SUMMARY
MicroRNAs (miRNAs) are an integral component of the metazoan genome and affect posttranscriptional repression of target messenger RNAs. The extreme phylogenetic conservation of certain miRNAs suggests their ancient origin and crucial function in conserved developmental processes. We demonstrate that highly conserved miRNA-183 orthologs exist in both deuterostomes and protostomes and their expression is predominant in ciliated ectodermal cells and organs. The miRNA-183 family members are expressed in vertebrate sensory hair cells, in innervated regions of invertebrate deuterostomes, and in sensilla of Drosophila and C. elegans. Thus, miRNA-183 family member expression is conserved in possibly homologous but morphologically distinct sensory cells and organs. The results suggest that miR-183 family members contribute specifically to neurosensory development or function, and that extant metazoan sensory organs are derived from cells that share genetic programs of common evolutionary origin.
INTRODUCTION
Ciliated neurosensory cells mediate photoreception, electro-reception, chemosensation and mechanosensation in sensory organs of eukaryotic organisms. Although ciliated sensory organs can have elaborate and highly specialized morphologies, homologous developmental processes and molecular mechanisms contribute to their cellular architecture and function (Dahl et al. 1997; van Heyningen and Williamson 2002; Boekhoff-Falk 2005; Fritzsch et al. 2006). A number of transcription factors have been identified that are crucial for the development of neurosensory cells. These include at the organ level Pax2/5/8 for mechanosensory development and Pax6 for photosensory development (Czerny et al. 1997; Dahl et al. 1997; Pfeffer et al. 1998; Kavaler et al. 1999; Kozmik et al. 2003). At the cellular level, Atonal is required for mechanosensory, photosensory, and chemosensory development in Drosophila (Jarman et al. 1993, 1995; Gupta and Rodrigues 1997), and its vertebrate orthologs Atoh1 (Math1) and Atoh7, are respectively, required for development of mechanosensory hair cells in the ear (Bermingham et al. 1999) and for retina ganglia cells (Fritzsch and Piatigorsky 2004). This conservation of developmental mechanisms at the level of some genes shown to be crucial through knockout analysis is not sufficient to explain the apparent morphological similarities and variation across phyla. Nevertheless, it is possible that the major sensory organs come about through developmental reorganizations such as placodal formation to combine single sensory cells into complex organs such as the eye or ear (Gehring 2005; Fritzsch et al. 2007).
miRNAs are regulators of target genes involved in normal development and disease such as cancer (Engels and Hutvagner 2006; Garzon et al. 2006; Kloosterman and Plasterk 2006). The ability of miRNAs to effect cellular expression profiles (Chen et al. 2004; Lim et al. 2005), influence cell differentiation (Chen et al. 2004; Kanellopoulou et al. 2005; Wang et al. 2007), and impact morphogenesis (Giraldez et al. 2005; Harfe et al. 2005; Harris et al. 2006) suggest they can be as important as are highly conserved protein factors. Among the hundreds of phylogenetically conserved vertebrate miRNAs, several exhibit sequence conservation in evolutionarily distant organisms (Sempere et al. 2006; Prochnik et al. 2007). The expression of certain miRNAs may be highly conserved among certain cell types such as neurons or muscle (Lagos-Quintana et al. 2002; Aboobaker et al. 2005), but specific organ or cellular expression domains can vary considerably from organism to organism (Aboobaker et al. 2005; Ason et al. 2006; Darnell et al. 2006). Such observations suggest that some conserved miRNAs might possess distinct biological roles in different phyla as a result of target gene evolution and/or differential regulation of miRNA expression.
In vertebrates, the expression domain of conserved mi-RNA-183 (miR-183) family members appears to be restricted to ciliated neurosensory epithelial cells and certain cranial and spinal ganglia (Wienholds et al. 2005; Kloosterman et al. 2006). Vertebrate miR-183 family members (miR-183, miR-96, and miR-182) are co-expressed from a 1 to 4 kb segment of intergenic DNA between the Nrf1 and Ube2h genes. In zebrafish the miRNAs are detected in the eye, nose epithelium, and sensory hair cells of the ear and neuromasts (Wienholds et al. 2005), and injection of miR-183 and miR-200 family members in zebrafish embryos have been demonstrated to impact development and affect neuromast migration (Ason et al. 2006). Additionally, we have previously demonstrated expression of miR-183 family members in mouse eye and sensory hair cells of the ear (Weston et al. 2006). Considering the ancestry and importance of neurosensory cells in eukaryotic organisms, we reasoned that miR-183 family members might be more widely distributed amongst evolutionarily divergent eukaryotic organisms. Conducting a broader analysis of miR-183 family member expression, we demonstrate an exceptional conservation of miRNA expression in ciliated sensory organs amongst both vertebrate and invertebrate organisms. Results suggest that miR-183 family members represent one of the most highly conserved factors in neurosensory development, and that neurosensory organs thus share a common molecular and possibly cellular evolutionary origin.
