the other time period. Serum proteins, glucose, globulins, and A:G ratio were equally variable in cloned and control pigs. Both T3 and cortisol were unique in that there was not reduced variability at Week 15, but there was reduced variability at Week 27. Although the variation in blood profiles showed two different patterns, with some traits showing reduced variability and others showing the same or increased variability, examination of the range of values for all traits at 15 and 27 wk of age (Table 2 and Table 3) indicated that values for control and cloned pigs overlapped.
FIG. 1.
Variation in blood parameters between cloned and control pigs. Blood was collected for age-matched cloned and control pigs at 15 and 27 wk of age, and the following parameters measured: albumin, phosphorus, creatinine, total T3, ALT (=SGPT), ALP, calcium, BUN, serum protein, glucose, aspartate aminotransferase (AST), globulins, A:G ratio, and cortisol. Values of <1 indicate reduced variability for cloned pigs. *Significance was determined by an F-test, P < 0.05.
TABLE 2.
Blood profiles* in cloned and control pigs at 15 wk of age.
| Parameter | Clone 1 | Control 1 | Clone 2 | Control 2 | Clone 1 + 2 | Control 1 + 2 |
|---|---|---|---|---|---|---|
| Creatinine (mg/dl) | 1.08 ± 0.21 | 1.63 ± 1.32 | 0.95 ± 0.24 | 1.53 ± 0.59 | 1.02 ± 0.22 | 1.58 ± 0.95 |
| (0.9−1.4) | (0.9−3.6) | (0.7−1.2) | (0.8−2.1) | (0.7−1.4) | (0.8−3.6) | |
| ALP (U/L) | 213.40 ± 10.31 | 210.50 ± 9.71 | 202.75 ± 11.53 | 260.00 ± 28.83 | 208.67 ± 11.60 | 235.25 ± 33.12 |
| (206−226) | (201−223) | (192−218) | (225−294) | (192−226) | (201−294) | |
| BUN (mg/dl) | 10.18 ± 1.73 | 11.75 ± 2.16 | 9.08 ± 0.85 | 7.40 ± 1.23 | 9.69 ± 1.45 | 9.58 ± 2.84 |
| (7.7−11.9) | (8.9−13.9) | (8.0−10.0) | (6.3−7.7) | (7.7−11.9) | (6.3−11.9) | |
| ALT (SGPT)† (U/L) | 47.40 ± 5.41 | 58.25 ± 8.27 | 46.00 ± 2.71 | 48.25 ± 7.81 | 46.78 ± 4.24 | 53.25 ± 9.16 |
| (46−56) | (52−70) | (44−50) | (41−58) | (46−56) | (41−70) | |
| Albumin (g/dl) | 4.18 ± 0.08 | 4.38 ± 0.17 | 4.25 ± 0.17 | 4.23 ± 0.28 | 4.21 ± 0.13 | 4.40 ± 0.21 |
| (4.1−4.3) | (4.2−4.6) | (4.0−4.3) | (4.1−4.7) | (4.0−4.3) | (4.1−4.7) | |
