Abstract
Chemoenzymatic routes toward complex glycoconjugates often depend on the availability of sugar-1-phosphates. Yet the chemical synthesis of these vital components is often tedious, whereas natural enzymes capable of anomeric phosphorylation are known to be specific for one or only a few monosaccharides. Herein we describe the application of directed evolution and a high-throughput multisugar colorimetric screen to enhance the catalytic capabilities of the Escherichia coli galactokinase GalK. From this approach, one particular GalK mutant carrying a single amino acid exchange (Y371H) displayed a surprisingly substantial degree of kinase activity toward sugars as diverse as d-galacturonic acid, d-talose, l-altrose, and l-glucose, all of which failed as wild-type GalK substrates. Furthermore, this mutant provides enhanced turnover of the small pool of sugars converted by the wild-type enzyme. Comparison of this mutation to the recently solved structure of Lactococcus lactis GalK begins to provide a blueprint for further engineering of this vital class of enzyme. In addition, the rapid access to such promiscuous sugar C-1 kinases will significantly enhance accessibility to natural and unnatural sugar-1-phosphates and thereby impact both in vitro and in vivo glycosylation methodologies, such as natural product glycorandomization.
Keywords: galactokinase, glycorandomization, in vitro, evolution enzyme
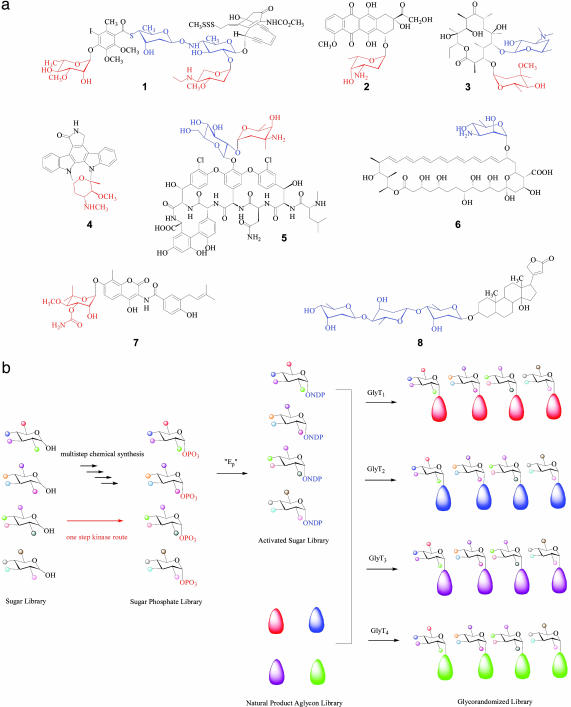
Many clinically important medicines are derived from glycosylated natural products, the d- or l-sugar substituents of which often dictate their overall biological activity. This paradigm is found throughout the anticancer and antiinfective arenas with representative clinical examples (Fig. 1a), including enediynes (calicheamicin, 1), polyketides (doxorubicin, 2; erythromycin, 3), indolocarbazoles (staurosporine, 4), nonribosomal peptides (vancomycin, 5), polyenes (nystatin, 6), coumarins (novobiocin, 7), and cardiac glycosides (digitoxin, 8) (1–7). Given the importance of the sugars attached to these and other biologically significant metabolites, extensive effort has been directed in recent years toward altering sugars as a means to enhance or alter natural product-based therapeutics by both in vivo and in vitro approaches (ref. 8 and references therein). Among these, in vitro glycorandomization (IVG) makes use of the inherent or engineered substrate promiscuity of nucleotidylyltransferases and glycosyltransferases to activate and attach chemically synthesized sugar precursors to various natural product scaffolds (6, 7, 9–15), the advantage of which is the ability to efficiently incorporate highly functionalized “unnatural” sugar substitutions into the corresponding natural product scaffold (Fig. 1b). In a recent demonstration of IVG, >50 analogs of 5 were generated, some of which displayed enhanced and distinct antibacterial profiles from the parent natural product (13).
Fig. 1.
(a) Representative examples for natural product glycosides used as therapeutics: calicheamicin (1), doxorubicin (2), erythromycin (3), staurosporine (4), vancomycin (5), nystatin (6), novobiocin (7), and digitoxin (8). The attached sugars are highlighted in color with red indicating l-configured, sugars and blue representing d-sugars. (b) Schematic for natural product IVG. Ep, α-d-glucopyranosyl phosphate thymidylyltransferase; GlyTn different glycosyltransferases.
