Abstract
Rearranged Ig genes undergo diversification in sequence and structure initiated by the DNA deaminase, AID. Ig genes must be transcribed for diversification to occur, but whether there are additional requirements for cis-activation has not been established. Here we show, by chromatin immunoprecipitation, that the regulatory factor E2A associates with the rearranged IgλR gene in the chicken DT40 B cell line, which carries out constitutive Ig gene diversification. By analysis of a DT40 derivative in which polymerized lactose operator tags the rearranged λR gene, we show that E2A must function in cis to promote diversification, and that stimulation of diversification in cis depends on the E2A activation domains. By direct imaging, we show that λR/E2A colocalizations are most prominent in G1 phase of cell cycle. We further show that expression of the E2A antagonist Id1 prevents λR/E2A colocalizations in G1 phase, and impairs diversification but not transcription of λR. Thus, E2A acts in cis to promote Ig gene diversification, and G1 phase is the critical window for E2A action.
Introduction
Immunoglobulin (Ig) gene diversification alters DNA sequence and structure at Ig loci that are already producing functional antibodies, to increase repertoire diversity and respond dynamically to infection by pathogens (1–4). In antigen-activated mammalian B cells, somatic hypermutation introduces nontemplated point mutations into rearranged and expressed variable (V) regions, and class switch recombination (CSR) juxtaposes a new constant region to the expressed V region. In chicken and other fowl, gene conversion expands a limited pre-immune repertoire by using upstream pseudo-V (ψV) gene segments as templates for mutagenesis of rearranged and expressed V regions.
The regulated changes in genomic sequence and structure that take place at the Ig loci reflect both targeting of DNA damage to these genes, and escape from faithful repair. Somatic hypermutation, CSR and gene conversion are all initiated by the B cell-specific enzyme, activation-induced deaminase (AID) (5–8). AID deaminates cytosine to uracil, with clear preference for single-stranded DNA (9–11). Transcription is prerequisite for diversification, which may reflect preference of AID for single-stranded substrates. Uracil in DNA is a common lesion, which can be repaired faithfully by highly conserved and efficient pathways (12). However, the Ig loci can escape from faithful repair and undergo repair by error-prone pathways (13).
E2A, a member of the E family of bHLH proteins, is a critical regulator of many aspects of lymphocyte development (14–19). E proteins dimerize to bind to the E box motif, CANNTG, and their function is antagonized by Id proteins, which heterodimerize with E proteins to prevent DNA binding. In activated murine B cells, E2A regulates CSR as well as expression of the gene that encodes AID (20–22). There may be functional overlap between E2A and the related HEB and E2-2 proteins, which are also regulated by Id interactions and which may promote CSR in the absence of E2A (18). In chicken B cells, inactivation of the E2A gene impairs Igλ gene diversification but not transcription (23, 24); while, conversely, ectopic expression of E47 (one of two functionally equivalent isoforms encoded by E2A) promotes Igλ gene diversification, but does not affect Igλ transcript levels (25).
The possibility that E2A might regulate Ig gene diversification by binding to sites in cis was first suggested by evidence that multimerized E-boxes stimulate hypermutation but not transcription of an Ig transgene in mice (26). This possibility has been further supported by the demonstration that multimerized E-boxes can promote Ig gene diversification but not transcription in chicken B cells (27). However, clear resolution of the question of whether E2A acts directly at the Ig genes to promote diversification has been difficult, for several reasons. E-boxes function as sites for E2A-dependent regulation only in specific contexts, so the presence of an E-box does not guarantee E2A function at a site. The loose consensus and frequent occurrence of E-box motifs precludes mutational analysis of each individual site. In addition, at some loci E2A is recruited by protein-protein rather than protein-DNA interaction, so an E-box is not always prerequisite for E2A-dependent regulation (28).
We have now established that E2A acts in cis at the Ig genes to promote diversification, in experiments which take advantage of derivatives of the constitutively diversifying chicken B cell line, DT40, in which the rearranged Igλ allele is tagged with polymerized lactose operator (DT40 PolyLacO-λR). By chromatin immunoprecipitation (ChIP), we show that E2A associates with the rearranged but not unrearranged Igλ allele in the parental line, DT40. We demonstrate that, in DT40 PolyLacO-λR cells, diversification is accelerated upon expression of an E47-LacI fusion protein, which effectively tethers E47 to λR; and that the stimulatory effect of E47-LacI expression is not evident in cells cultured with IPTG, so binding in cis is necessary to promote diversification. The activation domains of E47 are required for acceleration of diversification. By direct imaging of the rearranged λR gene in DT40 PolyLacO-λR GFP-LacI cells, we show that λR/E2A colocalizations predominate in G1 phase; and that expression of the E2A antagonist, Id1, impairs diversification and diminishes λR/E2A colocalizations specifically in G1 phase, but does not affect λ transcript levels or localization of λR to active transcription factories. We conclude that E2A acts in cis in G1 phase to promote Ig gene diversification.
Materials and Methods
Cell culture and gene targeting
DT40 and its derivative cell lines were maintained and transfected as described (29). DT40 PolyLacO-λR was generated by gene targeting with a construct, pPolyLacO-ψVλ, which carried a 3.8-kb PolyLacO fragment and homology arms designed for insertion between ψVλ17 and ψVλ20. To generate this targeting construct, pBluescript KS (Stratagene) was engineered to contain two modified loxP recombination sites (7) at the XbaI-SpeI and BamHI-PstI sites; a Histidinol selection marker (subcloned from a plasmid provided by Andrew Scharenberg, University of Washington, Seattle, Washington) was inserted at the BamHI site; and the 3.8 kb PolyLacO fragment was subcloned from pAFS59-13 (provided by Aaron Straight, Stanford University, Stanford, CA) into the EcoRV site. A 2 kb fragment containing ψVλ20-ψVλ23 and a 3.5 kb fragment containing ψVλ13-ψVλ17 were amplified from DT40 genomic DNA using DyNAzyme (Finnzymes) and cloned into blunted NotI and XhoI sites, respectively. The construct was verified by restriction analyses and partial sequencing. Plasmids carrying PolyLacO were propagated in recombination-deficient E. coli strains Stbl2 (Invitrogen) or SURE2 (Stratagene) to maintain repeat stability.
To insert PolyLacO in the ψVλ array, wild-type DT40 cells were transfected with the pPolyLacO-ψVλ construct; candidate clones were screened by Southern blotting; and genomic organization was verified by PCR. A naturally-occurring polymorphism at ψVλ17 (30) was used to distinguish integration at the rearranged or unrearranged λ allele, and random integration. The loxP-flanked selection marker was deleted by transient transfection with a Cre recombinase expression plasmid, pCre-IRES-GFP (provided by Keith Fournier, Fred Hutchinson Cancer Research Center, Seattle, Washington). Single integration was also confirmed by Southern blotting.
ChIP analysis
Chromatin was prepared and immunoprecipitated as described (31–33) using anti-E2A antibody (ab11176; Abcam) or control IgG. Semiquantitative PCR was performed with FastStart Taq DNA polymerase (Roche), using previously described primers for VλR and VλU (33); and primers 5’-ATTGCGCATTGTTATCCACA-3’ and 5’-TAAGCCCTGCCAGTTCTCAT-3’ for ovalbumin (Ova). PCR products were quantitated with ImageQuant software (Amersham). Enrichment was calculated as the ratio of the amplicon of interest to the Ova amplicon, normalized to the ratio from control IgG, e.g.: Enrichment VλR = (anti-E2A [VλR/Ova])/(IgG [VλR/Ova]).