| Phosphorus (mg/dl) | 10.26 ± 0.55 | 10.425 ± 1.00 | 10.33 ± 0.26 | 11.08 ± 0.53 | 10.29 ± 0.42 | 10.75 ± 0.82 |
| (9.6−10.4) | (9.5−11.8) | (10.1−10.6) | (10.6−11.8) | (9.6−10.6) | (9.5−11.8) | |
| Calcium (mg/dl) | 11.44 ± 0.23 | 11.90 ± 0.22 | 11.13 ± 0.10 | 11.08 ± 0.50 | 11.30 ± 0.24 | 11.49 ± 0.57 |
| (11.1−11.7) | (11.7−12.2) | (11.0−11.2) | (10.4−11.5) | (11.0−11.7) | (10.4−12.2) | |
| Serum protein (g/dl) | 6.10 ± 0.26 | 5.90 ± 0.33 | 6.65 ± 0.13 | 6.28 ± 0.21 | 6.34 ± 0.35 | 6.09 ± 0.32 |
| (5.7−6.4) | (5.9−6.3) | (6.5−6.8) | (6.0−6.5) | (5.7−6.8) | (5.9−6.5) | |
| Glucose (mg/dl) | 98.60 ± 13.60 | 109.50 ± 4.65 | 103.00 ± 3.7 | 122.25 ± 19.69 | 100.56 ± 10.03 | 115.88 ± 14.89 |
| (78−113) | (105−116) | (101−108) | (109−151) | (101−113) | (105−151) | |
| Globulins (g/dl) | 1.92 ± 0.19 | 1.53 ± 0.17 | 2.40 ± 0.27 | 1.85 ± 0.13 | 2.13 ± 0.33 | 1.69 ± 0.22 |
| (1.6−2.1) | (1.3−1.7) | (2.2−2.8) | (1.7−2.0) | (1.6−2.8) | (1.3−2.0) | |
| A : G ratio | 2.19 ± 0.22 | 2.89 ± 0.25 | 1.79 ± 0.24 | 2.41 ± 0.29 | 2.01 ± 0.30 | 2.65 ± 0.36 |
| (2.05−2.56) | (2.69−3.23) | (1.43−1.95) | (2.15−2.71) | (1.43−2.56) | (2.15−3.23) | |
| Total T3 (ng/dl) | 73.22 ± 14.21 | 90.37 ± 21.21 | 68.12 ± 2.73 | 100.59 ± 14.96 | 70.95 ± 10.05 | 95.48 ± 17.85 |
| (60.99−92.99) | (74.12−119.51) | (65.34−71.57) | (86.53−120.07) | (60.09−92.99) | (74.12−120.07) | |
| Cortisol (g/dl) | 4.46 ± 2.85 | 7.63 ± 2.43 | 6.93 ± 1.27 | 5.50 ± 2.09 | 5.56 ± 2.52 | 6.56 ± 2.39 |
| (1.2−8.9) | (5.5−10.9) | (5.1−7.4) | (3.1−7.9) | (1.2−8.9) | (3.1−10.9) |
Mean ± SD (range in parentheses).
ALT, Alanine aminotransferase (=SGPT).
TABLE 3.
Blood profiles* in cloned and control pigs at 27 wk of age.
| Parameter | Clone 1 | Control 1 | Clone 2 | Control 2 | Clone 1 + 2 | Control 1 + 2 |
|---|---|---|---|---|---|---|
| Creatinine (mg/dl) | 1.02 ± 0.08 | 1.00 ± 0.12 | 1.23 ± 0.10 | 1.50 ± 0.25 | 1.11 ± 0.14 | 1.25 ± 0.32 |
| (0.9−1.1) | (0.9−1.1) | (1.1−1.3) | (1.2−1.8) | (0.9−1.3) | (0.9−1.8) | |
| ALP (U/L) | 87.00 ± 6.75 | 84.25 ± 28.25 | 118.00 ± 8.98 | 151.50 ± 43.41 | 100.78 ± 17.89 | 117.88 ± 49.54 |
| (80−95) | (56−123) | (109−128) | (101−196) | (80−128) | (56−196) | |
| BUN (mg/dl) | 10.26 ± 1.44 | 6.58 ± 0.84 | 9.88 ± 1.26 | 9.13 ± 2.17 | 10.09 ± 1.29 | 7.85 ± 2.04 |
