Abstract
Alterations in RNA levels are frequently reported in brain of subjects diagnosed with autism, schizophrenia, depression and other psychiatric diseases, but it remains unclear whether the underlying molecular pathology involves changes in gene expression, as opposed to alterations in mRNA processing. Pre-clinical studies have revealed that stress, drugs, and a variety of other environmental factors lead to changes in RNA levels in brain via epigenetic mechanisms, including modification of histone proteins. A number of site-specific modifications of the nucleosome core histones—including the trimethylated forms of histone H3 lysines K4, K9, and K27—are of particular interest for postmortem research, because these marks differentiate between active and inactive chromatin, and appear to remain relatively stable during tissue autolysis. Therefore, histone methylation profiling at promoter regions could provide important clues about mechanisms of gene expression in human brain during development and in disease. Intriguingly, mutations within the genes encoding the H3K9-specific methyltransferase, EHMT1, and the H3K4-specific histone demethylase, JARID1C/SMCX, have been linked to mental retardation and autism, respectively. In addition, the H3K4-specific methyltransferase, MLL1, is essential for hippocampal synaptic plasticity and may be involved in cortical dysfunction of some cases of schizophrenia. Together, these findings emphasize the potential significance of histone lysine methylation for orderly brain development and also as a molecular toolbox to study chromatin function in postmortem tissue.
Keywords: gene expression, autism, psychosis, affective disorder, epigenetic, methylation
Introduction
Quantification of mRNAs extracted from human postmortem brain is a feasible and widely used approach to study the molecular genetic changes associated with normal development, aging, and chronic neuropsychiatric disease. Earlier studies examined select gene transcripts using in situ hybridization histochemistry; later the relative ease of quantitative, real-time polymerase chain reaction (qRT-PCR) and the availability of microarrays and other “high-throughput” tools lead to an exponential growth of the field (1, 2). Analysis of mRNA levels turned out to be particularly useful for the study of neuropsychiatric disorders such as schizophrenia and depression that otherwise lack a defining neuropathology. While no one gene transcript has been definitively linked to these diseases, it is now thought that psychosis involves alterations in gene expression, including dysregulation of transcripts involved in cellular metabolism (3-5), GABAergic (6, 7) and glutamatergic neurotransmission (8, 9), and myelination and other oligodendrocyte functions (10-12). However, very little attention has been given to the molecular mechanisms that contribute these observed mRNA alterations. Specifically, it is not known whether the alterations in a specific transcript are brought about by a change in production (i.e., gene transcription), or alternatively, a mechanism involving mRNA turnover and catabolism. Therefore, in order to determine whether transcription itself is altered, additional molecular assays, in combination with more traditional approaches (quantification of mRNA and protein), would be extremely useful for the postmortem field.
In the past decade, the study of epigenetic regulation of gene expression in the brain, particularly within the context of neuropsychiatric disorders, has blossomed. Animal models have allowed scientists to study the impact of stress, drugs and other environmental factors associated with such diseases on epigenetic modifications and how they are associated with mRNA levels (13-16). The application of epigenetic approaches within the field of experimental neurobiology has important implications for the postmortem field, because study of chromatin modifications make it possible to study one aspect of transcriptional regulation in human brain tissue.
The purpose of this review will be to provide a selective overview of histone and DNA modifications that appear amenable for postmortem studies, followed by a brief discussion of technical aspects and limitations of chromatin immunoprecipitation assays (ChIP) on brain tissue. Next, we will discuss studies which have utilized ChIP to study epigenetic regulation of gene expression in human brain. Finally, we highlight a subset of histone methylation enzymes that have been linked to neurodevelopmental disease. We predict that histone methylation profiling at defined genomic loci will become a valuable tool to examine epigenetic control of gene expression in normal and diseased human brain.
Epigenetic modifications associated with the transcriptional process
Histone modifications
The nucleosome as the elementary unit of chromatin is comprised of 146 bp of DNA wrapped around an octamer of 4 different histone proteins (H2A, H2B, H3 and H4) (17). Both the nucleosome core histones and some of their variants (H3.1, H3.3 etc) are subject to covalent modifications of specific residues located primarily at the amino-terminal tail; these include lysine acetylation, methylation, SUMOylation, and ubiquitinylation; arginine methylation; serine phosphorylation; and proline isomerization (18). Some of these modifications are commonly associated with activation (e.g. acetylation) or repression (e.g., SUMOylation) but other types (e.g., methylation) have very different effects on gene transcription depending upon the specific position of the histone tail residue and also on the presence or absence of various chromatin remodeling complexes in the particular cell type under investigation (18). There is additional complexity, because the lysine side chains can carry up to three methyl groups. These mono-, di- and tri-methylated forms of specific lysine residues show cell-specific regulation during development (19) and are differentially distributed across chromatin fibers (20). For example, tri-methylated lysine residue 4 of histone H3 localizes to gene promoter regions, where it is associated with transcriptional activation, while mono-methylation of the same histone residue appears to be enriched at enhancer sequences further removed from transcription start sites (21). In the case of the human genome, an even more striking example is provided by the methylation mark of lysines 9 and 27 of histone H3, and lysine 20 of H4, for which the mono-methylated forms are linked to gene activation, while di- or tri-methylation of the same residues are associated with repression (22). Table 1 provides a brief overview of the specific histone lysine residues that are subject to methylation modifications and their positive or negative association with transcription. While there is emerging evidence specifying which of these modifications are associated with transcriptional activation and which are associated with repression, the precise role(s) of these epigenetic marks for the transcriptional process itself is an area of active investigation. For more complete discussion of post-translational histone modifications, see (18, 23-25).