Assays of sIgM-loss, cell cycle, and cell cycle-dependence of Id1 levels
The sIgM-loss assay was carried out as described (29, 34), and results compared using the Mann-Whitney U test with the R software package (http://www.r-project.org). For cell-cycle profiles based on DNA content, 1 × 106 exponentially growing cells were suspended in 0.1% Triton X-100, treated with 100 µg/ml RNase A and 50 µg/ml propidium iodide (PI), and analyzed as described (29). To assess cell cycle-dependence of Id1 protein levels, DT40 cells were transfected with expression constructs of Id1-GFP or its derivative using a Nucleofector (Amaxa). At 24 hr posttransfection, cells were fixed with cytofix/cytoperm solution (BD Bioscience); treated with 100 µg/ml RNase A and 5 µg/ml PI; and GFP intensity and DNA content determined on a FACScan flow cytometer (BD Biosciences).
Expression constructs
The E47-LacI expression construct was generated by subcloning of E47 cDNA from the S003 E47 plasmid (35) (provided by Cornelis Murre, University of California, San Diego, California) into the XbaI-BsrGI site of the p3’SS-GFP-LacI plasmid (provided by Andrew Belmont, University of Illinois, Urbana, Illinois). Activation domain (AD)-deleted mutants, E47ΔAD1-LacI lacks residues 2–99; E47ΔAD2-LacI lacks residues 325–432; and E47ΔAD1/2-LacI lacks both AD domains. These mutants were made by first generating fragments carrying deletions by PCR amplification, and then exchanging these fragments with the corresponding regions of the parental E47-LacI plasmid. A DNA binding mutant, E47BM-LacI, which carries an R558K mutation in the basic region, was made by QuikChange site-directed mutagenesis (Stratagene). The Id1 expression construct (36) was provided by Barbara Christy (University of Texas, San Antonio, Texas). Id1-GFP was constructed by subcloning of Id1 cDNA into the XhoI site of a pEGFP-N1 vector (Clontech); and its derivative Id1DBM-GFP, which carries R119G and L122V mutations in its canonical D-box motif (residues 119–126, RxxLxxxN), was generated from Id1-GFP by QuikChange mutagenesis.
RT-PCR, Western blotting
For RT-PCR assays, Igλ transcript was amplified with primers 5’-GTCAGCAAACCCAGGAGAAAC-3’ and 5’-AATCCACAGTCACTGGGCTG-3’; AID and β-actin were with primers as described (7); E47 and E12 with published primers (37); HEB with 5’-TCAAATCTGATGGGGAAAGC-3’ and 5’-ACTGGGACATGTGGGAAGAG-3’; and E2-2 with 5’-AATGACCTGAGCGCTCCTAA-3’ and 5’-CTGTGGGAATGTAGGGAGGA-3’. For Western blotting, whole cell lysates (50 µg) were resolved by SDS-PAGE, blotted with anti-Id1 antibody (JC-FL; Santa Cruz) and detected using FluorChem HD2 (Alpha Innotech).
Immunofluorescence staining and image analysis
To image PolyLacO, DT40 PolyLacO-λR cells were transfected with the GFP-LacI expression construct, p3’SS-GFP-LacI (from Andrew Belmont, University of Illinois, Urbana, Illinois), which encodes LacI engineered to contain SV40 nuclear localization signals and lacking a sequence necessary for tetramer formation (38); or its derivative, RFP-LacI, in which GFP was replaced with RFP (DsRed-monomer; Clontech). For immunostaining, cells (~3 × 105) were deposited onto glass slides using Cytospin3 (800 rpm, 4 min; Shandon), fixed with 2% paraformaldehyde for 20 min, and stained as described previously (29). Primary antibodies used were: anti-E2A (ab11176, 1:200; Abcam); anti-Pol II C-terminal domain phosphorylated at Ser5 (ab5131, 1:500; Abcam). Alexa Fluor 488- or 594-conjugated anti-IgG (Molecular Probes) was used as secondary antibodies. Fluorescent images were acquired using the DeltaVision deconvolution microscopy system (Applied Precision) and processed and analyzed with softWoRx (Applied Precision) and Imaris softwares (Bitplane). Fraction of colocalization was analyzed with Pearson’s χ2 test. Nuclear radii were calculated as the average of at least two independent measurements of diameter, divided by two. Cell cycle dependence of mean nuclear radius was determined independently for each cell line, and proved to be relatively invariable. Standard values used to correlate nuclear radius to cell cycle were: G1, r < 4 µm; G2, r ≥ 5.2 µm.
Results
E2A associates with the rearranged but not unrearranged Igλ gene
Despite the considerable evidence for the importance of E2A in Ig gene diversification, this factor had not been shown to associate directly with the Ig genes. To test association of E2A with Igλ, we used anti-E2A antibodies to immunoprecipitate chromatin from the chicken DT40 B cell line. This line was derived from a bursal lymphoma and carries out constitutive diversification of both Ig heavy and light chain genes by gene conversion. A search of the 11 kb chicken λ light chain locus identified more than 50 matches to the E2A consensus, CANNTG: 17 in the region between Vλ and ψVλ1, the most proximal of the upstream pseudogenes; 2 in the matrix attachment region (MAR) in the J-C intron; and 6 in the 3’ enhancer. In DT40 B cells, the functional allele has undergone VJ recombination early in B cell development, which deletes a 1.8-kb region to join the V and J segments, while the inactive λ allele is unrearranged (Figure 1A), allowing the two alleles to be readily distinguished by PCR. Following ChIP, recovery of the rearranged and unrearranged λ alleles was assayed relative to a control gene, ovalbumin. This showed that E2A was 17.6-fold enriched at the rearranged VλR allele, but not at the unrearranged VλU allele (Figure 1B). Thus E2A associates directly with the rearranged VλR allele.
FIGURE 1. E2A associates with the rearranged Igλ locus.