MATERIALS AND METHODS
Genomic sequence analysis
miRBase (Griffiths-Jones et al. 2006) contains the sequences of conserved vertebrate miR-183 family members (miR-183, miR-96, and miR-182) and protostome homologs. miR-183 family members are conserved in at least sixteen vertebrate species including human (Homo sapiens; hsa), mouse (Mus musculus; mmu), chicken (Gallus gallus; gga), frog (Xenopus tropicalis; xtr), and zebrafish (Danio rerio; dre). Protostome homologs were identified by BLASTN sequence alignment to miR-183. Orthologs include miR-263 or miR-263b from five species of arthropods including fruit fly (Drosophila melanogaster; dme), and miR-228 from two species of nematodes including Caenorhabditis elegans (cel).
Invertebrate deuterostome miR-183 family members were identified in the sea squirt (Ciona intestinalis; cin) genome (Dehal et al. 2002) and purple sea urchin (Strongylocentrotus purpuratus; spu) genome (Sodergren et al. 2006) by a combination of BLASTN sequence alignment, BLASTP sequence alignment to mouse Nrf1 or Ube2h to establish synteny, and manual searching of local genomic sequence for alignment to miR-183 family member sequences. miR-183 family members or homologs identified as miR-183-like and miR-96-like reside adjacent to an Nrf1 homolog in the sea squirt genome or a Ube2h homolog in the sea urchin genome. Moreover, sequences are predicted using MFOLD (Zuker 2003) to form precursor hairpin structures consistent with miRNA biogenesis (Supplementary Fig. S1). In neither genome were miR-182 homologs identified.
Organisms for which no genomic data are available but evidence for miRNA expression is indicated by in situ hybridization or RT-PCR include spotted salamander (Ambystoma maculatum; ama), sea lamprey (Petromyzon marinus; pma), atlantic hagfish (Myxine glutinosa; mgl), acorn worm (Saccoglossus kowalevskii; sko), and green sea urchin (Strongylocentrotus droebachiensis; sdr).
In situ hybridization (ISH)
Whole-mount ISH detecting miRNA expression was performed as described previously (Weston et al. 2006) using locked nucleic acid (LNA) probes (Integrated DNA Technologies, Coralville, IA, USA) labeled with digoxigenin (DIG; Roche, Indianapolis, IN, USA). Briefly, tissues fixed in PBS containing 4% PFA were defatted with ethanol and digested with proteinase K before hybridization of 12 pmol LNA probe, washing and RNase A digestion, and probe detection using alkaline phosphatase (AP) conjugated sheep anti-DIG Fab fragment and BM Purple AP Substrate (Roche). Tissues were whole mounted in glycerol or sectioned (20 μm) in frozen OCT medium (Sakura Finetek, Torrance, CA, USA) and mounted in VECTASHIELD containing DAPI (Vector Labs, Burlingame, CA, USA). Samples were viewed by differential interference contrast and epifluorescence microscopy on a Nikon Eclipse 800 microscope.
Drosophila tissue samples were additionally treated with 1 U/ml chitinase (Sigma, St. Louis, MO, USA) for 60 min at room temperature before proteinase K digestion. Negative control samples lacked DIG-labeled LNA probe, but were otherwise prepared identically. LNA probe sequences for miR-183, spu-miR-183-like, and miR-263b are antisense to the miRNA sequences shown in Fig. 1B and contain LNA modifications at every third nucleotide position from the 5′ end. Such LNA probes have previously been shown to provide exceptional miRNA hybridization specificity that is sensitive to 1 or 2 nucleotide mismatches (Kloosterman et al. 2006). Salamander skin was kindly provided by W. Thoreson, acorn worms by C. Lowe, and sea lamprey and atlantic hagfish ears by B. Fritzsch. Green Sea urchins were purchased live from the Marine Biological Laboratory (Woods Hole, MA, USA), and wild-type fruit fly cultures were purchased from Nebraska Scientific (Omaha, NE, USA).