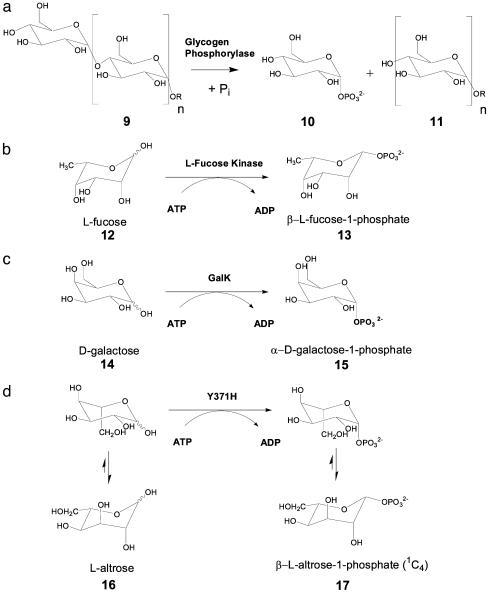
As starting materials, sugar phosphates play a key role in the entire IVG process. Thus, the ability to rapidly construct sugar phosphate libraries would directly contribute to the efficiency of IVG. Compared to the existing chemical synthetic methods for anomeric phosphorylation (14–15), single step enzymatic (kinase) routes bypass required multistep synthetic manipulations and could be coupled to IVG in a single reaction vessel. Known C-1 phosphorylating enzymes are limited to mainly three types (Fig. 2): the glycogen phosphorylases (16), which convert glycogen (9) into d-glucose-1-phosphate (10); fucokinases, which transfer a phosphate from ATP to the anomeric position of l-fucose (12) to provide β-l-fucose-1-phosphate (13) (17); and the galactokinases (GalK), which catalyze the formation of α-d-galactose-1-phosphate (Gal-1-P, 15) from d-galactose (14) and ATP. Previous studies have revealed that GalK from various sources have a limited substrate scope (18–21), and in all C-1 kinases studied thus far, a strict adherence to either d-sugars (GalK and glycogen phosphorylases) or l-sugars (as in fucokinase) was observed. Thus, to apply any of these kinases for generating a randomized sugar phosphate library, their monosaccharide substrate promiscuity must first be enhanced.
Fig. 2.
Reactions catalyzed by anomeric kinases. (a) Glycogen phosphorylase. (b) Fucokinase. (c) Galactokinase. (d) Proposed phosphorylation of l-altrose accomplished by the evolved GalK mutant Y371H.
Two general routes for altering enzyme substrate specificity are currently available. Structure-based engineering relies on knowledge of an enzyme's three-dimensional structure and an explicit molecular-level understanding of substrate recognition. An example of structure-based engineering as applied to IVG includes increasing the substrate scope of nucleotidylyltransferases used (Fig. 1b,Ep). The application of Ep rational engineering led to active site mutants capable of accepting a variety of substrates not used by the wild-type enzyme (9–11). The alternative to rational engineering is enzyme evolution, a process primarily dependent on the availability of a selection or high-throughput screen for the desired enhanced or altered enzymatic properties (22–24). With respect to carbohydrate enzymology, recent applications include the tagatose-1,6-bisphosphate aldolase modified by in vitro evolution toward an unnatural stereoselectivity (25), an evolved N-acetylneuraminic acid aldolase for l-sialic acid synthesis (26), or a 2-deoxy-d-ribose-5-phosphate aldolase with an expanded substrate range after site-directed mutagenesis (27). Usually, in vitro evolution strategies include error-prone PCR for gene diversification (28, 29) and/or locating critical amino acid residues for saturation mutagenesis (30) or, more prominently, shuffling of fragmented diversified genes or gene families according to a number of different protocols (31–36). Subsequently, the diversified proteins are subjected to a screen.
Although the first structure for a sugar C-1 kinase (GalK from Lactococcus lactis) recently emerged (37), the extreme variability in solution structures among anticipated monosaccharide library members and the availability of a specific high-throughput sugar anomeric kinase colorimetric screen (21) prompted an initial evolutionary approach. As a model system, we selected the well characterized Escherichia coli galactokinase GalK (38) and focused the evolutionary approach toward significant C-5 (e.g., l-sugar variants) and C-6 (e.g., deoxy, amino, and uronic acid derivatives) alterations in an attempt to probe and elucidate the specific enzymatic architecture responsible for restricting substrate substitution at C-5 and C-6. Herein we describe the application of directed evolution and a high-throughput multisugar colorimetric screen to enhance the catalytic capabilities of the E. coli GalK. From this approach, one particular GalK mutant carrying a single amino acid exchange (Y371H) displayed a surprisingly substantial degree of kinase activity toward sugars as diverse as d-galacturonic acid, d-talose, l-altrose, and l-glucose, all of which failed as wild-type GalK substrates. Furthermore, this mutant provides enhanced turnover of the small pool of sugars converted by the wild-type enzyme. Comparison of this mutation to the recently solved structure of L. lactis GalK begins to provide a blueprint for further engineering of this vital class of enzyme. In addition, the rapid access to such promiscuous sugar C-1 kinases will significantly enhance accessibility to natural and unnatural sugar-1-phosphates and thereby impact both in vitro and in vivo glycosylation methodologies, such as natural product glycorandomization.