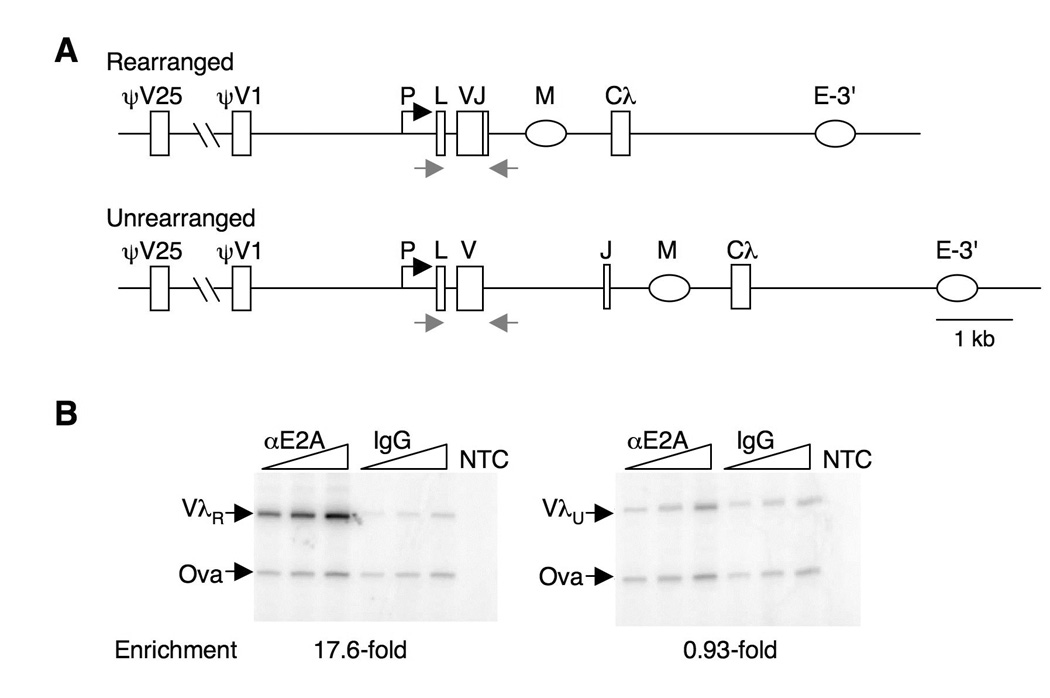
A. Schematic of the rearranged and unrearrranged Igλ loci in the chicken B cell lymphoma line, DT40. Shown are promoter (P), leader (L), variable (V), joining (J) and constant (Cλ) regions; the putative matrix attachment region (M) in the J-C intron; the 3’-enhancer (E-3’); and the most proximal (ψV1) and distal (ψV25) of the upstream nonfunctional pseudo-variable regions which are templates for gene conversion. The rearranged and unrearranged alleles can readily be distinguished by PCR, using primers indicated by arrows.
B. ChIP analysis of E2A enrichment at the rearranged λR and unrearranged λU loci in DT40 cells, relative to ovalbumin gene control amplicon (Ova). Fold enrichment is shown below. NTC, no template control.
E2A acts in cis to regulate Igλ diversification
To ask if E2A must bind in cis to promote diversification, we took advantage of a derivative of DT40, DT40 PolyLacO-λR, in which polymerized lactose operator (PolyLacO) has been inserted in the ψVλ array by homologous gene targeting (Figure 2A), allowing factors expressed as fusions with lactose repressor (LacI) to be tethered to the rearranged λR allele and released by culture with IPTG (33). Cell cycle distribution and clonal rates of Ig gene conversion were comparable in DT40 PolyLacO-λR and wild-type DT40 (Figure 2B, C). DT40 PolyLacO-λR cells were stably transfected with a plasmid expressing the E47 isoform of E2A fused to LacI (E47-LacI), or a control plasmid expressing green fluorescent protein fused to LacI (GFP-LacI). The E47 isoform was used because it has previously been shown to promote Ig gene diversification upon ectopic expression (25). Cell cycle distribution was comparable in GFP-LacI and E47-LacI transfectants, although cultures of the latter line contained some sub-G1 (apoptotic) cells (Figure 2D). Levels of Igλ transcripts were unaltered in the E47-LacI transfectants (Figure 2E), as predicted by published results showing that E2A does not regulate Ig gene expression in chicken B cells (24, 25). Levels of AID transcripts were approximately 3-fold higher in the E47-LacI transfectants (Figure 2E), as reported by others (25).
FIGURE 2. E2A acts in cis to regulate Igλ gene diversification.
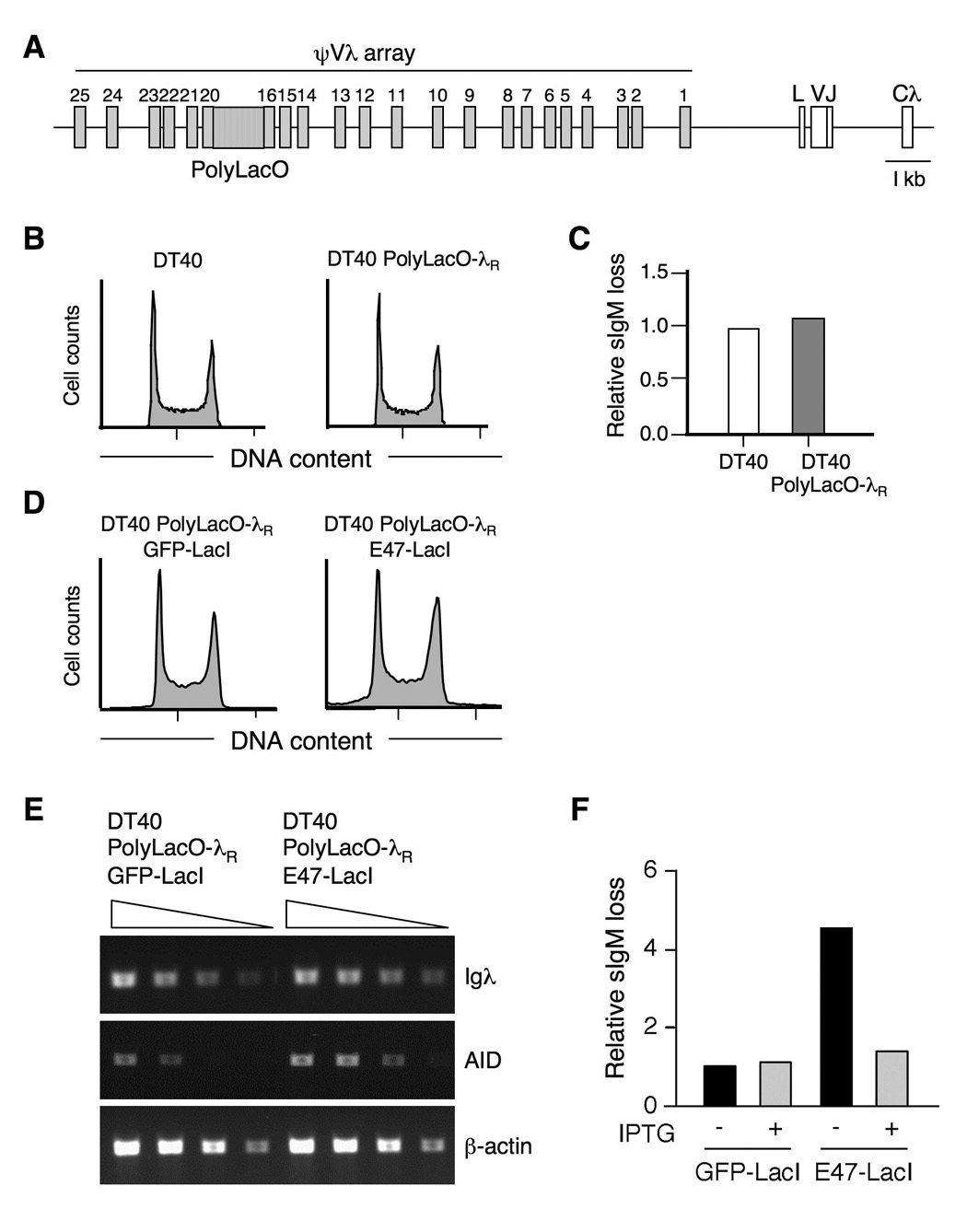
A. Schematic of the PolyLacO-tagged rearranged Igλ locus. PolyLacO is integrated between ψV17-20; other notions as in Figure 1A.