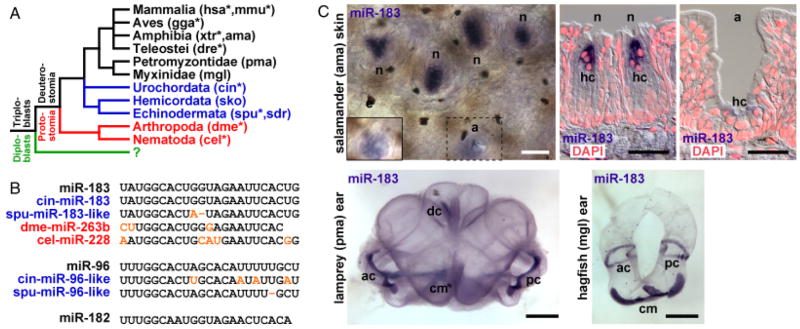
Fig. 1.
Phylogenetic conservation of miR-183 family members and expression in vertebrate sensory organs. (A) Representative bilaterian organisms that contain miR-183 family members include vertebrates (black) among invertebrate deuterostomes (blue) and protostomes (red). Asterisks indicate species for which certain miR-183 family members are supported by genomic sequence data (see Materials and Methods). The question mark indicates that no miR-183 family members have been identified among diploblasts (green). (B) Sequence alignment of conserved vertebrate miR-183 family members (black) with those identified among other deuterostomes (blue) and protostomes (red). Nucleotide substitutions or deletions (−) are colored orange. (C) ISH detecting miR-183 demonstrates expression in vertebrate hair cells (hc). Depicted are salamander skin neuromasts (n) and ampillary (electroreceptive) organs (a) in whole-mount and cross-section superimposed upon false-colored DAPI-labeled nuclei (red). Hair cell staining in whole-mount ampillary organ (dashed box) is shown in a different focal plane in the inset (solid box). Whole-mount lamprey and hagfish ears show hair cell staining in the common macula (mc), anterior crista (ac), posterior crista (pc), and dorsal crista (dc; lamprey only). Scale bars indicate 100 μm for salamander skin and 1 mm for lamprey and hagfish ears.
Immunocytochemistry (ICC)
Antibodies for acetylated tubulin (Sigma), BDNF (R&D Systems, Minneapolis, MN, USA), Prox1 (Covance Research Products, Denver, PA, USA), and MyoVIIa (Proteus Biosciences, Ramona, CA, USA) were used for ICC as described previously (Matei et al. 2005; Pauley et al. 2006). Briefly, tissues fixed in PBS containing 4% PFA were defatted with ethanol, blocked with goat serum, and incubated for 1 h with primary antibody. Secondary antibody was conjugated to either Alexa 543 or Alexa 648. Tissues were mounted in glycerol and viewed using a Zeiss LMS 510 confocal microscope.
RT-PCR
Total RNA was isolated from green sea urchin (sdr) digestive gland, gonad, and tube feet using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). Spu-miR-183-like primary transcript was detected as a 170-bp-PCR product using primers 5′-TCGTTCACCTGTGCTTGACTTC and 5′-TGATG GTGAATGCAGCACTTCAC, and compared with detection of U6 snRNA as a 101-bp-PCR product using primers 5′-GCTTC GGCAACACATCTATTAAAATTG and 5′-AAAAATATGG AACGCTTCACGATTTTG. For each PCR reaction, ~ 100 ng total RNA digested with DNase I (Invitrogen, Carlsbad, CA, USA) was reversed transcribed using 2 pmol of specific primer and SuperScript III Reverse Transcriptase (Invitrogen). RT products were used to seed PCR reactions containing 1 μM each primer, 10 mM Tris-HCl pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 200 μM each dNTP, and 1 U FastStart Taq DNA polymerase (Roche). Reactions were incubated at 95°C for 4 min followed by 15 cycles (for U6 snRNA) or 35 cycles (for spu-miR-183-like primary transcript) consisting of incubation at 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s. PCR products were analyzed by agarose gel electrophoresis and SYBR Green I (Sigma) fluorescence with 100 bp ladder (Invitrogen). The identity of the RT-PCR product from tube feet was confirmed by cycle sequencing.
