The viability of all living organisms depends on the stability of natively folded proteins, and a vast experimental literature has been devoted to probing this stability in vitro using chemical denaturants. Guanidinium chloride (GdmCl) and urea are the two chemical denaturants most frequently used to disrupt native structure in studies of protein folding, GdmCl being the significantly stronger denaturant of the two.1 The microscopic mechanisms by which these cosolvents cause denaturation, however, remain a matter of some controversy. Classic approaches to the question have either focused on the increased solubility of hydrophobic amino acids in denaturant solution2,3 or else have posited a certain number of binding sites for the cosolvent molecules exposed in the unfolded protein.4 Some studies in silico, meanwhile, have seen a role for denaturants in attenuating the drive to bury hydrophobic side chains,5,6 while others have pointed to the importance of favorable electrostatic interactions between cosolvent molecules and those polypeptide atoms that are more exposed to the solvent in the denatured state.7,8 Here, we add a new perspective to this debate by examining the impact of chemical denaturants on the phenomenon of dewetting. In the current context, dewetting refers to the process sometimes also known as capillary evaporation, by which water trapped between nanoseparated hydrophobic surfaces is driven by confinement to undergo pronounced evaporation.9 Using molecular dynamics simulations of hydrophobic plates immersed in water and denaturant solutions, we demonstrate that both urea and GdmCl stabilize the liquid phase against dewetting, thereby diminishing the attraction between hydrophobic surfaces. Our findings indicate that urea and guanidinium could act, at least in part, by stabilizing more expanded polypeptide conformations against hydrophobic collapse.
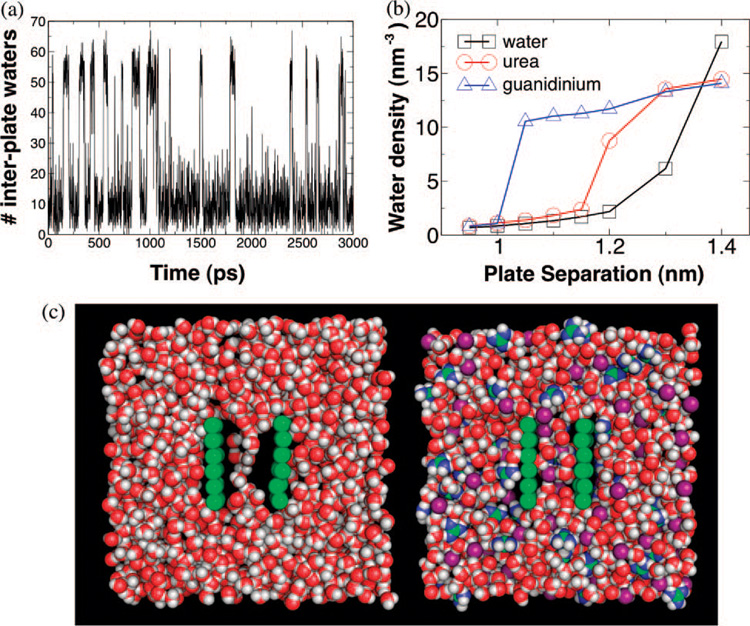
Dewetting has previously been observed in all-atom simulations of ellipsoidal hydrophobic plates held at subnanometer separations9,10 and is thought to be important for the collapse of hydrophobic polymers,11 the assembly of some macromolecular complexes,12 and the operation of certain membrane ion channels.13 To test the effect of denaturants on dewetting, we set up a simple system in which the phenomenon could be observed in silico. Hydrophobic plates were simulated at various separations in water, 5MGdmCl, or 5Murea (see Supporting Information). The pure water system demonstrated a dewetting transition brought on by increased confinement of the liquid phase between the plates. At a plate separation of 1.4 nm, water readily permeated the volume between the plates, forming a dense liquid. At 1.3 nm, however, the liquid and vapor phases became nearly comparable in stability (within roughly 2 kT), and the system vacillated between these two free energy minima (Figure 1a,c). At even shorter distances, the vapor phase became dominant. We found that both urea and GdmCl bring about a quantitative change in the energetic balance between vapor and liquid phases in the volume between plates.
Figure 1.
(a) Time trace of the number of water molecules between plates immersed in water at 1.3 nm separation. The interplate water density fluctuates between a liquid phase and a vapor phase of nearly comparable stability. (b) Interplate water density is calculated as a function of plate separation for hydrophobic plates in pure water (black squares), 5 M urea (red circles), and 5 M GdmCl (blue triangles). Both cosolvents stabilize the liquid phase at the plate separation where pure water dewets, with GdmCl having a stronger effect than urea. (c) Snapshots of the simulated system for pure water at 1.3 nm plate separation (left) and for 5 M GdmCl at 1.2 nm plate separation (right). Carbon and plate atoms are colored green; hydrogens, white; oxygens, red; chloride ions, purple; and nitrogens, blue.