| (8.9−11.6) | (5.8−7.4) | (8.9 ± 11.7) | (6.4−11.7) | (8.9−11.7) | (5.8−11.7) | |
| ALT (SGPT)† (U/L) | 38.40 ± 1.67 | 37.75 ± 11.15 | 38.50 ± 3.70 | 40.00 ± 5.83 | 38.44 ± 2.55 | 38.88 ± 8.32 |
| (36−40) | (22−47) | (34−42) | (34−48) | (34−42) | (22−48) | |
| Albumin (g/dl) | 4.00 ± 0.29 | 3.85 ± 0.65 | 4.28 ± 0.10 | 4.45 ± 0.21 | 4.12 ± 0.26 | 4.15 ± 0.55 |
| (3.6−4.3) | (3.0−4.4) | (4.2−4.4) | (4.2−4.7) | (3.6−4.3) | (3.0−4.7) | |
| Phosphorus (mg/dl) | 8.12 ± 0.72 | 7.75 ± 1.20 | 7.55 ± 0.19 | 7.75 ± 0.49 | 7.87 ± 0.60 | 7.75 ± 0.85 |
| (7.0−8.87) | (6.1−8.9) | (7.4−7.8) | (7.2−8.3) | (7.0−8.8) | (6.1−8.9) | |
| Calcium (mg/dl) | 10.80 ± 0.10 | 10.35 ± 0.58 | 12.38 ± 0.13 | 12.35 ± 0.33 | 11.50 ± 0.84 | 11.35 ± 1.12 |
| (10.7−10.9) | (9.5−10.8) | (12.2−12.5) | (11.9−12.7) | (10.7−12.5) | (9.5−12.7) | |
| Serum protein (g/dl) | 7.24 ± 0.27 | 7.45 ± 0.55 | 6.60 ± 0.34 | 6.55 ± 0.10 | 6.96 ± 0.44 | 7.00 ± 0.60 |
| (7.0−7.7) | (7.0−8.2) | (6.2−7.0) | (6.4−6.6) | (6.2−7.7) | (6.4−8.2) | |
| Glucose (mg/dl) | 83.40 ± 7.67 | 94.75 ± 6.85 | 91.25 ± 2.75 | 103.50 ± 5.45 | 86.89 ± 7.03 | 99.13 ± 7.40 |
| (70−88) | (87−101) | (88−94) | (98−107) | (70−94) | (87−107) | |
| Globulins (g/dl) | 3.24 ± 0.31 | 3.60 ± 0.60 | 2.33 ± 0.34 | 2.10 ± 0.14 | 2.83 ± 0.57 | 2.85 ± 0.89 |
| (2.9−3.6) | (2.7−4.0) | (2.2−2.8) | (1.9−2.2) | (2.2−3.6) | (1.9−4.0) | |
| A : G ratio | 1.25 ± 0.19 | 1.11 ± 0.37 | 1.87 ± 0.26 | 2.13 ± 0.25 | 1.52 ± 0.69 | 1.62 ± 0.62 |
| (1.03−1.48) | (0.75−1.63) | (1.50−2.10) | (1.91−2.47) | (1.03−2.10) | (0.75−2.47) | |
| Total T3 (ng/dl) | 50.34 ± 10.34 | 34.88 ± 22.38 | 46.43 ± 8.96 | 53.10 ± 12.54 | 48.60 ± 9.37 | 43.99 ± 19.41 |
| (43.41−54.63) | (15.00−64.18) | (36.71−51.77) | (36.45−66.87) | (36.71−54.63) | (15.00−66.87) | |
| Cortisol (g/dl) | 4.52 ± 1.31 | 7.63 ± 2.36 | 4.65 ± 2.44 | 1.70 ± 0.68 | 4.58 ± 1.76 | 4.66 ± 3.55 |
| (3.2−6.7) | (5.3−10.0) | (3.7−8.3) | (0.9−2.4) | (3.2−8.9) | (0.9−10.0) |
Mean ± SD (Range in parentheses).
ALT, Alanine aminotransferase (=SGPT).
Additional Phenotypic Traits
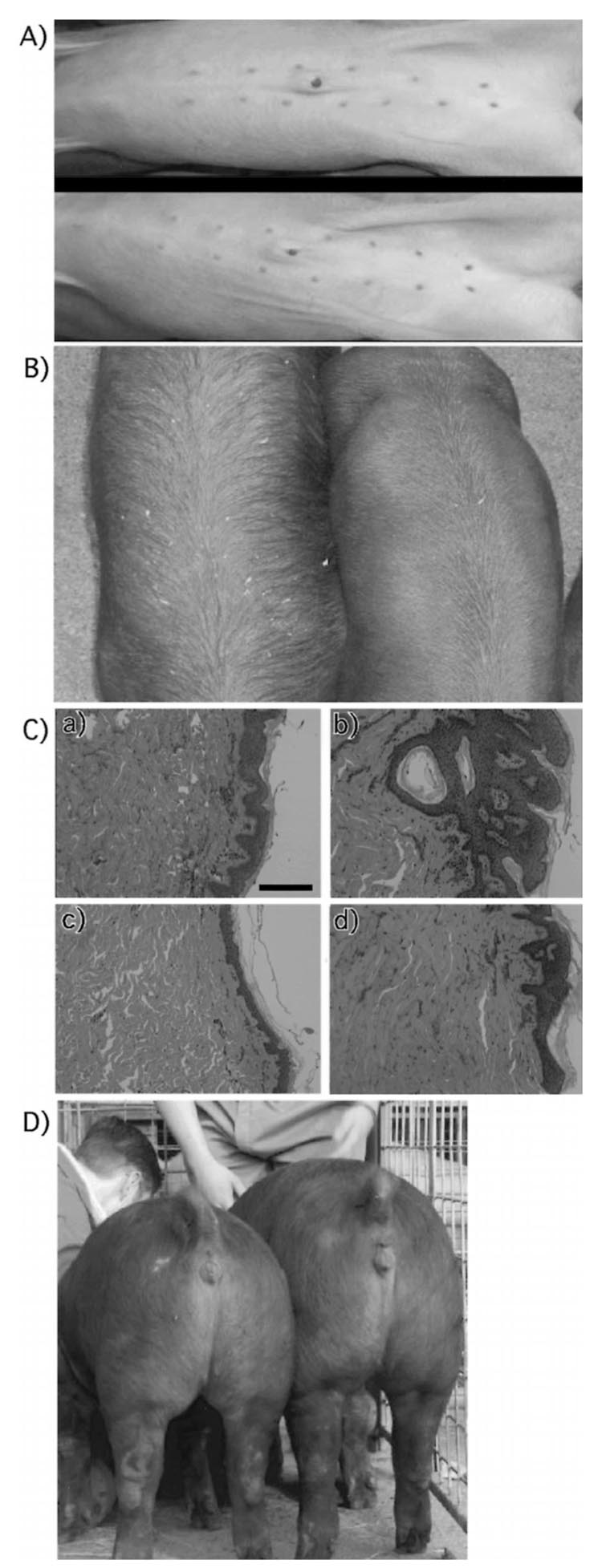