Generation and analysis of transgenic C. elegans
The miR-228p::GFP reporter was generated by primer overlap-based PCR fusion of the miR-228 promoter to GFP as described previously (Hobert 2002). Approximately 2.2 kb of the 3 kb intergenic region immediately upstream of miR-228 (chromosome IV; nucleotides 559802-5562018) was used for the reporter construct consistent with similar promoter analyses in C. elegans (Dupuy et al. 2004). Primers used for miR-228 promoter amplification from C. elegans genomic DNA were 5′-TGCAATGCGGGAAGA GACGA and 5′-GCTCCACCGATCCCCTCCGGAGATAA GGAGGAAAATGTCTCGCC, for GFP amplification from pPD95.75 were 5′-TCCGGAGGGGATCGGTGGAGCATGAG TAAAGGAGAAGAACTTTTCACTGG and 5′-GCCCGTACG GCCGACTAGTAGG, and for nested amplification were 5′-TTCTGTGTTGAGTTTGCATTGTCC and 5′-TAGGAAAC AGTTATGTTTGGTATATTGGG. GR1560 (mgEx731 [miR-228p::GFP, ttx-3p::dsRed]) was generated by injecting N2 worms with 10 ng/μl pooled PCR products from six separate reactions with 30 ng/μl pBluescript and 10 ng/μl ttx-3p::dsRed as a co-injection marker. Transgenic young adult worms were viewed following paralysis in 50 mM sodium azide by phase contrast and fluorescence microscopy using an Olympus IX70 microscope.
RESULTS AND DISCUSSION
Analysis of available miRNA and genomic sequence data suggest that miR-183 family members or homologs are present throughout vertebrate and invertebrate deuterostomes and protostomes that are separated by ~ 600 million years of evolutionary divergence (Sempere et al. 2006) (Fig. 1A; see Materials and Methods). In the urochordate Ciona intestinalis (sea squirt), miRNA-183 and a homologous miR-96 sequence (cin-miR-183 and cin-miR-96-like) are present adjacent to an Nrf1 homolog (Fig. 1B). The conservation of miR-183 family sequences and genomic synteny extends also to the echinoderm Strongylocentrotus purpuratus (purple sea urchin), where homologous miR-183 and miR-96 sequences (spu-miR-183-like and spu-miR-96-like) are present adjacent to a Ube2h homolog. Moreover, miRNA sequences homologous to miR-183 have been identified in the protostomes Drosophila melanogaster (fruit fly) and Caenorhabditis elegans (nematode worm) as miR-263b and miR-228, respectively (Fig. 1B).
Using in situ hybridization (ISH) we examined the expression of miR-183 family members in representative bilaterian organisms. miR-183 expression is detected in vertebrate hair cells of spotted salamander neuromasts (Ambystoma maculatum), sea lamprey ear (Petromyzon marinus), and atlantic hagfish ear (Myxine glutinosa; Fig. 1C). Weaker expression is also detected in hair cells of salamander ampillary (electro-receptive) organs (Fig. 1C). Combined with prior detection of miR-183 family members in zebrafish and mouse (Wienholds et al. 2005; Weston et al. 2006), the data demonstrate that miR-183 family members are expressed in most ciliated neurosensory epithelial cell types in vertebrate organs of divergent function that mediate photoreception, electroreception, chemosensation and mechanosensation.
Although invertebrate deuterostomes lack defined ectodermal sensory organs, we examined whether miR-183 family member expression might be detected and associated with putative neurosensory regions. Analysis of the hemichordate Saccoglossus kowalevskii (acorn worm) by ISH for miR-183 demonstrates detection in certain epithelial cells throughout the organism (Fig. 2A). Considering the evolutionary relationship of hemichordates with echinoderms, it is interesting to note that acorn worm fails to demonstrate the same pattern of detection by ISH for the echinoderm miR-183 homolog, spu-miR-183-like (Supplementary Fig. S2A). Moreover, detection of vertebrate miR-183 expression in mouse cochlear hair cells by probe for miR-183 but not spu-miR-183-like demonstrates the discriminatory properties of the probes (Supplementary Fig. S2B). These data suggest that despite the apparent evolutionary relationship of hemichordates and echinoderms, acorn worm appears to have conserved vertebrate miR-183 sequence or a more highly related sequence than have echinoderms.