For a separation of 1.3 nm, the liquid phase was stable for both urea and GdmCl solutions (Figure 1b,c). By 1.2 nm, however, 5 M urea solution began to dewet, while the solution with the same concentration of GdmCl remained stable (Figure 1b) with a mixture of both guanidinium and chloride ions present between the plates. Only near 1.0 nm separation did the GdmCl solution finally withdraw from the volume confined between the plates. It should moreover be noted that the suppression of dewetting we observed for denaturants is not an effect that is general to small cosolutes. In separate simulations we performed on plates in a KCl solution, the presence of the ions proved to promote dewetting relative to pure water (see Supporting Information).
Although urea and GdmCl both demonstrated an ability to inhibit the onset of dewetting, the two cosolvents apparently differ somewhat at a microscopic level in how they achieve this feat. By calculating the mean-squared cosine of the angle between the normal vectors of the planes of denaturant molecules and that of the hydrophobic plates, it was possible to quantify any orientational bias near the plates. We found that guanidinium ions tend to orient their flat, relatively less-polar faces toward the hydrophobic plates when between the plates, and even when up to 1 nm away from them (Figure 2). This result was understandable given past evidence from both simulation and experiment that the flat faces of guanidinium ions are locally water-depleted,6,14 which may facilitate hydrophobic stacking of pairs of cations.6 In contrast, urea molecules, which have essentially the same size and shape, showed less bias toward flattened orientations and oriented significantly only when close enough to the hydrophobic surfaces for sterics to favor flattening (Figure 2). The difference between the two cosolvents is most notable halfway between the plates, where guanidinium exhibits a pronounced orientational bias while urea exhibits none at all.
Figure 2.
(a) The mean-squared projection () of the normal vector to the plane of cosolvent molecules onto the z-axis (which is normal to the plates) is plotted as a function of the z-coordinate in a simulation with an interplate distance of 1.3 nm. Only molecules in the same x—y region as the plates were counted. Urea (red circles) shows relatively little orientational bias (unless very near the plates), whereas guanidinium ions (blue triangles) tend to adopt an orientation parallel to the plate surface. (b) A schematic illustration of a cosolvent molecule (blue) and its orientation with respect to nearby plates (gray).
Urea molecules were found, however, to be visibly more prone to aggregate and form transient, nanosized clumps,15 with the result that the urea density fluctuations between the plates were much higher (Figure S1 in Supporting Information). This greater tendency toward aggregation is unsurprising given that, unlike guanidinium cations, urea molecules have no net charge to produce a repulsion between them. Urea molecules are, moreover, able to form hydrogen bonds with each other, which helps to explain the lesser tendency of urea to flatten out against the plate surface: orientational diversity favors the formation of a more extensive network of hydrogen bonds.
Past computational studies of urea and GdmCl have had difficulty quantitatively reconciling the cosolvents’ polarity, solubility, and apparent affinity for charged species with other data, which range from calculations of hydrogen bond strengths in simulation5 to measurements of experimental transfer free energies,2,3 suggesting that denaturants weaken hydrophobic effects. Here, we have presented direct quantitative evidence for the attenuation of the hydrophobic effect, manifested as dewetting, by the denaturants. Since dewetting normally occurs when the free energy cost of forming an interface between the liquid and the plates overwhelms the free energy contributed by bulk pressure forcing liquid into the region between the plates,10 the stabilization of water under tighter confinement by denaturant molecules demonstrates their capacity to reduce the cost of interface formation. The effect of guanidinium is achieved through interaction of its dehydrated6,14 flat face with hydrophobic surfaces; the guanidinium cation appears to act like a middle man between the nonpolar surface to which it binds and the neighboring water molecules. While it is possible that urea may also play the same role to some extent, the less polar cosolvent’s greater tendency to clump together forces us to consider the possibility that the effect of urea on dewetting is due to the formation of transient, aggregated droplets15 that allow the liquid phase to remain stable even as it becomes locally depleted of water.