B. Cell cycle profile of DT40 and DT40 PolyLacO-λR cells.
C. Accumulation of sIgM-loss variants by DT40 and DT40 PolyLacO-λR cells. Frequencies of sIgM-loss variants in 24 subclones from each line were quantitated by flow cytometry following 6 wk clonal expansion. Frequencies shown were normalized to DT40; mean sIgM-loss frequencies were 0.8% and 0.9%, respectively.
D. Cell cycle profile of DT40 PolyLacO-λR GFP-LacI and DT40 PolyLacO-λR E47-LacI cells.
E. RT-PCR analysis of expression of Igλ, AID or β-actin mRNAs in DT40 PolyLacO-λR GFP-LacI and DT40 PolyLacO-λR E47-LacI transfectants. Triangles indicate 30, 10, 3 and 1x relative concentrations of cDNA templates.
F. Mean sIgM-loss of independent clonal DT40 PolyLacO-λR GFP-LacI (n = 13) and DT40 PolyLacO-λR E47-LacI (n = 19) transfectants, cultured for 3 wk in the absence or presence of 100 µM IPTG. Values were normalized to DT40 PolyLacO-λR GFP-LacI cells cultured without IPTG.
To ask if E2A regulates diversification directly, via binding to Igλ, we cultured independent E47-LacI (n = 19) or GFP-LacI (n = 13) transfectants in the presence and absence of IPTG, and determined clonal diversification rates using the sIgM-loss fluctuation assay (29, 33, 34). This assay scores inactivating mutations regardless of whether they occur by gene conversion, point mutation, deletion or insertion, and thus quantitates initiating events independent of the outcome of mutagenesis. This analysis showed that the clonal rate of diversification was 4.5-fold higher in E47-LacI transfectants relative to GFP-LacI controls (P = 0.019, Mann-Whitney U test; Figure 2F). Culture with IPTG causes LacI to be released from PolyLacO (33), so in cells cultured with IPTG E47-LacI does not bind specifically to the Igλ locus, but is present throughout the nucleus. Culture with IPTG had no effect on GFP-LacI control transfectants, but reduced diversification rates in DT40 PolyLacO-λR E47-LacI cells to background levels (Figure 2F). Thus E47-LacI promotes diversification by acting in cis.
Accelerated diversification depends upon the E2A activation domains
We then asked which functions of E2A are necessary to promote Ig gene diversification by determining the effect on diversification rate of mutations in either activation domain of E2A, AD1 or AD2; or in the DNA binding domain. We generated DT40 PolyLacO-λR derivatives stably expressing E47-LacI mutants lacking AD1 (E47ΔAD1-LacI), AD2 (E47ΔAD2-LacI) or both (E47ΔAD1/2-LacI) (Figure 3A). Transfectants of E47ΔAD1-LacI were isolated with efficiencies comparable to the control transfectants, and transfectants of E47ΔAD2-LacI and E47ΔAD1/2-LacI at a slightly lower efficiency. Neither of these mutants accelerated Ig gene diversification (Figure 3B). Thus, function of E47 in Ig gene diversification depends upon the E2A activation domains.
FIGURE 3. Accelerated diversification depends upon the E2A activation domains.
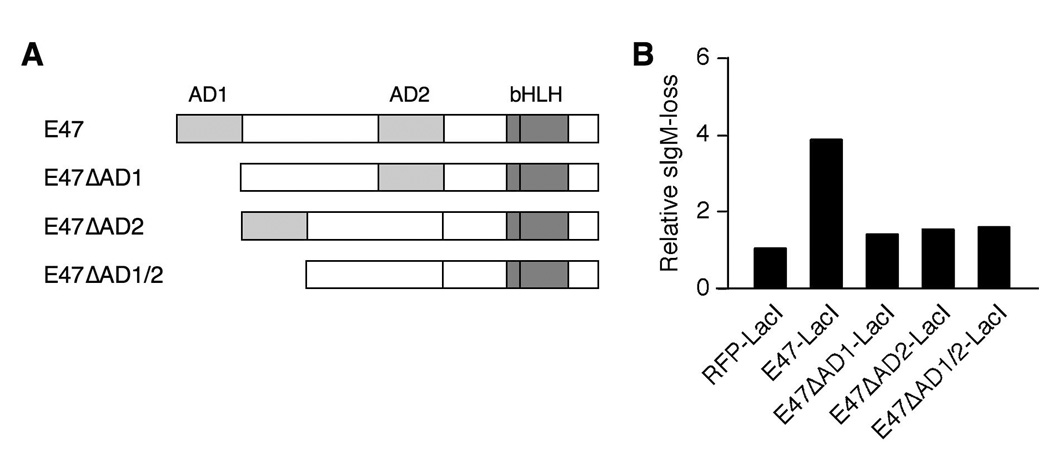
A. Schematic of wild-type E47 and its mutants. AD1, activation domain 1; AD2, activation domain 2; bHLH, basic-helix-loop-helix.
B. Mean sIgM-loss of independent clonal DT40 PolyLacO-λR cells expressing RFP-LacI (n = 17), E47-LacI (n = 10), E47ΔAD1-LacI (n = 22), E47ΔAD2-LacI (n = 9), or E47ΔAD1/2-LacI (n = 7), analyzed 3 wk posttransfection. The result shown is representative of two independent experiments. Values were normalized to DT40 PolyLacO-λR RFP-LacI cells.
We also attempted to generate transfectants expressing an E47-LacI mutant carrying a mutation in the basic region of E47 (R558K) which inactivates DNA binding (39). However, in each of two independent transfections few transfectants were recovered (about 15% compared to the control), suggesting this mutation may have a dominant negative effect on cell survival. The effect of this mutation was not further anlayzed.
E2A localizes to IgλR in G1 phase of cell cycle
In human B cells, receptor crosslinking in G1 phase of cell cycle can initiate somatic hypermutation, producing identifiable mutations within 90 minutes (40). Thus it was of interest to determine the stage of cell cycle in which E2A acts at Igλ. This is commonly done by sorting cells stained with Hoechst 33342 for DNA content to enrich for cells in G1, S, or G2/M stage of cell cycle. However, while the rearranged and diversifying λR gene can readily be imaged as a bright dot in DT40 PolyLacO-λR cells expressing GFP-LacI (33), following Hoechst 33342 staining and cell sorting, the fraction of cells exhibiting a clear fluorescent signal from the tagged gene diminished from the 90–95% routinely observed in unsorted cells to approximately 45%. As such a loss in signal could bias results, we therefore determined cell cycle stage by a different approach. Analysis of Hoechst 33342-stained and sorted cells showed that nuclear size was significantly smaller in G1 phase than in G2/M phase cells (e.g. Figure 4A). We therefore asked if nuclear radius (r) could be used to establish the stage of cell cycle, by measuring nuclear radii of G1 cells (n = 55) and G2 cells (n = 55) from an exponentially growing DT40 PolyLacO-λR population which had been stained with Hoechst 33342 and sorted based on DNA content. Mean nuclear radii of G1 cells was 3.8 ± 0.3 µm; and of G2 cells, 5.4 ± 0.5 µm (Figure 4B). Comparison of the ratios of G1: S: G2 cells as determined by nuclear radius (3: 6: 1) and staining (2.7: 5.5: 1.7) further validated this approach. Thus, G1 cells were identified experimentally as r < 4 µm; and G2 cells, r > 5.2 µm.