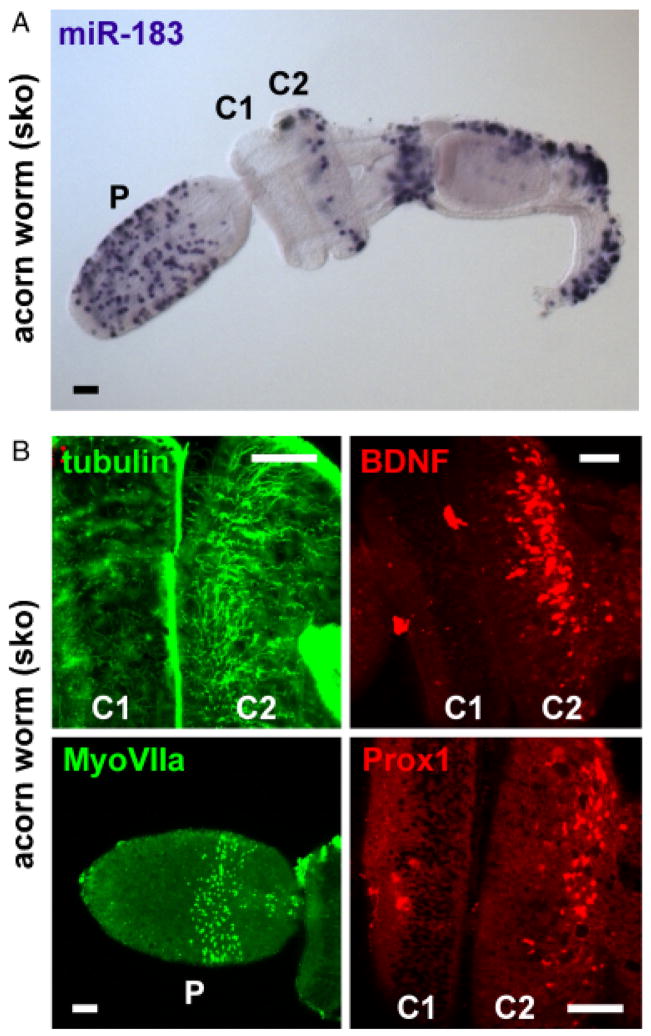
Fig. 2.
miR-183 family member expression in acorn worm is associated with putative sensory regions. (A) ISH detecting miR-183 expression in epithelial cells of the acorn worm proboscis (P), second collar region (C2), and dorsolateral regions posterior to the gill slits and ciliated band. (B) Association of miR-183 expression with putative sensory regions of acorn worm proboscis and collar. Clockwise from upper left: tubulin ICC demonstrates specific innervation of C2, BDNF ICC shows neurotrophin expression in C2, and Prox1 and MyoVIIa ICC respectively demonstrate strongest expression in C2 and proboscis. Scale bars indicate 50 μm.
Further analysis of the acorn worm demonstrates that regions of miR-183 detection, especially in epithelial cells of the proboscis and second collar region, overlap with a number of markers indicative of neurosensory domains in other organisms. miR-183 detection in the proboscis partially coincides with myosin VIIa (MyoVIIa) detection by immunocytochemistry (ICC; Fig. 2B), where MyoVII is well conserved among animals (Goodson and Dawson 2006), MyoVIIa is specifically expressed in vertebrate ciliated cells (Hasson et al. 1995; Wolfrum et al. 1998), and mutation causes hearing loss in fruit fly and mammals (Todi et al. 2005). miR-183 detection in the second collar region is specifically associated with innervation of the collar region as indicated by tubulin ICC commonly used to label cytoskeletal processes such as nerve fibers and cilia (Fig. 2B; Matei et al. 2005; Pauley et al. 2006). Innervation logically follows the detected expression pattern of brain-derived neurotrophic factor (BDNF; Fig. 2B), which is highly conserved among deuterostomes including sea urchin (Burke et al. 2006; Hallbook et al. 2006) and is expressed by mature hair cells in vertebrate ear (Farinas et al. 2001; Fritzsch et al. 2005). Additionally, miR-183 expression and innervation of the second collar region correspond to the detected expression domain of Prox1 (Fig. 2B), which is expressed in sensory epithelial development and in supporting cells of the vertebrate ear (Bermingham-McDonogh et al. 2006). These data demonstrate that the expression domain of miR-183 is largely associated with those of known neurosensory markers, and extends further the predilection of miR-183 family member expression in putative invertebrate neurosensory regions.
In the echinoderm S. droebachiensis (green sea urchin), ISH for the miR-183 homolog, spu-miR-183-like, demonstrates detection in the sucker cup rim of tube feet (Fig. 3A). This region of the tube foot also exhibits innervation and the presence of cilia by ICC for tubulin (Fig. 3B), and is consistent with a general sensory function for echinoderm tube feet (Burke et al. 2006). Although expression is relatively diffuse and weak compared to miR-183 expression in other deuterostomes, RT-PCR shows primary spu-miR-183 transcript (pri-spu-miR-183) is specifically expressed in tube feet compared to gonad or digestive gland (Fig. 3C). Sequencing of the RT-PCR product obtained from green sea urchin tube feet confirmed the identity of the pri-miRNA transcript, where a single G to U transversion resides in the terminal loop of the precursor hairpin compared to that for purple sea urchin (Supplementary Fig. S1). Interestingly, Pax6 gene expression typically associated with sensory organ develop in vertebrates (Dahl et al. 1997; van Heyningen and Williamson 2002) has previously been demonstrated in acorn worm (Lowe et al. 2003) and sea urchin tube feet (Czerny et al. 1997; Burke et al. 2006). These data suggest that invertebrate deuterostomes have employed homologous genetic programs for sensory organ development within different morphological contexts to develop putative neurosensory cells. Moreover, some starfish develop terminal tube feet into photoreceptive ocelli, indicating the potential of these structures to form neurosensory organs (Bullock and Horridge 1965).