Materials and Methods
Materials. E. coli strains XL1-Blue and BL21-Gold (DE3) were purchased from Stratagene. The template plasmid pGalK has been described (21). Expression vector pET15b was from Novagen. All reagent-grade chemicals and enzymes were purchased from Promega, Sigma, Fisher, or Fluka.
Chemical Synthesis of d- and l-idose. The syntheses of d- and l-idose followed preparations described in refs. 39–41.
Gene Diversification, Library Preparation, and Characterization. For the gene library used, error-prone PCR was accomplished under the following conditions: 25 mM MgCl2/0.1 mM MnCl2/0.2 mM each dATP and dGTP/1.0 mM each dCTP and dTTP/500 pg of template plasmid pGalK/40 pmol of each primer 5′-CTTGGTTATGCGGGTACTGC-3′ and 5′-TCCCGCGAAATTAATACGAC-3′/5 units of TaqDNA-polymerase in the buffer supplied with the enzyme, in a total volume of 100 μl. Thermocycle parameters were as follows: initial denaturation, 5 min, 94°C; amplification, 30 cycles, 94°C for 0.5 min, 54°C for 0.5 min, and 72°C for 1.5 min; terminal hold, 5 min at 72°C. The amplification products were digested with BamHI/XbaI, purified on an agarose gel (0.8% w/vol), eluted by using the QIAquick Gel Extraction kit (Qiagen, Valencia, CA), and ligated into appropriately digested pET15b to directly transform E. coli XL1-Blue. Plasmids were isolated from randomly picked colonies (≈20 representatives for each library generated) and sequenced on an ABI 310 automatic DNA sequencer (Perkin–Elmer). On verifying the desired mutation rate, all transformants were pooled and cultured overnight. The collectively recovered plasmids were used to transform E. coli BL21-Gold (DE3), and the library was processed as described below.
Bacterial Fermentation. E. coli was grown in LB and supplemented with ampicillin (100 μg ml-1 final) under standard conditions. For expression of GalK enzyme library members, individual transformants were grown as 1-ml miniature cultures in 96-deep well blocks overnight as seed cultures, then replicated in fresh medium (2%, vol/vol). Afterward, 15% (vol/vol) glycerol was added, and the culture was mixed and stored at -80°C. The replicated cultures were grown to an OD600 ≈0.7; protein expression was induced by adding 1 mM isopropyl β-d-thiogalactoside (final) for 2 h. The cultures were then harvested by centrifugation for 10 min at 3,000 × g. The cell paste was frozen at -20°C. After the harvested expression cultures were thawed, the biomass of each well was resuspended in 50 μl of NPI buffer (50 mM NaH2PO4/300 mM NaCl, pH 8.0) to which was then added 70 μl of NPI buffer supplemented with 1 μl of Lysonase (Novagen) for lysis. Cell debris was collected by centrifugation (10 min at 3,000 × g), and 20 μl of the clear supernatant, which contained an average of ≈0.5 μg of the expressed GalK variant, was used for each kinase assay.
Library Screening. Enzymatic reaction mixtures and assays were set up and read in 96-well format on a Biomek FX Liquid Handling Workstation (Beckman Coulter) fitted to a FLUOstar OPTIMA plate reader (BMG Labtechnologies, Durham, NC). The in vitro enzymatic reaction mixtures and assays followed the previously published protocol (21), which was modified slightly for automated liquid handling: 150 μl of sugar solution (8 mM final) and 12 μl of ATP/Mg2+ solution (20 mM/5 mM final) were mixed and preincubated at 37°C for 5 min; then, 20 μl of the cleared supernatant was added. The reaction mixture was then incubated at 37°C for an additional 2 h. To assay the phosphorylation reactions, 50 μlofthe enzymatic reaction mixture was taken at time 0 and, after the 2-h incubation, mixed with 100 μl of 3,5-dinitrosalicylic acid (DNS) reagent, heated at 100°C for 5 min, and immediately chilled on ice for 2 min. Assays were run as endpoint measurements after the decrease in absorption (λ = 575 nm, ε = 758 M-1·cm-1), as described in ref. 21.