FIGURE 4. Nuclear radius correlates with cell cycle.
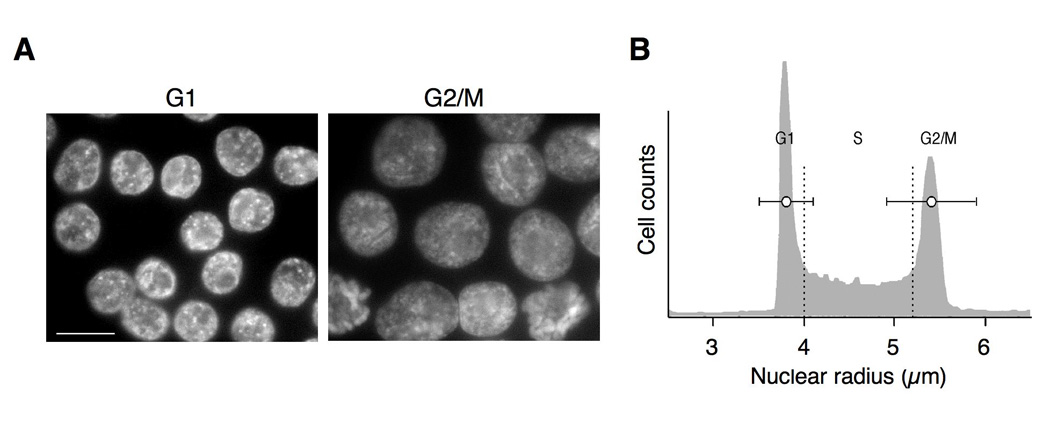
A. Representative images of G1- and G2/M-enriched cells. G1 (left) and G2/M (right) cells were stained with Hoechst 33342 (10 µM; Molecular Probes) then sorted based on DNA content. Bar, 10 µm.
B. Representative cell cycle profile of DT40 PolyLacO-λR cells. Mean radii ± s.d. are shown as horizontal bars within the representative profile. Dotted vertical lines indicate cut-offs for G1 and G2 used in experimental analyses: G1, r < 4 µm; G2, r > 5.2 µm.
Colocalizations of λR/E2A were readily identified by deconvolution microscopic analysis of DT40 PolyLacO-λR GFP-LacI cells stained with anti-E2A antibodies (e.g. Figure 5A). λR/E2A colocalizations were evident in 26% of asynchronous cells (n = 227). Analysis of the cell cycle distribution of colocalizations showed that 45% of λR/E2A colocalizations occurred in G1 phase, 38% in S phase, and 17% in G2 phase cells (Figure 5B). Thus, there was an apparent excess of λR/E2A colocalizations in G1 phase (45%) relative to the fraction (25%) of G1 phase cells (P < 0.0001, χ2 test).
FIGURE 5. E2A localizes to λR in G1 phase of cell cycle.
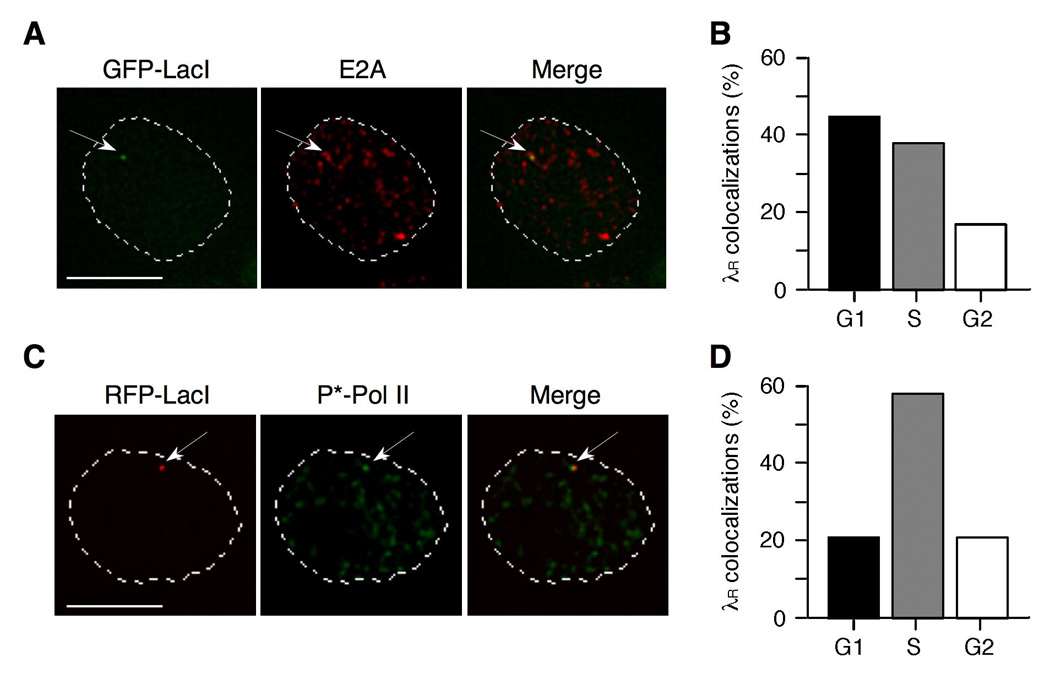
A. Representative image of colocalization of λR and E2A in DT40 PolyLacO-λR GFP-LacI cells. Nuclear perimeter as determined by DAPI staining is outlined by the dashed white line. Bar, 5 µm.
B. Fraction of λR/E2A colocalizations occurring in each phase of cell cycle in DT40 PolyLacO-λR GFP-LacI cells.
C. Representative image of colocalization of λR and active Pol II (P*-Pol II) in DT40 PolyLacO-λR RFP-LacI cells. Notations as in panel A.
D. Fraction of λR/P*-Pol II colocalizations occurring in each phase of cell cycle in DT40 PolyLacO-λR RFP-LacI cells.
We also determined the cell cycle-dependence of λR transcription, identifying active transcription factories by staining with antibody to phosphorylated Ser5 in the C-terminal domain of RNA polymerase II (P*-Pol II), a modification characteristic of elongating Pol II molecules (41). In asynchronous cell populations of DT40 PolyLacO-λR RFP-LacI cells, numerous active transcription factories could be identified throughout the nucleus, and λR/P*-Pol II colocalizations were readily observed in 19% of cells (n = 392; e.g. Figure 5C). Analysis of the cell cycle distribution of λR/P*-Pol II colocalizations showed that 21% of colocalizations occurred in G1 phase; 58% in S phase; and 21% in G2 phase cells (Figure 5D). This is comparable to the cell cycle distribution. Thus, λR is transcribed throughout the cell cycle, but λR/E2A colocalizations predominate in G1 phase.
Id1 expression inhibits Ig gene diversification and diminishes λR/E2A colocalizations
To ask if λR/E2A colocalizations in G1 phase are critical to diversification, we determined the effect of Id expression on these colocalizations. Id antagonizes E2A, and expression in DT40 B cells of Id1 or Id3 has previously been shown to diminish Ig gene diversification (25). We generated stable DT40 PolyLacO-λR RFP-LacI Id1 transfectants, confirmed Id1 expression by Western blotting (Figure 6A), and showed that Id1 expression did not alter the cell cycle profile (Figure 6B). Assays of sIgM-loss verified that Id1 expression diminished the clonal rate of Ig gene diversification in DT40 PolyLacO-λR RFP-LacI Id1 (P < 0.001, Mann-Whitney U test; Figure 6C), as predicted.