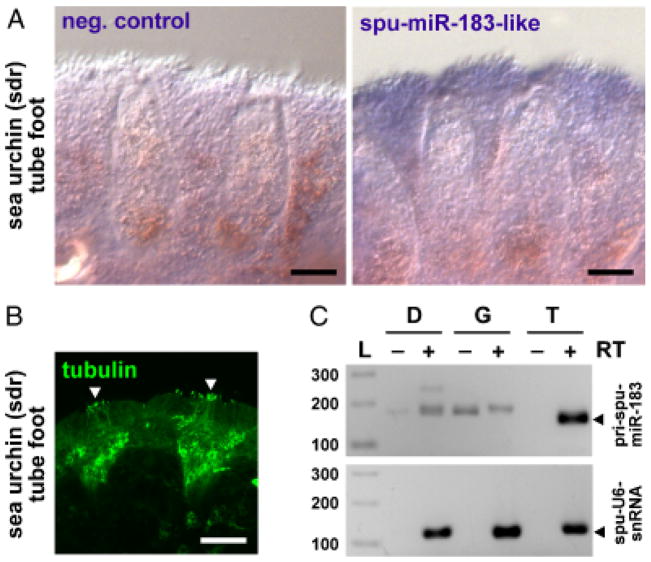
Fig. 3.
miR-183 family member expression in sea urchin tube feet. (A) ISH detecting moderate expression of spu-miR-183-like in the sucker cup rim of green sea urchin tube feet compared to negative control lacking probe. (B) Tubulin ICC demonstrates innervation of spu-miR-183-like positive regions containing cilia (arrowheads). Scale bars indicate 100 μm. (C) spu-miR-183-like-primary transcript expression is specific to tube feet. Depicted is primary transcript detection relative to U6 snRNA by RT-PCR of total RNA isolated from sea urchin digestive gland (D), gonad (G), and tube feet (T) in reactions prepared with (+) or without (−) reverse transcriptase (RT) along side 100 bp ladder (L).
The presence of miR-183 homologs in protostomes suggest a considerably wider phylogenetic distribution and more ancient evolutionary origin for miR-183 family members in bilaterian organisms, implying a similar role in neurosensory development or function. In D. melanogaster (fruit fly), ciliated sensory organs are distributed throughout the organism but are abundant in mechanosensory organs such as the haltere and Johnston’s (auditory) organ (antennal segment 2; A2), and in the chemosensory antennal segment 3 (A3) (Stocker 1994; Boekhoff-Falk 2005). ISH detecting miR-263b expression in adult fruit fly demonstrates expression in campaniform sensilla of the haltere, scolopidia of the Johnston’s organ, and in A3 (Fig. 4A), although specific cell types are not easily resolved. These data are consistent with the prior detection of miR-263b in sensory organ precursors of fruit fly embryo (Aboobaker et al. 2005).
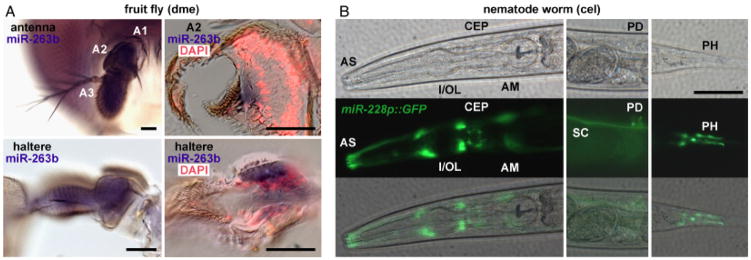
Fig. 4.
miRNA-183 family member expression in sensory organs of protostomes. (A) ISH detecting miR-263b expression in fruit fly mechanosensory and chemosensory antennal segments 2 and 3, respectively (A2 and A3; top left), and haltere (bottom left). Cross-sectional views of the Johnston’s organ (A2; top right) and haltere (bottom right) indicate miR-263b expression in scolopidia and campaniform sensilla, respectively, superimposed upon false-colored DAPI-labeled nuclei (red). (B) miR-228 expression in C. elegans indicated by miR-228p::GFP transgene expression. miR-228 expression is indicated in cells provisionally identified as sheath and/or socket cells of phasmid (PH), posterior deirid (PD), and anterior sensilla (AS) including inner/outer labial (I/OL), cephalic (CEP), and amphid (AM). Phase contrast (top) and fluorescence (center) images are superimposed (bottom) for each lateral view of the anterior (left), middle (center), and posterior (right) region of the worm. Seam cell (SC) expression is also indicated. Scale bars indicate 50μm.