Characterization of GalK Mutants. The GalK mutant Y371H was overexpressed and purified following the procedure described for wild-type GalK (21). The fractions containing homogenous Y371H GalK were collected, concentrated, and quantified by using a Bradford protein assay (42). The DNS assay was used to assess the substrate specificity of the purified GalK mutant (Y371H) as described (21). Standard curves for each sugar were prepared as described (21). To determine the kinetic data for each active monosaccharide substrate, the sugar concentration was varied over a range of 1–16 mM under saturating ATP (15 mM). Change in absorbance at 575 nm as a function of time was obtained by using the DNS assay, and the initial velocity was determined by the slope of the linear phase in the progress curve. The kinetic data were analyzed by using ENZYME KINETICS MODULE software (SSPS, Chicago) as described (21).
Preparative Phosphorylation of l-altrose and Product Characterization. l-altrose (21.6 mg, 0.12 mmol) was dissolved in 15 ml of 50 mM sodium phosphate buffer (pH 7.5). To this solution, ATP (125 mg, 0.23 mmol) and MgCl2·6H2O (15.3 mg, 0.07 mmol) were added, the mixture was incubated at 37°C for 5 min, the reaction was initiated via the addition of enzyme (Y371H) to a final concentration of 150 μg ml-1, and reaction progress was monitored by TLC. After completion, the mixture was diluted 5-fold in ddH2O and applied to a 200-ml anion exchange column (Q-Sepharose Fast Flow, Amersham Pharmacia). The column was eluted with a gradient of 0–400 mM NaCl, and the active fraction was collected and evaporated under reduced pressure. The crude product was desalted on a P-2 gel filtration column (The Nest Group, Southport, MA) to give 16.1 mg of purified product (yield: 52%). [α]D = 3.5° (c = 1, H2O) 1H NMR (2H2O): 5.48 (dd, J = 8.6, 1.8 Hz, 1H), 4.14 (dd, 5.3, 3.3 Hz, 1H), 3.99 (m, 1H), 3.98 (dd, J = 4.3, 1.8 Hz, 1H), 3.91 (d, J = 3.3, 1H), 3.89 (m, 1H), 3.84 (dd, J = 12.1, 7.8 Hz, 1H); 13C NMR (2H2O): 94.10, 76.23, 70.70, 69.99, 65.68, 62.21; 31P NMR (2H2O): 3.77; MS: calculated for C6H13O9P 260.0, found m/z 259.0 [M - H]-.
Results and Discussion
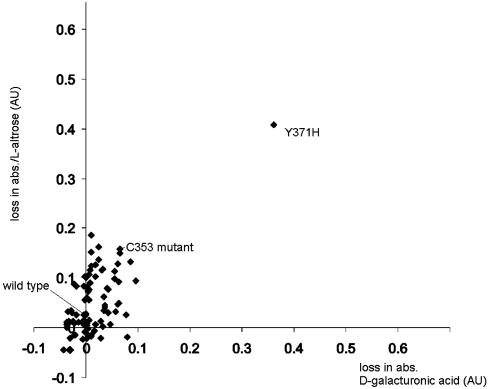
Directed Evolution of GalK with Expanded Specificity. The cloned wild-type galK gene from E. coli was subjected to random mutagenesis by error-prone PCR performed over the entire gene. The level of sequence alteration was adjusted to an average of 1.5 aa substitutions per enzyme molecule and verified by the DNA sequencing of corresponding genes. However, error-prone PCR usually results in a more or less strong mutational bias (28, 29, 43) and is therefore not a truly random process. Under the conditions selected, the library contained a transition/transversion ratio of ≈3.0, the transitions outnumbering the transversions, and the AT→GC to GC→AT ratio was found to be 2.4. An evaluation of 3,500 GalK variants from this library was conducted to determine the ability of the variants to accommodate an expanded spectrum of sugar substrates. Our principal goal was kinase activity toward l-configured sugars (C-5 alteration) or to those with altered substituents at C-6. Unlike most assays in high-throughput campaigns that screen or select toward a single substrate or substrate mimic, our recently developed DNS assay is universally applicable to all reducing sugars (21). Consequently, simultaneous screens for a relaxed enzyme specificity were carried out with a set of appropriately selected sugar substrates rather than a one-dimensional single substrate screen of each library member. To first focus on C-5 and C-6, the set included a single C-5 challenge [l-altrose (Table 1; 16, the C-5-epimer of d-galactose)] and three C-6 challenges [6-deoxy-d-galactose (22, d-fucose), 6-amino-6-deoxy-d-galactose (23), and d-galacturonic acid (24)]. Of this set, only d-fucose (22) showed any turnover with wild-type GalK, albeit 2.7% of the turnover rate observed for d-galactose (14) (21). The graphical display of a typical screening result is given in Fig. 3. Surprisingly, after only one round of evolution, two GalK variants appeared independently and phosphorylated the C-5/C-6 set of sugars with roughly similar efficiency. Subsequent DNA sequencing of both GalK variant genes revealed that they were identical in their sequence and carried a single, forward mutation, a T→C transition at position 1,111 of the wild-type reading frame, which translates into a tyrosine-371-to-histidine replacement.