We also measured number of teats, skin type, and hair growth pattern of the pigs to determine the degree of variation. Although there was a strong genetic influence on number of teats, with all cloned pigs except one having a 6 + 6 distribution, there was one cloned pig that had a 6 + 7 distribution. We have seen this variation in number of teats in other clones generated by us but not used in this study (Fig. 2A).
FIG. 2.
Phenotypic variation among control genetically identical cloned pigs. A) Variation in number of teats. All but one of the cloned pigs had 12 teats, arranged 6 on the right and 6 on the left. B) Hair growth pattern variation. The cloned pig on the left has elongated and more sparse hair than the other two clones, with a tendency for dirt (shavings) to get trapped in the hair. C) Histology of porcine skin from cloned (b and d) and control (a and c) pigs at 25 weeks of age. All samples indicated normal morphology with well-organized epidermal and dermal layers, except for one cloned pig (b) that showed irregular epidermal hyperplasia and hyperkeratosis. Results from four representative animals are shown. Magnification ×10. D) Variation in body weight in cloned pigs is reflected by two 27-wk-old cloned pigs originating from the same nuclear donor cell line and born to the same recipient mother.
A unique hair growth pattern was also observed in one of the cloned pigs in the clone 1 group (Fig. 2B). The unusual hair growth pattern prompted us to examine in more detail the histological organization of the skin in the control and cloned pigs. With one exception, the morphology of the skin did not show any unusual variation among the pigs. The exception was one cloned pig that had morphology indicative of hyperkeratosis (Fig. 2C). To ensure that this observation was not due to a sampling error a second sample from this animal was analyzed, and similar results were obtained.
Uterine/environmental and/or epigenetic factors can produce different effects in genetically identical animals. The two cloned pigs in Figure 2D represent the largest and the smallest (by size) in one group of clones. The smaller pig was smaller at birth and never achieved the weight/size of the other cloned pigs in that litter.