In C. elegans (nematode worm), mechanosensory and chemosensory functions are associated with anterior, deirid, and phasmid sensilla (Altun and Hall 2005). Using an extra-chromosomal miR-228 promoter-driven green fluorescent protein reporter construct (miR-228p::GFP), miR-228 expression is indicated in inner/outer labial, cephalic, and amphid sensilla, the posterior deirid, and in phasmid sensilla of the young adult worm (Fig. 4B). Additionally, expression is observed in seam cells. Based on morphology of GFP-positive cells in sensilla, expression is provisionally identified in sheath and/or socket cells rather than ciliated neurons. This expression in protostome glial or supporting cell types stands in contrast to miR-183 detection in vertebrate ciliated epithelial cells (i.e., sensory hair cells). Thus, miR-183 family members appear to be differentially utilized within similar sensory organs among phylogenetically disparate organisms as well as within functionally distinct sensory organs of individual organisms (e.g., eye, nose, and ear of zebrafish; Wienholds et al. 2005).
The phylogenetic distribution of miR-183 family members is thus demonstrated to include orthologs in deuterostomes and protostomes that exhibit an exceptional conservation of expression in ciliated sensory organs. While these results suggest that miR-183 family member functions might be deduced through scrutiny of homologous predicted miRNA target genes, the precise functions of miR-183 family members might vary among morphologically distinct sensory organs and remain to be demonstrated. Interestingly, expression mapping of co-regulated genes in C. elegans topographically localizes miR-228 gene expression (sjj_T12E12.5) among other genes with sensory functions, particularly chemoreceptors (Kim et al. 2001). In zebrafish, injection of miR-183 family members perturbs neuromast migration (Ason et al. 2006), suggesting a distinct role for the miRNAs in development of vertebrate mechanosensory organs. Moreover, the human miRNA-183 family locus lies within a chromosomal segment identified as a nonsyndromic autosomal dominant deafness locus (DFNA50) within which a causative gene has not yet been identified (Modamio-Hoybjor et al. 2004).
The conservation of miR-183 family members in organisms diverged by ~ 600 million years of evolution suggest their importance in the development or function of ciliated neurosensory organs and supports the notion of a common ancestry for mechanosensation and audition in particular (Boekhoff-Falk 2005; Fritzsch et al. 2006). Further examination of such highly conserved miRNAs that likely influence neurosensory cell development in homologous sensory organs will be key to understanding genetic programs that might be manipulated to affect cellular identity in prospective therapeutic strategies for regenerating sensory hair cells and restoring hearing.
Supplementary Material
miR-183 family members and homologs identified in the sea squirt genome (cin) and purple sea urchin genome (spu). Shown are predicted miRNA precursor hairpin structures with homologous miRNA sequences reported in Fig. 1B highlighted in red. Genomic scaffold segments corresponding to depicted sequences are indicated. The highlighted position in the terminal loop of the precursor hairpin for spu-miR-183-like (blue) denotes the site of G to U transversion in the sequence determined for green sea urchin (sdr).
Discrimination by probes for miR-183 and spu-miR-183-like. (A) ISH detecting expression of miR-183 in acorn worm compared to probe for spu-miR-183-like and negative control lacking probe. Scale bars indicate 100 μm. (B) ISH detecting expression of miR-183 in mouse cochlea compared to probe for spu-miR-183-like. miR-183 expression is specifically detected in the single row of inner hair cells (IHC) and three rows of outer hair cells (OHC). Scale bars indicate 20 μm.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1525-142X.2007.00217.x (This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Acknowledgments
This work was supported by U.S. Public Health Service grants P20RR018788 (G. A. S.) and R01DC005590 (B. F.), and Nebraska State Fund LB692 (B. F.). The research was conducted in a facility constructed with support from the National Center for Research Resources, National Institutes of Health, Research Facilities Improvement Program grant C06RR017417, and utilized the Nebraska Center for Cell Biology confocal microscope facility supported by the National Science Foundation EPSCoR grant EPS-0346476. We thank Wallace Thoreson and Christopher Lowe for providing tissues for ISH, and Liqin (Julie) Zhu for assistance with ICC.