Table 1. Kinetic data for the 10 active substrates of GalK variant Y371H.
—, No conversion.
Fig. 3.
Representative quantitative data for a set of GalK variants, illustrating screen for d-galacturonic acid (x axis) and l-altrose (y axis). The higher the loss in absorption [shown in absorption units (AU)], the more active the enzyme variant.
Recently, the structure of the L. lactis galactokinase was published (37). Despite a rather low sequence homology to the E. coli GalK (36% identity, 53% similarity), these two kinases clearly share three characteristic footprint motifs (44), and all amino acid residues found within the catalytic center of the L. lactis galactokinase (R36, E42, D45, D183, and Y233) are invariably present in its E. coli homolog (R28, E34, D37, D174, and Y223, respectively) and embedded in highly conserved sequence environments. For this reason, we speculate that the equivalent residues also form the catalytic apparatus in the E. coli enzyme. Surprisingly, residue 371 in the E. coli wild-type enzyme, found to be essential for widened substrate specificity and activity toward l-configured sugars, does not appear as part of its deduced active site. In the L. lactis galactokinase, the Cα of the equivalent amino acid Y385, which is located within the C-terminal domain β-strand K, is ≈20 Å from the anomeric carbon of the substrate when bound in the active site. In the L. lactis galactokinase crystal structure, Y385 is located in close proximity to C353 (C339 in E. coli GalK). The tyrosine phenolic oxygen is located ≈5.5 Å from the cysteine sulfur atom. Interestingly, during our screen, a mutant on position 339 was discovered with a similarly expanded substrate profile, yet this mutant also showed low catalytic activity regardless of the sugar (data not shown). This finding suggests that a potential Y385–C353 (Y371–C339 in E. coli GalK) side-chain interaction may play a role in stabilizing this structure, thereby dictating substrate specificity. Although an induced-fit model has not been previously put forth for this class of enzyme, such “gate-keeping” interactions are known to occur in other enzymes devoted to carbohydrate metabolism, such as hexokinase (45).
Characterization of the GalK Variant (Y371H). To determine the substrate specificity of the Y371H GalK variant, a sugar library of 20 putative substrates was tested with the purified enzyme. For each sugar, both the DNS assay and TLC were used to monitor the reaction progress; control assays in the absence of enzyme or sugar were performed in parallel. The mutant GalK demonstrated the ability to turn over compounds 14, 16, and 18–25 (Table 1), strikingly expanding the overall substrate scope compared with wild-type E. coli GalK. The kinetic parameters of the mutant enzyme with all active substrates (14, 16, and 18–25) were determined by using the DNS assay and a comparison to wild-type GalK activity. These kinetic studies also revealed, as expected, that the evolved enzyme remains an efficient catalyst with d-galactose (kcat = 220 min-1, Km = 5.6 mM) and displays remarkably enhanced kcat values for all of the previously known substrates for wild-type GalK (14, 18, 19, 21, and 22), the affinity for which is slightly reduced in all cases.
Whereas most in vitro evolution projects require repeated rounds of random mutation and/or recombination to generate the desired activity, a leap in GalK catalytic activity and substrate selectivity was accomplished in the initial round of random mutagenesis. Other recent similar examples of single forward mutations leading to a catalytic shift include the Arabidopsis thaliana cycloartenol synthase, yeast lanosterol synthase (46–49), and the adipyl acylase evolved from a Pseudomonas glutaryl acylase (50). From an analysis of the GalK substrate specificity profiles, one can begin to construct a loose structure-activity requirement for both wild-type enzyme and the corresponding Y371H mutant. Specifically, wild-type GalK displays a stringent requirement for the substrate galactose architecture from C-3 through C-6 and is capable of limited flexibility toward substitution at C-2. Yet it is interesting to note that these stringent requirements, with the exception of the extensive contacts at C-4, are not readily apparent in the L. lactis GalK active site structure. In contrast to wild-type GalK, the Y371H mutation primarily retains only the stringent requirement for the C-4 galactose architecture with an enhanced substrate flexibility at all other positions of the sugar (for example, d-glucose, 4-deoxy-d-glucose, and d-mannose are not substrates, unpublished data). Remarkably, with essentially all substrates accepted, an enhancement of catalytic efficiency ranging from 5- to 22-fold was observed in the Y371H mutant. The only exception was the wild-type substrate d-galactose, for which the catalytic efficiency was decreased slightly in comparison to wild-type GalK, albeit kcat in this case was also increased 2-fold.