Epigenetic Analysis
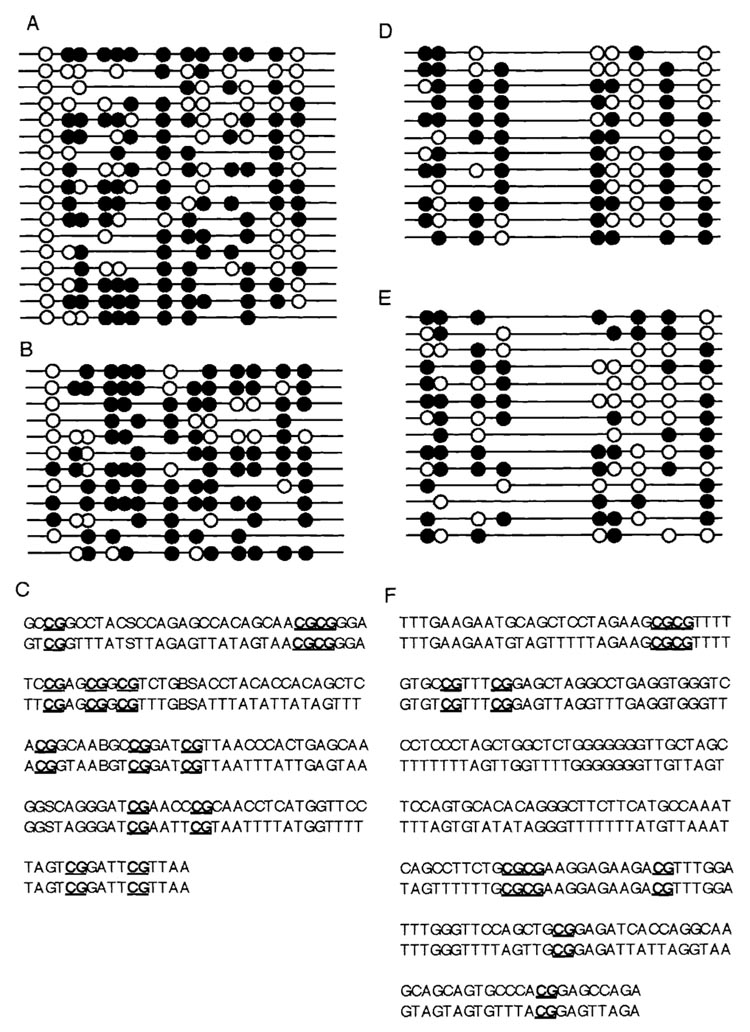
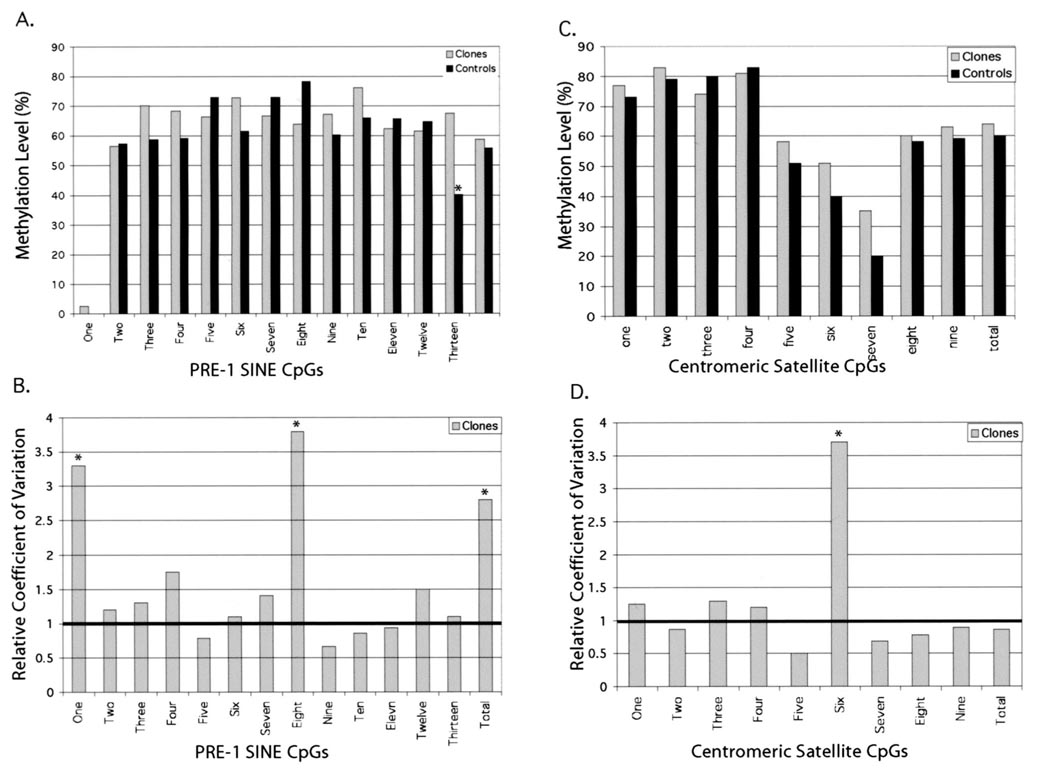
The cloned pigs used in this study were genetically identical, as previously determined by analysis of microsatellite markers [16]. In the present study, we extended our genetic analysis to include the methylation status of two repeat elements located in the porcine genome: centromeric satellites and the PRE-1 SINE region. Figure 3 shows the regions analyzed and the conserved CpGs identified. Data are shown for one cloned and one control pig for both the PRE-1 SINE and centromeric satellite regions. We compared the degree of variation among control and among cloned pigs in the degree of methylation of the different CpGs. The overall degree of methylation did not differ between cloned and control pigs for either of the regions, but a more localized analysis revealed a region of PRE-1 SINE (region 13) that was hypomethylated in control pigs (Fig. 4). When the degree of variation in each of these regions was compared, there was an increase in the variability of CpGs 1 and 8 in the PRE-1 SINE region and CpG 6 in the centromeric region.