References
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun ZF, Hall DH. WormAtlas. 2005 http://www.wormatlas.org/handbook/contents.htm.
- Ason B, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, Hayashi T. Expression of Prox1 during mouse cochlear development. J Comp Neurol. 2006;496:172–186. doi: 10.1002/cne.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhoff-Falk G. Hearing in Drosophila: development of Johnston’s organ and emerging parallels to vertebrate ear development. Dev Dyn. 2005;232:550–558. doi: 10.1002/dvdy.20207. [DOI] [PubMed] [Google Scholar]
- Bullock TH, Horridge GA. Structure and Function in the Nervous Systems of Invertebrates. Freeman & Company; San Francisco: 1965. [Google Scholar]
- Burke RD, et al. A genomic view of the sea urchin nervous system. Dev Biol. 2006;300:434–460. doi: 10.1016/j.ydbio.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Czerny T, Bouchard M, Kozmik Z, Busslinger M. The characterization of novel Pax genes of the sea urchin and Drosophila reveal an ancient evolutionary origin of the Pax2/5/8 subfamily. Mech Dev. 1997;67:179–192. doi: 10.1016/s0925-4773(97)00119-6. [DOI] [PubMed] [Google Scholar]
- Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Dupuy D, et al. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels BM, Hutvagner G. Principles and effects of micro-RNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- Farinas I, et al. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. J Neurosci. 2001;21:6170–6180. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel K, Pauley S, Soukup GA. Molecular Evolution of the vertebrate mechanosensory cell and ear. Int J Dev Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, et al. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Beisel KW. Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res. 2006;1091:151–171. doi: 10.1016/j.brainres.2006.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Piatigorsky J. Ancestry of photic and mechanic sensation? Science. 2004;308:1113–1114. [PubMed] [Google Scholar]
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Dawson SC. Multiplying myosins. Proc Natl Acad Sci USA. 2006;103:3498–3499. doi: 10.1073/pnas.0600045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta BP, Rodrigues V. Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila. Genes Cells. 1997;2:225–233. doi: 10.1046/j.1365-2443.1997.d01-312.x. [DOI] [PubMed] [Google Scholar]
- Hallbook F, Wilson K, Thorndyke M, Olinski RP. Formation and evolution of the chordate neurotrophin and Trk receptor genes. Brain Behav Evol. 2006;68:133–144. doi: 10.1159/000094083. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci USA. 1995;92:9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. Atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaler J, Fu W, Duan H, Noll M, Posakony JW. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development. 1999;126:2261–2272. doi: 10.1242/dev.126.10.2261. [DOI] [PubMed] [Google Scholar]
- Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, et al. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;55:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- Matei V, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modamio-Hoybjor S, et al. A novel locus for autosomal dominant nonsyndromic hearing loss, DFNA50, maps to chromosome 7q32 between the DFNB17 and DFNB13 deafness loci. J Med Genet. 2004;41:e14. doi: 10.1136/jmg.2003.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool B Mol Dev Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- Sodergren E, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Todi SV, Franke JD, Kiehart DP, Eberl DF. Myosin VIIA defects, which underlie the Usher 1B syndrome in humans, lead to deafness in Drosophila. Curr Biol. 2005;15:862–868. doi: 10.1016/j.cub.2005.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–301. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wolfrum U, Liu X, Schmitt A, Udovichenko IP, Williams DS. Myosin VIIa as a common component of cilia and microvilli. Cell Motil Cytoskeleton. 1998;40:261–271. doi: 10.1002/(SICI)1097-0169(1998)40:3<261::AID-CM5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miR-183 family members and homologs identified in the sea squirt genome (cin) and purple sea urchin genome (spu). Shown are predicted miRNA precursor hairpin structures with homologous miRNA sequences reported in Fig. 1B highlighted in red. Genomic scaffold segments corresponding to depicted sequences are indicated. The highlighted position in the terminal loop of the precursor hairpin for spu-miR-183-like (blue) denotes the site of G to U transversion in the sequence determined for green sea urchin (sdr).
Discrimination by probes for miR-183 and spu-miR-183-like. (A) ISH detecting expression of miR-183 in acorn worm compared to probe for spu-miR-183-like and negative control lacking probe. Scale bars indicate 100 μm. (B) ISH detecting expression of miR-183 in mouse cochlea compared to probe for spu-miR-183-like. miR-183 expression is specifically detected in the single row of inner hair cells (IHC) and three rows of outer hair cells (OHC). Scale bars indicate 20 μm.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1525-142X.2007.00217.x (This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.