In light of experiments pointing to a dominant contribution of solvation free energy changes for the polypeptide backbone to destabilization by urea,16 and of evidence that polypeptides collapse in water even in the absence of hydrophobic sidechains,17 it is reasonable to expect that denaturants could disrupt the native structure by stabilizing more expanded conformations that expose more backbone to the solvent. The work presented here, however, should not be misunderstood to contradict past observations that suggest polar interactions may also be important to the denaturation mechanism. Indeed, it is quite plausible that GdmCl and urea are both so effective as denaturants because of the combination of effects they bring to bear on a protein,18,19 striking a balance between interacting with hydrogen bonding partners in the polypeptide while also excluding water from the more hydrophobic parts of the chain.
Future simulations will probe the relative importance of, and possible cooperation between, these two mechanisms of denaturation in protein folding. At the same time, the origins of differences between the effects of different cosolvents demand more precise characterization. The differences in orientational tendencies of urea and guanidinium observed here may, for example, have an effect on each denaturant’s ability to solvate aromatic versus aliphatic side chains. In addition, while the current work presents a quantitative method for testing the modulation of hydrophobic interactions by chemical denaturants, it can readily be used for probing the strength of the hydrophobic effect in the presence of other osmolytes,20,21 some of which (such as trimethylamine n-oxide) actually stabilize native proteins.
Acknowledgment
J.E. thanks the Fannie and John Hertz Foundation for financial support and Imran Haque for helpful comments. The research of G.H. is made possible in part by the generosity of the Perlman Family, as well as by financial support of the NIH (Grant No. 1R01GM080515). V.P. acknowledges support from the NIH (PN1 EY016525-02 and U54 GM072970) and NSF (EF-0623664 and CNS-0619926).
Footnotes
Supporting Information Available: Simulation details and simulations with potassium chloride as a solute. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pace CN. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 2.Whitney PL, Tanford C. J. Biol. Chem. 1962;237:1735–1737. [PubMed] [Google Scholar]
- 3.Nozaki Y, Tanford C. J. Biol. Chem. 245;1969:1648–1652. [PubMed] [Google Scholar]
- 4.Schellman JA. Biopolymers. 1989;26:549–559. doi: 10.1002/bip.360260408. [DOI] [PubMed] [Google Scholar]
- 5.Stumpe M, Grubmüller H. J. Am. Chem. Soc. 129;2007:16126–16131. doi: 10.1021/ja076216j. [DOI] [PubMed] [Google Scholar]
- 6.Mason PE, Neilson GW, Enderby JE, Saboungi M-L, Dempsey CE, MacKerell AD, Jr, Brady JW. J. Am. Chem. Soc. 2004;126:11462–11470. doi: 10.1021/ja040034x. [DOI] [PubMed] [Google Scholar]
- 7.Mountain R, Thirumalai D. J. Am. Chem. Soc. 2003;125:1950–1957. doi: 10.1021/ja020496f. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien EP, Dima RI, Brooks B, Thirumalai D. J. Am. Chem. Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- 9.Wallqvist A, Berne BJ. J. Phys. Chem. 1995;99:2893–2899. [Google Scholar]
- 10.Huang X, Margulis CJ, Berne BJ. Proc. Nat. Acad. Sci. U.S.A. 2003;100:11953–19958. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller TF, Vanden-Eijinden E, Chandler D. Proc. Nat. Acad. Sci. U.S.A. 2007;104:14559–14564. doi: 10.1073/pnas.0705830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P, Huang X, Zhou R, Berne BJ. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 13.Anishkin A, Sukharev M. Biophys. J. 2004;86:2883–2895. doi: 10.1016/S0006-3495(04)74340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason PE, Neilson GW, Dempsey CE, Barnes AC, Cruickshank JM. Proc. Nat. Acad. Sci. U.S.A. 2003;100:4557–4561. doi: 10.1073/pnas.0735920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stumpe M, Grubmüller H. J. Phys. Chem. B. 2007;111:6220–6228. doi: 10.1021/jp066474n. [DOI] [PubMed] [Google Scholar]
- 16.Auton M, Bolen W. Proc. Nat. Acad. Sci. U.S.A. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Möglich A, Joder K, Kiefhaber T. Proc. Nat. Acad. Sci. U.S.A. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennion BJ, Daggett V. Proc. Nat. Acad. Sci. U.S.A. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camilloni C, Rocco AG, Eberini I, Gianazza E, Broglia RA, Tiana G. Biophys. J. 2008;94:4654–4662. doi: 10.1529/biophysj.107.125799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Street TO, Bolen DW, Rose GD. Proc. Nat. Acad. Sci. U.S.A. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harries D, Rösgen J. Methods Cell Biol. 2008;84:679–735. doi: 10.1016/S0091-679X(07)84022-2. [DOI] [PubMed] [Google Scholar]