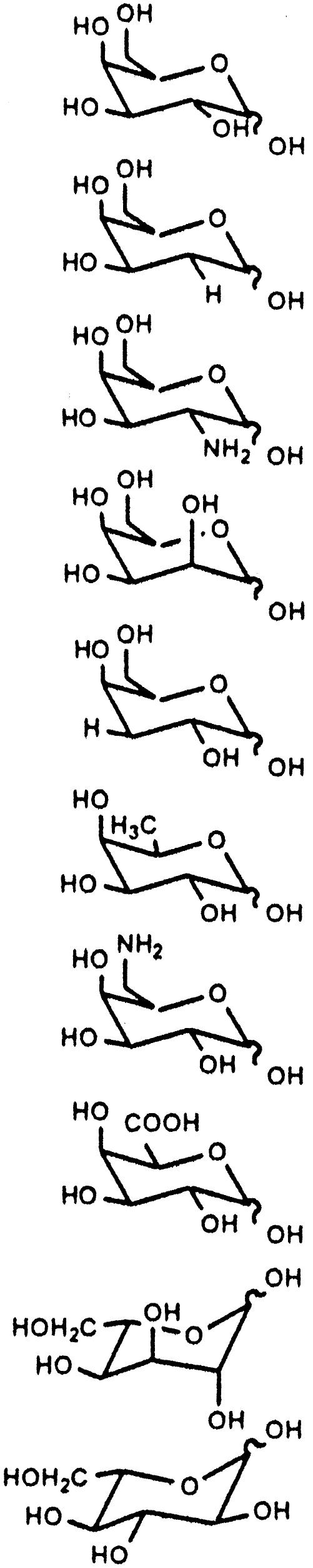
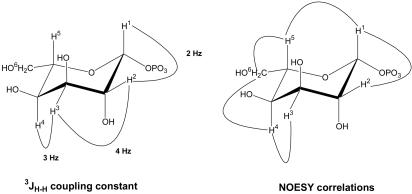
Confirmation of l-sugar Conversion. The substrate specificity studies have demonstrated GalK variant Y371H to be a d/l-unspecific sugar kinase. To confirm the evolved enzyme retains regio- and stereoselectivity with l-sugar substrates, a representative l-sugar reaction product was further characterized. Specifically, a small-scale preparative phosphorylation reaction was performed with l-altrose (21.6 mg, 0.12 mmol). The DNS assay indicated 91% of l-altrose conversion within 4 h. Product isolation was readily achieved by anion exchange chromatography, and the yield of purified product was ≈52%. The purified product was characterized by 1H and 13C NMR, from which H–H coupling and NOESY data confirmed the product to be β-l-altrose-1-phosphate in a 1C4 conformation (Fig. 4). In particular, 3JH–H coupling data showed two typical axial–equatorial couplings (H1–H2 and H3–H4) and one equatorial–equatorial coupling (H2–H3). NOESY data also revealed the anticipated correlations consistent with this structure (H1–H2, H1–H5, H3–H4, H4–H6, and H5–H6). Based on these data, we propose the Y371H mutant must bind and phosphorylate l-altrose in the same 4C1 conformation as d-galactose (Fig. 2 c and d) (37), the product of which subsequently rapidly equilibrates to the more stable 1C4 conformation on its release from the enzyme. The fact that gluco-configured sugars failed as substrates further supports this hypothesis.
Fig. 4.
3JH–H coupling patterns and NOESY correlations for the GalK Y371H product β-l-altrose-1-phosphate.
Implications for in Vitro Glycorandomization. Apart from total synthesis, current approaches to alter glycosidic structures include combinatorial biosynthesis or in vitro biocatalysis. Combinatorial biosynthesis primarily relies on in vivo diversification by means of the genetic engineering of involved sugar biosynthetic pathways (8). However, combinatorial biosynthesis is significantly limited by enzyme specificity, which substantially biases the ultimate extent of accessible diversity. In contrast, IVG presents a significant advantage by providing a truly unbiased library of activated sugars to use for drug-lead glycosylation. The present advent of kinase-enhanced IVG not only simplifies the upstream availability of sugar-1-phosphates for IVG but also potentially opens the door to in vivo applications of glycorandomization. Specifically, the expression of a tandem promiscuous sugar-1-kinase (GalK) and nucleotidylyltransferase (Ep) in a given organism should present the prospect of generating a library of NDP-sugars in situ. As such, this work provides the foundation for eventually “glycorandomizing” a variety of clinically important secondary metabolites in vivo to rapidly enhance drug discovery efforts.
Acknowledgments
We are grateful to the University of Wisconsin–Madison NMR facility and Dr. Thomas Stringfellow for the analysis of sugar phosphates and helpful discussions. This work was supported in part by National Institutes of Health Grants GM58196, CA84374, and AI52218. We gratefully acknowledge a postdoctoral fellowship to D.H. from the Deutsche Forschungsgemeinschaft. J.S.T. is an Alfred P. Sloan Research Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IVG, in vitro glycorandomization; DNS, 3,5-dinitrosalicylic acid.