FIG. 3.
Analysis of CpG methylation of PRE-1 SINE and centromeric satellite CpG islands by bisulfite sequencing. A) Methylation status of conserved CpGs in the PRE-1 SINE regions. Results from one representative control pig. B) Methylation of PRE-1 SINE CpGs in one representative cloned pig. C) Conserved CpGs present in the PRE-1 SINE region. Upper strand is the sequence of unmodified DNA; lower strand is the sequence of DNA after bisulfite modification. All CpGs are assumed to be protected by methyl groups. D) Methylation status of conserved CpGs in centromeric satellite region of one representative control pig. E) Methylation status of conserved centromeric satellite CpGs in one representative cloned pig. F) Conserved CpGs in centromeric satellite region. Upper strand is unmodified DNA; lower strand is bisulfite-modified DNA. Solid circles indicate methylated CpG; open circles indicate unmethylated CpG.
FIG. 4.
CpG methylation in PRE-1 SINE and centromeric satellite regions in cloned and control pigs. A) Percent methylation of each of the 13 conserved CpGs in the selected PRE-1 SINE region. B) Relative variation in methylation at each of the conserved CpGs. Column values with an asterisk are significantly different from controls at P < 0.05. C) Percent methylation at nine conserved CpGs in the selected region of the centromeric satellite region. D) Relative methylation variation in the centromeric satellite region.
DISCUSSION
The initial expectation for cloned animals was that a group of animals with identical genetic constitution (clones) would have reduced variability associated with all traits when compared with offspring from naturally bred females. The aim of these experiments was to determine to what extent healthy adult clones are affected by the cloning process and whether all phenotypes or traits are affected equally. This information is critical for those proposing to use clones to reduce variability in experimental animals and for those attempting to generate cloned pets with identical phenotypes. Overall, results of the present study indicated a high degree of variation among cloned pigs in some of the specific traits examined. Our experimental design is unique in that it allowed comparison of the degree of variation between clones and controls; we used age-, breed-, and sex-matched clones and control animals housed under identical conditions. Such an experimental design allows the differentiation between environmental effects and effects due to the cloning process itself.
To ensure that the results obtained were not affected by mitochondrial background, we sequenced the D-loop regions of both cloned and control pigs. As previously described [16], oocytes used to generate these clones were purchased from a commercial supplier that collects oocytes from slaughterhouse material. Thus, oocyte populations represent multiple maternal sources and potentially different mitochondrial types. To separate any potential mitochondrial effects from the epigenetic effects due to methylation differences, the polymorphic hypervariable D-loop mitochondrial region of clones and controls was sequenced. The D-loop region is the site of replication, has a mutation frequency 1000-fold greater than that of the rest of the mitochondrial genome [17], and therefore is a good indicator of the variability in the remainder of the mitochondrial genome. Little variability among animals was detected, with some cloned and control pigs having identical mitochondrial genotypes in the D-loop regions. This finding indicates that the mitochondrial backgrounds for cloned and control pigs were very similar and unlikely to result in major phenotypic differences. Conservation of mitochondrial background is not surprising, based on the high degree of selection within maternal lines in commercial swine herds and the low degree of mitochondrial variation in swine breeds in general [18].
Examination of body weight (Table 1) indicated that cloning did not reduce the degree of variation associated with this trait, and, with the exception of one cloned pig, values for cloned and control pigs overlapped and were in the normal range for swine at that age. Although the environment can have a significant effect on the weight of an animal, both control and cloned pigs were maintained in the same environment, with the expectation of greater variability in control than in cloned pigs because of higher genetic variation.
When the phenotypic analysis was extended to blood parameters, two overall classes of traits were observed. In one class, there was reduced variation in cloned pigs, and in the other class there was as much variation in cloned as in control pigs. Because all pigs were maintained in the same environment, these results suggest that those traits showing reduced variability in cloned pigs are under tight genetic control. In some cases, the variability within traits for cloned pigs, as determined by relative CVs, was less than half that for control pigs (Fig. 1). This finding is remarkable considering the small number of animals available for this study. Because all pigs were subjected to the same environment, it is unlikely that this reduction in variability is due to an environmental factor; it is more likely due to reduced genetic variability in the clones. Other phenotypes, however, were highly variable among the cloned pigs, suggesting that these traits are more susceptible to aberrations introduced during the cloning process or to an overriding effect of some environmental factor on genetic variability of the clones (either the microenvironment or the initial uterine environment).