FIGURE 6. Id1 expression inhibits Ig gene diversification and λR/E2A colocalizations.
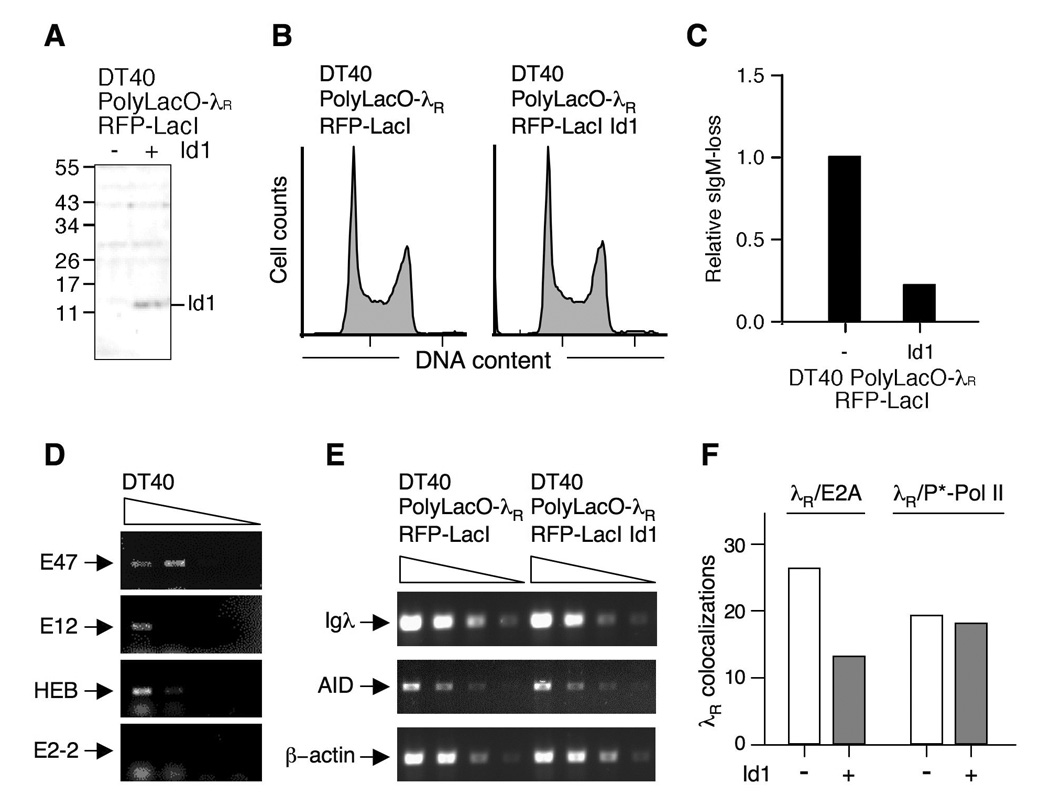
A. Western blot assaying Id1 expression in DT40 PolyLacO-λR RFP-LacI (left) and DT40 PolyLacO-λR RFP-LacI Id1 cells (right). Marker polypeptide sizes (kDa) at left.
B. Cell cycle profile of DT40 PolyLacO-λR RFP-LacI and DT40 PolyLacO-λR RFP-LacI Id1 cells.
C. Mean sIgM-loss of independent clonal DT40 PolyLacO-λR RFP-LacI Id1 transfectants (n = 6), cultured for 6 wk. Values were normalized to DT40 PolyLacO-λR RFP-LacI cells.
D. RT-PCR analysis of mRNA expression of E proteins in DT40 cells. Arrows indicate predicted sizes of products; smudge below (HEB, E2-2) is due to unincorporated primers.
E. Id1 expression does not alter Igλ or AID transcript levels. RT-PCR analysis of expression of Igλ, AID or β-actin control mRNA in DT40 PolyLacO-λR RFP-LacI and DT40 PolyLacO-λR RFP-LacI Id1 cells.
F. Id1 expression diminishes λR/E2A colocalizations. Colocalizations of λR/E2A and λR/P*-Pol II in asynchronous populations of DT40 PolyLacO-λR RFP-LacI cells and a derivative stably expressing Id1 (Id1 − and +, respectively).
E proteins target many sites in B cells, and ectopic expression of Id proteins can also affect other E proteins. We used RT-PCR to identify the E proteins present in DT40, assaying transcripts of the two isoforms of E2A, E47 and E12; as well as two other E proteins associated with lymphocytes, HEB and E2-2. We detected transcripts of E47, E12 and HEB, but not E2-2 (Figure 6D). We then asked whether Id1 expression affected levels of Igλ or AID transcripts, and showed that it did not (Figure 6E). Thus, basal levels of AID expression may not be mediated by E proteins; or E proteins may regulate AID expression via another mechanism, such as protein-protein interaction.
Next, we compared λR/E2A colocalizations in DT40 PolyLacO-λR RFP-LacI and its Id1-expressing derivative, by staining with anti-E2A antibodies. λR/E2A colocalizations were evident in 13% of asynchronous DT40 PolyLacO-λR RFP-LacI Id1 cells (n = 90), compared to 26% of DT40 PolyLacO-λR RFP-LacI cells (P = 0.0030, χ2 test; Figure 6F). λR/P*-Pol II colocalizations were identified in 18% of DT40 PolyLacO-λR RFP-LacI Id1 cells stained with antibodies to active transcription factories (n = 290), comparable to the parental line (19%, P = 0.80, χ2 test; Figure 6F). Thus, expression of Id1 diminishes λR colocalizations with E2A but not P*-Pol II.
Id1 expression diminishes λR/E2A colocalizations in G1 phase
Quantification of λR/E2A colocalizations with respect to cell cycle showed that, in DT40 PolyLacO-λR RFP-LacI cells, 45% of colocalizations occurred in G1 phase; 38% in S phase; and 17% in G2 phase; and in DT40 PolyLacO-λR RFP-LacI Id1 cells, 10% of colocalizations occurred in G1 phase; 58% in S phase; and 32% in G2 phase. Thus, the most pronounced effect of Id1 expression was in G1 phase, when the fraction of λR/E2A colocalizations diminished from 45% in the parental line to 10% in Id1 transfectants (P < 0.0001, χ2 test). As total colocalizations were diminished two-fold in Id1 transfectants, it was useful to plot those results in terms of the entire cell population. As shown in Figure 7A, Id1 expression reduced λR/E2A colocalizations from 12% to 1.4% in G1 phase, or about nine-fold; but had a much more modest effect in S phase or G2. In contrast, the cell cycle profile of λR/P*-Pol II colocalizations was comparable in Id1 transfectants and the parental line: 21%, 58% and 21% of colocalizations occurred in G1, S and G2/M phase in the parental cell line, and 24%, 60% and 16% in Id1 transfectants. As shown in Figure 7B, in terms of the entire cell population, Id1 expression had essentially no effect in any stage of cell cycle. The absence of effect of Id1 expression on λR/P*-Pol II colocalizations is consistent with undiminished Igλ transcript levels in DT40 PolyLacO-λR RFP-LacI Id1 transfectants (Figure 6D). Thus, Id1 expression affects the cell cycle distribution of colocalizations of λR with E2A, but not with P*-Pol II.