References
- 1.Weymouth-Wilson, A. C. (1997) Nat. Prod. Rep. 14, 99-110. [DOI] [PubMed] [Google Scholar]
- 2.Potier, P. (1999) Actual. Chim. 11, 9-11. [Google Scholar]
- 3.Thorson, J. S. & Vogt, T. (2003) in Carbohydrate-based Drug Discovery, ed. Wong, C.-W. (Wiley–VCH, Weinheim, Germany), Vol. II, pp. 685-711. [Google Scholar]
- 4.Kren, V. & Martinkova, L. (2001) Curr. Med. Chem. 8, 1303-1328. [DOI] [PubMed] [Google Scholar]
- 5.Kren, V. (2001) in Glycoscience: Chemistry and Chemical Biology, eds. Fraser-Reid, B. O., Tatsuta, K. & Thiem, J. (Springer, Heidelberg, Germany), Vol. 3, pp. 2471-2529. [Google Scholar]
- 6.Thorson, J. S., Hosted, T. J., Jr., Jiang, J., Biggins, J. B. & Ahlert, J. (2001) Curr. Org. Chem. 5, 139-167. [Google Scholar]
- 7.Albermann, C., Soriano, A., Jiang, J., Vollmer, H., Biggins, J. B., Barton, W. A., Lesniak, J., Nikolov, D. B. & Thorson, J. S. (2003) Org. Lett. 5, 933-936. [DOI] [PubMed] [Google Scholar]
- 8.Mendez, C. & Salas, J. (2001) Trends Biotechnol. 11, 449-456. [DOI] [PubMed] [Google Scholar]
- 9.Barton, W. A., Biggins, J. B., Jiang, J., Thorson, J. S. & Nikolov, D. B. (2002) Proc. Natl. Acad. Sci. USA 99, 13397-13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton, W. A., Biggins, J. B., Lesniak, J., Jeffrey, P. D., Jiang, J., Rajashankar, K. R., Thorson, J. S. & Nikolov, D. B. (2001) Nat. Struct. Biol. 8, 545-551. [DOI] [PubMed] [Google Scholar]
- 11.Thorson, J. S., Barton, W. A., Hoffmeister, D., Albermann, C. & Nikolov, D. B. (2003) Chembiochem, in press. [DOI] [PubMed]
- 12.Jiang, J., Albermann, C. & Thorson, J. S. (2003) Chembiochem 4, 443-446. [DOI] [PubMed] [Google Scholar]
- 13.Fu, X., Albermann, C., Jiang, J., Liao, J., Zhang, C. & Thorson, J. S. (2003) Nat. Biotechnol., in press. [DOI] [PubMed]
- 14.Jiang, J., Biggins, J. B. & Thorson, J. S. (2001) Angew. Chem. Intl. Ed. Engl. 40, 1502-1505. [PubMed] [Google Scholar]
- 15.Jiang, J., Biggins, J. B. & Thorson, J. S. (2000) J. Am. Chem. Soc. 122, 6803-6804. [Google Scholar]
- 16.Johnson, L. N. & Barford, D. (1990) J. Biol. Chem. 265, 2409-2412. [PubMed] [Google Scholar]
- 17.Park, S. H., Pastuszak, I., Drake, R. & Elbein, A. D. (1998) J. Biol. Chem. 273, 5685-5691. [DOI] [PubMed] [Google Scholar]
- 18.Lavine, J. E., Cantlay, E., Roberts, C. T., Jr., & Morse, D. E. (1982) Biochim. Biophys. Acta 717, 76-85. [DOI] [PubMed] [Google Scholar]
- 19.Dey, P. M. (1983) Eur. J. Biochem. 136, 155-159. [DOI] [PubMed] [Google Scholar]
- 20.Thomas, P., Bessell, E. M. & Westwood, J. H. (1974) Biochem. J. 139, 661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, J., Fu, X., Jia, Q., Shen, J., Biggins, J. B., Jiang, J., Zhao, J., Schmidt, J. J., Wang, P. G. & Thorson, J. S. (2003) Org. Lett. 5, 2223-2226. [DOI] [PubMed] [Google Scholar]
- 22.Bornscheuer, U. T. & Pohl, M. (2001) Curr. Opin. Chem. Biol. 5, 137-143. [DOI] [PubMed] [Google Scholar]
- 23.Petrounia, I. P. & Arnold, F. H. (2000) Curr. Opin. Biotechnol. 11, 325-330. [DOI] [PubMed] [Google Scholar]
- 24.Tao, H. & Cornish, V. W. (2002) Curr. Opin. Chem. Biol. 6, 858-864. [DOI] [PubMed] [Google Scholar]