Results for two related parameters, T3 and cortisol, were unusual. At 15 wk, variation was similar in cloned and control pigs, but at 27 wk variation was lower cloned pigs, although mean values were not significantly different. Thus, there was no difference between the controls and clones in overall T3 and cortisol concentrations at 27 wk (Table 3); however, cloned pigs showed a reduction in the variability of these hormone concentrations. This reduction may be due to an adaptation of the cloned pigs to the environment or to the sampling process, which was more homogenous than that for the controls, suggesting that adaptability to stress responses has a strong genetic component. However, as for all other phenotypic traits examined, both cloned and control pigs were subjected to the same environmental stimuli and were balanced with respect to sex, breed, and age.
Comparison of phenotypes based on blood parameters and body weights indicated two classes of traits: those not affected by cloning and having decreased variability in cloned compared with control pigs and those in which there was equal variability in cloned and control pigs. One interpretation of these results is that some traits, under tight genetic control and not heavily influenced by epigenetics, show a drastic reduction in variability as expected for genetically identical animals. Other traits, however, either because of epigenetic disregulation introduced during the cloning process or susceptibility to very strong environmental effects not accounted for, exhibit a high degree of variability regardless of genotypic similarities. These findings indicate that cloned animals can indeed be utilized when researchers want to reduce the size of an experimental group of animals, but only if the phenotype being examined has reduced variability in clones. For other phenotypes, cloned animals will have no advantage over controls.
In addition to the blood profiles, clonal differences were observed in skin type, hair growth pattern, and number of teats, supporting the observation that cloning creates variation that is independent of genetic background. For number of teats, a recent whole-genome scan for quantitative trait loci (QTL) in swine revealed that three loci, two of which were imprinted, significantly affect this trait [17]. Because cloning is known to affect imprinted genes [3], the differences in number of teats in cloned pigs could be due to aberrant expression of the imprinted QTL loci associated with this trait.
These additional trait differences reinforce our previous observations that the cloning procedure can affect traits in some but not all clones. Although it is difficult to differentiate between epigenetic disregulation and environmental effects on blood parameters, it is unlikely that environmental effects that cannot be controlled would have modified traits such as number of teats, hair growth patterns, and skin types.
To determine whether epigenetic disregulation, as represented by CpG methylation, was involved in phenotypic variability in cloned pigs, the methylation status of two repeats, one located in euchromatic regions (PRE-1 SINE) and one located in heretochromatic regions (centromeric satellite), was determined. We detected a differences in methylation status for one of the CpGs in the euchromatic regions and an increase in variation in degree of methylation in cloned pigs. Although it is not possible to prove cause and effect between hypermethylation of a region of the PRE-1 SINE marker and variation in any of the phenotypes measured, the results indicate that the cloning process creates both an abnormal methylation pattern, as determined by hypermethylation of CpG 13 in PRE-1 SINE, and a random variable methylation pattern in several regions, as determined by increased variability in percent methylation in several CpG groups of PRE-1 SINE and one CpG in the centromeric satellite region.
The methylation differences between cloned animals and offspring from naturally bred animals contrast with differences previously described [11–13, 19], in that the differences we observed were relatively small. Previous epigenetic analyses have been carried out in animals or tissues with obvious developmental defects, whereas we compared cloned pigs with no apparent developmental defects. Thus, the epigenetic disregulation in normal ‘‘healthy’’ clones should be lower than that in clones that die in utero or are born with severe defects. In short, our results reinforce the hierarchical effects of the nuclear transfer process, with both generalized effects, as reflected by the hypermethylation of CpG 13 of the PRE-1 SINE in clones, and clone-specific effects, as reflected by increased variability in the methylation of the PRE-1 SINE.