FIGURE 7. Id1 expression diminishes λR/E2A colocalizations in G1 phase.
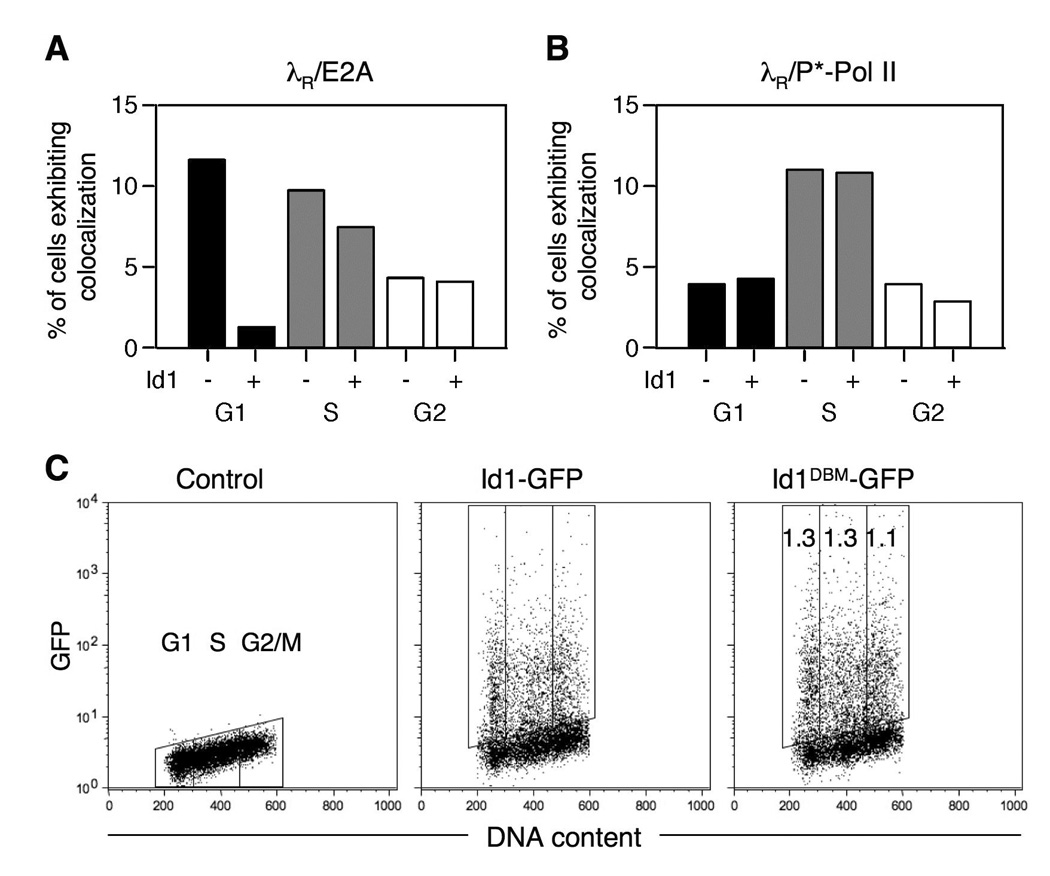
A. Effect of Id1 expression on cell cycle-dependence of λR/E2A colocalizations. The Colocalizations in DT40 PolyLacO-λR RFP-LacI cells and a derivative stably expressing Id1 (Id1 − and +, respectively) in each stage of cell cycle are graphed, showing the percent of total cells in which colocalizations were evident. Note that Id1 expression diminished total colocalizations by about half.
B. Effect of Id1 expression on cell cycle-dependence of λR/P*-Pol II colocalizations. Details as in Figure 7A.
C. Fluorescence analysis of Id1-GFP levels in the course of cell cycle. DT40 cells were transiently transfected with Id1-GFP or its derivative carrying mutations in the D-box, which determines cell cycle-dependent instability; and GFP intensity and DNA content analyzed at 24 hr posttransfection. Fold increases of mean GFP fluorescence intensity in Id1DBM-GFP relative to Id1-GFP is shown in the rightmost panel.
Id proteins are unstable, and they are targeted for degradation by a D-box motif conserved among Id family members (42). This raised the possibility that cell cycle-dependent regulation of Id1 levels could account for cell cycle-dependence of λR/E2A colocalizations. To test this, we monitored expression levels in the course of cell cycle of wild-type Id1 and a derivative carrying a mutation in the D-box mutant, Id1DBM, predicted to be unaffected by cell cycle-dependent destabilization. These proteins were transiently expressed as GFP fusions in DT40 cells, and at 24 hr posttransfection GFP intensity was determined with respect to cell cycle, as determined by PI staining. There was no apparent correlation of Id1-GFP levels with cell cycle, nor any difference between levels of Id1-GFP or Id1DBM-GFP in the course of cell cycle (Figure 7C). Thus, cell cycle-dependence of Id1 levels does not account for diminished λR/E2A colocalizations in Id1 transfectants in G1 phase. Instead, G1 phase appears to be the critical window in which E2A promotes diversification.
Discussion
We have shown that E2A must act in cis to promote Ig gene diversification, and that G1 phase is the critical window in which E2A functions in this process. Our experiments have examined λ genes tagged with PolyLacO and imaged by binding to GFP-LacI or RFP-LacI. This provides a powerful approach for studying gene diversification. The tagged locus is visible in >90% of fixed cells, enabling analysis of colocalizations with factors involved in diversification. The ability to tether potential regulators and release by culture with IPTG makes it possible to study the effects of a factor at the Igλ locus independent of its other targets. This is especially useful for a factor like E2A, which functions at the top of a large and complex regulatory hierarchy (43).
Our results establish that E2A directly regulates Ig gene diversification by physical association with the Ig loci. ChIP provided clear evidence for association of E2A with the rearranged λR allele in DT40 B cells. That E2A must function in cis was established by showing that the acceleration in diversification resulting from tethering E2A (E47-LacI fusion) to the Igλ allele in DT40 PolyLacO-λR cells was not evident in cells cultured with IPTG, which releases LacI from LacO. E2A is best-known as a transcriptional regulator, but E2A function in diversification did not reflect transcriptional activation at Igλ, as levels of Igλ transcripts were not altered by ectopic expression of E47, confirming results of others (23, 25, 26). Function in diversification did require the E2A activation domains, AD1 and AD2. Moreover, expression of the E2A antagonist, Id1, diminished diversification and λR/E2A colocalizations, but did not affect λR/P*-Pol II colocalizations, or Igλ transcript levels. Thus, E2A is not required to recruit λR to transciption factories.