- 25.Williams, G. J., Domann, S., Nelson, A. & Berry, A. (2003) Proc. Natl. Acad. Sci. USA 100, 3143-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada, M., Hsu, C. C., Franke, D., Mitchell, M., Heine, A., Wilson, I. & Wong, C.-H. (2003) Bioorg. Med. Chem. 11, 2091-2098. [DOI] [PubMed] [Google Scholar]
- 27.DeSantis, G., Liu, J., Clark, D. P., Heine, A., Wilson, I. A. & Wong, C.-H. (2003) Bioorg. Med. Chem. 11, 43-52. [DOI] [PubMed] [Google Scholar]
- 28.Leung, D. W., Chen, E. & Goeddel, D. V. (1989) Technique 1, 11-15. [Google Scholar]
- 29.Cadwell, R. G. & Joyce, G. F. (1992) PCR Methods Appl. 2, 28-33. [DOI] [PubMed] [Google Scholar]
- 30.Liebeton, K., Zonta, A., Schimossek, K., Nardini, M., Lang, D., Dijkstra, B. W., Reetz, M. T. & Jaeger, K.-E. (2000) Chem. Biol. 7, 709-718. [DOI] [PubMed] [Google Scholar]
- 31.Stemmer, W. P. C. (1994) Nature 370, 389-391. [DOI] [PubMed] [Google Scholar]
- 32.Zhao, H., Giver, L., Shao, Z., Affholter, J. A. & Arnold, F. H. (1998) Nat. Biotechnol. 16, 258-261. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi, M., Ohnishi, K. & Harayama, S. (1999) Gene 236, 159-167. [DOI] [PubMed] [Google Scholar]
- 34.Coco, W. M., Levinson, W. E., Crist, M. J., Hektor, H. J., Darzins, A., Pienkos, P. T., Squires, C. H. & Monticello, D. J. (2001) Nat. Biotechnol. 19, 354-359. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki, K. (2002) Nucleic Acids Res. 30, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zha, D., Eipper, A. & Reetz, M. T. (2003) Chembiochem 4, 34-39. [DOI] [PubMed] [Google Scholar]
- 37.Thoden, J. B. & Holden, H. M. (2003) J. Biol. Chem. 278, 33305-33311. [DOI] [PubMed] [Google Scholar]
- 38.Debouck, C., Riccio, A., Schumperli, D., McKenney, K., Jeffers, J., Hughes, C., Rosenberg, M., Heusterspreute, M., Brunel, F. & Davison, J. (1985) Nucleic Acids Res. 13, 1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc-Muesser, M., Defaye, J., Horton, D. & Tsai, J.-H. (1980) in Methods in Carbohydrate Chemistry, eds. Whistler, R. L. & BeMiller, J. N. (Academic, New York), Vol. 8, pp. 177-183. [Google Scholar]
- 40.Paulsen, H., Trautwein, W.-P., Espinosa, F. G. & Heyns, K. (1967) Chem. Ber. 100, 2822-2836. [Google Scholar]
- 41.Paulsen, H. & Herold, C. P. (1970) Chem. Ber. 103, 2450-2462. [Google Scholar]
- 42.Bradford, M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 43.Fromant, M., Blanquet, S. & Plateau, P. (1995) Anal. Biochem. 224, 347-353. [DOI] [PubMed] [Google Scholar]
- 44.Bork, P., Sander, C. & Valencia, A. (1993) Protein Sci. 2, 31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aleshin, A. E., Zeng, C., Bourenkov, G. P., Bartunik, H. D., Fromm, H. J. & Honzatko, R. B. (1998) Structure (London) 6, 39-50. [DOI] [PubMed] [Google Scholar]
- 46.Segura, M. J. R., Lodeiro, S., Meyer, M. M., Patel, A. J. & Matsuda, S. P. (2002) Org. Lett. 4, 4459-4462. [DOI] [PubMed] [Google Scholar]
- 47.Joubert, B. M., Hua, L. & Matsuda, S. P. (2000) Org. Lett. 2, 339-341. [DOI] [PubMed] [Google Scholar]
- 48.Herrera, J. B., Wilson, W. K. & Matsuda, S. P. (2000) J. Am. Chem. Soc. 122, 6765-6766. [Google Scholar]
- 49.Segura, M. J., Jackson, B. E. & Matsuda, S. P. (2003) Nat. Prod. Rep. 20, 304-317. [DOI] [PubMed] [Google Scholar]
- 50.Otten, L. G., Sio, C. F., Vrielink, J., Cool, R. H. & Quax, W. J. (2002) J. Biol. Chem. 277, 42121-42127. [DOI] [PubMed] [Google Scholar]