Severely affected clones are very likely to die in utero, whereas those that survive to birth, such as the cloned pigs used in this study, show a greater degree of variation in susceptible traits than expected for genetically identical animals. Overall, our results indicate that although cloning creates animals within the normal phenotypic range, it does affect some traits by increasing variability associated with that phenotype. This clonal variability may be useful for mapping epigenetically modified traits, either imprinted modifiers or genes susceptible to methylation effects. More importantly, the epigenetic disregulation introduced by the cloning process must be taken into account when considering the utilization of this technology for the generation of human stem cells, for the generation of companion animal clones, and for the generation of clones for experimental studies with the expectation of reduced genotypic and phenotypic variability among experimental animals.
ACKNOWLEDGMENTS
We thank Kenton Lillie and members of the laboratories of Drs. Piedrahita and Fuller Bazer for assistance with generation and maintenance of the experimental pigs. We are also grateful to Dr. Fuller Bazer for reviewing the manuscript.
Footnotes
This research was supported by NIH grant HL51587 to J.P. and a Texas A&M grant to T.F.
REFERENCES
- 1.Lanza RP, Cibelli JB, Faber D, Sweeney RW, Henerson B, Nevala W, West MD, Wettstein PJ. Cloned cattle can be healthy and normal. Science. 2001;294:1893–1894. doi: 10.1126/science.1063440. [DOI] [PubMed] [Google Scholar]
- 2.Hill JR, Edwards JF, Sawyer N, Blackwell C, Cibelli JB. Placental anomalies in a viable cloned calf. Cloning. 2001;3:83–88. doi: 10.1089/15204550152475581. [DOI] [PubMed] [Google Scholar]
- 3.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, III, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 4.Ohgane J, Wakayama T, Kogo Y, Senda S, Hattori N, Tanaka S, Yanagimachi R, Shiota K. DNA methylation variation in cloned mice. Genesis. 2001;30:45–50. doi: 10.1002/gene.1031. [DOI] [PubMed] [Google Scholar]
- 5.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 6.Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 7.Huynh K, Lee JT. Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr Opin Cell Biol. 2001;13:690–697. doi: 10.1016/s0955-0674(00)00272-6. [DOI] [PubMed] [Google Scholar]
- 8.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 9.Robertson KD. DNA methylation and chromatin—unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 10.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 11.Bourc’his D, Le D, Patin D, Niveleau A, Comizzoli P, Renard JP, Viegas-Pequignot E. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol. 2001;11:1542–1546. doi: 10.1016/s0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- 12.Dean W, Santos F, Stojkovic M, Zakhartchenko C, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang YK, Koo DB, Park JS, Choi YH, Kim NH, Chang WK, Lee KK, Han YM. Typical demethylation events in cloned pig embryos.Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem. 2001;276:39980–39984. doi: 10.1074/jbc.M106516200. [DOI] [PubMed] [Google Scholar]
- 14.Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lender ES, Golub TR, Jaenisch R. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc Natl Acad Sci U S A. 2002;99:2889–2894. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cibelli JB, Campbell KH, Seidel GE, West MD, Lanza RP. The health profile of cloned animals. Nat Biotechnol. 2002;20:13–14. doi: 10.1038/nbt0102-13. [DOI] [PubMed] [Google Scholar]
- 16.Walker SC, Shin T, Zaunbrecher GM, Romano JE, Johnson GA, Bazer FW, Piedrahita JA. A highly efficient method for porcine cloning by nuclear transfer using in vitro-matured oocytes. Cloning Stem Cells. 2002;4:105–112. doi: 10.1089/153623002320253283. [DOI] [PubMed] [Google Scholar]
- 17.Kim KI, Lee JH, Li K, Zhang YP, Lee SS, Gongora J, Morgan C. Phylogenetic relationships of Asian and European pig breeds determined by mitochondrial DNA D-loop sequence polymorphism. Anim Genet. 2002;33:19–25. doi: 10.1046/j.1365-2052.2002.00784.x. [DOI] [PubMed] [Google Scholar]
- 18.Hirooka H, de Koning DJ, Harlizious B, van Arendonk JA, Rattink AP, Groenen MA, Brascamp EW, Bovenhuis H. A whole-genome scan for quantitative trait loci affecting teat number in pigs. J Anim Sci. 2001;79:2320–2326. doi: 10.2527/2001.7992320x. [DOI] [PubMed] [Google Scholar]
- 19.Cezar GG, Bartolomei MS, Forsberg EM, First NL, Bishop MD, Eilertsen J. Genome-wide epigenetic alterations in cloned bovine fetuses. Biol Reprod. 2003;68:1009–1014. doi: 10.1095/biolreprod.102.010181. [DOI] [PubMed] [Google Scholar]