In addition to the direct cis-regulation of Ig diversification by E2A, documented above, E2A could also stimulate Ig gene diversification indirectly, for example by regulation of AID expression. The AID gene was shown to be a target of transcriptional regulation by E2A in murine B cells (21); and we and others (25) have shown that ectopic expression of E2A increases AID transcript levels in DT40 chicken B cells. This may contribute to or account for the modest increase in diversification evident in IPTG-cultured DT40 PolyLacO-λR E47-LacI cells relative to DT40 PolyLacO-λR GFP-LacI cells (Figure 2F). While E2A ablation has been reported not to diminish AID transcript levels in chicken B cells (24), other E proteins, such as HEB (Figure 6D), might ensure a minimum level of AID expression in the absence of E2A. Alternatively, since Id1 expression did not affect AID expression (Figure 6E), basal level AID expression may be mediated by factors other than E proteins.
Colocalizations of λR with E2A were most prominent in G1 phase. In contrast, localization of λR to active transcription factories was comparable to cell cycle distribution, suggesting that λR transcription occurs throughout the cell cycle. Id1 expression had the most pronounced effect on λR/E2A colocalizations in G1 phase, and relatively minor effects at other stages of cell cycle. This suggests that λR/E2A interaction in G1 is critical for initiation of diversification, but does not address whether associations of E2A at other stages of cell cycle (which were not impeded by Id1) might also be important for completion of diversification. Id1 levels were constant throughout cell cycle, so diminished λR/E2A colocalizations in G1 phase caused by ectopic expression of Id1 does not reflect cell cycle-dependence of Id1 levels. Id proteins heterodimerize with E proteins to inhibit DNA binding (14). Thus, the G1-specific effects of Id1 suggest that protein-protein interactions (e.g., Ref (28), rather than DNA binding, may be critical to E2A associations with Igλ at later stages of cell cycle. This raises the possibility of distinct modes of E2A association with Igλ during the cell cycle.
Additional lines of evidence support the view that diversification is initiated in G1 phase. Somatic hypermutation in the human BL2 cell line can be induced by in vitro stimulation that takes place only during G1 phase, and point mutations first become evident within 90 minutes of stimulation, when cells are still in G1 (40). In murine B cells activated for CSR, IgH colocalizations with NBS1 or γ-H2AX, participants in the switch recombination pathway, are prominent in G1 phase (44); and DNA breaks at the S regions can be detected in G1 phase (45). DNA breaks have also been identified in later stages of cell cycle in hypermutating human B cell lines (46), but these proved to be AID-independent (47).
We note that experiments thus far have not analyzed the course of diversification in a single cell. It is therefore possible that E2A may function in G1 phase to prepare a locus for events that occur later in cell cycle, or even during a subsequent cell cycle. E2A has been recently implicated in maintenance of histone H4 acetylation (24), and it is possible that E2A functions to establish a local chromatin environment favorable to AID attack or effective diversification in daughter cells.
Acknowledgments
We thank Greg Martin (Keck Imaging Center) for help with fluorescence microscopy; Michele Black, David Bell, Thong Pham and Mike Shen for help with cell sorting; Akiko Yabuki, Molly Weiner and David Bednarski for assistance in preliminary experiments; and all members of the Maizels laboratory for discussions.
Abbreviations used
- AD
activation domain
- AID
activation-induced cytidine deaminase
- ChIP
chromatin immunoprecipitation
- CSR
class switch recombination
- DAPI
4,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- Ig
immunoglobulin
- IPTG
isopropyl-β-D-thiogalactopyranoside
- LacI
lactose repressor
- PCR
polymerase chain reaction
- PI
propidium iodide
- PolyLacO
polymerized lactose operator
- r
nuclear radius
- RFP
red fluorescent protein
- RT-PCR
reverse transcriptase-PCR
- sIgM
surface IgM
- V
variable.
Footnotes
This work was supported by National Institutes of Health Grants R01 GM39799 and R01 GM41712 (to N.M.) and National Institutes of Health Predoctoral Training Programs (T32 GM07223 and T32 AG00057 to E.C.O.).
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Maizels N. Immunoglobulin gene diversification. Annu. Rev. Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 2.Martomo SA, Gearhart PJ. Somatic hypermutation: subverted DNA repair. Curr. Opin. Immunol. 2006;18:243–248. doi: 10.1016/j.coi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 4.Teng G, Papavasiliou FN. Immunoglobulin somatic hypermutation. Annu. Rev. Genet. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the Activation-Induced Deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 8.Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 9.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 11.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 12.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 13.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 14.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 15.Hagman J, Lukin K. Transcription factors drive B cell development. Curr. Opin. Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Murre C. Regulation and function of the E2A proteins in B cell development. Adv. Exp. Med. Biol. 2007;596:1–7. doi: 10.1007/0-387-46530-8_1. [DOI] [PubMed] [Google Scholar]
- 17.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 22.Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, Mori K, Yokota Y, Shimizu A. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoetz U, Cervelli M, Wang Y, Fiedler P, Buerstedde J. E2A Expression Stimulates Ig Hypermutation. J. Immunol. 2006;177:395–400. doi: 10.4049/jimmunol.177.1.395. [DOI] [PubMed] [Google Scholar]
- 24.Kitao H, Kimura M, Yamamoto K, Seo H, Namikoshi K, Agata Y, Ohta K, Takata M. Regulation of histone H4 acetylation by transcription factor E2A in Ig gene conversion. Int. Immunol. 2008;20:277–284. doi: 10.1093/intimm/dxm140. [DOI] [PubMed] [Google Scholar]
- 25.Conlon TM, Meyer KB. The chicken Ig light chain 3'-enhancer is essential for gene expression and regulates gene conversion via the transcription factor E2A. Eur. J. Immunol. 2006;36:139–148. doi: 10.1002/eji.200535219. [DOI] [PubMed] [Google Scholar]
- 26.Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 27.Kothapalli N, Norton DD, Fugmann SD. Cutting Edge: A cis-Acting DNA Element Targets AID-Mediated Sequence Diversification to the Chicken Ig Light Chain Gene Locus. J. Immunol. 2008;180:2019–2023. doi: 10.4049/jimmunol.180.4.2019. [DOI] [PubMed] [Google Scholar]
- 28.Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol. Cell. Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat. Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Humphries EH, Tjoelker L, Carlson L, Thompson CB. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol. Cell. Biol. 1990;10:3224–3231. doi: 10.1128/mcb.10.6.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 2005;15:470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 32.Larson ED, Cummings WJ, Bednarski DW, Maizels N. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol. Cell. 2005;20:367–375. doi: 10.1016/j.molcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Cummings WJ, Yabuki M, Ordinario EC, Bednarski DW, Quay S, Maizels N. Chromatin structure regulates gene conversion. PLoS Biol. 2007;5:e246. doi: 10.1371/journal.pbio.0050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 35.Engel I, Murre C. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc. Natl. Acad. Sci. USA. 1999;96:996–1001. doi: 10.1073/pnas.96.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- 37.Conlon TM, Meyer KB. Cloning and functional characterisation of avian transcription factor E2A. BMC Immunol. 2004;5:11. doi: 10.1186/1471-2172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belmont AS, Straight AF. In vivo visualization of chromosomes using lac operator-repressor binding. Trends Cell Biol. 1998;8:121–124. doi: 10.1016/s0962-8924(97)01211-7. [DOI] [PubMed] [Google Scholar]
- 39.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc. Natl. Acad. Sci. USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, Reynaud CA. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 41.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 42.Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. USA. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, Redon C, Ried T, Bonner WM, Honjo T, Nussenzweig MC, Nussenzweig A. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J. Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 46.Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 47.Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]