Table 1. Overview of histone lysine forms, including association with transcription.
Information on preferred localization is based on genome-wide studies, see ref. (18, 20, 22, 23, 25, 69, 70) for additional details. The role of H3-lysine 79 methylation is not yet completely understood.
| Histone Residue | Transcription |
|---|---|
| H3-lysine 4 (di- and tri-methyl) | Activation (Peak levels around transcription start sites) |
| H3-lysine 4 (mono-methyl) | Activation (Peak levels mostly at enhancer sequences) |
| H3-lysine 9 (di- and tri-methyl) | Repression (Peak levels in heterochromatin, DNA repeats, but also found at promoters and other sequences) |
| H3-lysine 9 (mono-methyl) | Activation |
| H3-lysine 27 (di- and tri-methyl) | Repression (Peak levels around transcription start sites) |
| H3-lysine 27 (mono-methyl) | Activation |
| H3-lysine 36 (tri-methyl) | Activation (Peak levels within gene coding and non-coding sequences) |
| H3-lysine 79 (tri-methyl) | Repression (?) |
| H4-lysine 20 (di- and tri-methyl) | Repression (Peak levels in heterochromatin, DNA repeats, but also found at promoters and other sequences) |
| H4-lysine 20 (mono-methyl) | Activation |
There is, perhaps not surprisingly, evidence for functional interaction between the different types of histone modifications. For example, binding of the basal transcription factor, TFIID, to tri-methylated H3-lysine 4, is enhanced by the acetylation of two lysine residues nearby, H3-lysines 9 and 14 (26). Further evidence for this potential cross-talk between different histone modifications is provided by recent reports that exposure of cultured neurons to sodium butyrate, a broadly acting inhibitor of histone deacetylase enzymes, not only results in generalized histone hyperacetylation but also up-regulates H3 lysine 4 methylation at a subset of GABAergic gene promoters (27, 28). Furthermore, chronic antidepressant treatment reportedly decreases hippocampal levels of di-methylated H3-lysine 27, a repressive chromatin mark, at a subset of brain-derived neurotrophic factor (Bdnf) gene promoters, in conjunction with increased histone H3 acetylation (29).
Which histone marker to choose for postmortem brain studies?
From the viewpoint of gene expression studies in postmortem brain, histone marks such as trimethyl-H3-lysine 4 (H3K4me3) are of particular interest. The H3K4me3 signal typically shows peak levels at the 5′_region of actively transcribed genes (its broader distribution at selected Hox gene clusters being a notable exception) (20), and shows strong positive correlations with transcription rates, activated RNA polymerase II occupancy, and histone acetylation levels (30). The H3K4me3 mark functions as the exclusive docking signal for plant homeo domain (PHD) finger proteins and other proteins involved in recruitment of chromatin remodeling complexes, thus providing its own layer of transcriptional regulation (18, 30). However, these insights are based on studies in cell lines and peripheral tissues, and whether the H3K4me3 tag exerts similar actions in brain chromatin warrants further investigation. Indeed, recent postmortem studies suggest that, in human brain, H3K4 methylation—including the tri-methylated form—is associated with transcriptional activation, as evidenced by its localization at sites of active promoters. For example, levels of both H3K4me3 and of the related mark, di-methyl H3K4, at glutamate receptor gene promoters showed a positive correlation with the corresponding RNAs in human cerebellar and prefrontal cortices (31). Furthermore, the H3K4me3 signal is extremely weak, or even non-detectable, at regulatory sequences of the (β)globin genes and other loci that remain silent in CNS cells (28, 31).
A useful complement to the open chromatin mark, H3K4me3, is provided by tri- and di-methylated forms of histone H3 lysines 9 and 27, which define inactive or repressed gene promoters (22). H3K27me3 mediates repression primarily via interaction with Polycomb group proteins (32, 33). On the other hand, H3K9me3 and H3K9me2 are thought to be important for heterochromatin formation, serving as a docking site for HP1 and other heterochromatin-associated proteins (34, 35). In addition, it has been suggested that H3K9 methylation exerts repression at gene promoters via recruitment of DNA methyltransferase enzymes, which catalyze methylation of CpG dinucleotides which then recruit methyl-CpG-binding proteins, and subsequently, repressive chromatin remodeling complexes (36). These principles may also apply to the human brain; for example, the GAA repeat expansion in intron 1 of the FXN gene (responsible for Friedreich ataxia) results in H3-lysine 9 and promoter DNA hypermethylation, in conjunction with a robust decline in FXN mRNA levels (37).
Taken aside sample-to-sample variability, the H3K4me3 and H3K27me3 modifications at gene promoters appear to be largely preserved for a certain period after death, as evidenced by a lack of correlation with tissue pH and autolysis times that were within a range (6-30 hours) representative for most of the specimens stored in brain banks (38). If these preliminary results on a handful of promoters could be extrapolated genome-wide, then at least some of the tri-methylated lysine marks of the nuclesome core histones appear amenable to analysis in postmortem tissue. However, not all histone modifications appear to be as stable during the postmortem interval. For example, one study found that immunoreactivity for acetylation at lysines 9 and 14 of histone H3, and of lysine 12 of H4 was correlated with brain tissue pH in bulk chromatin preparations from postmortem cerebral cortex (4). In the same study, immunoreactivity for methylated H3 lysine 4 and arginine 17 were not correlated with postmortem confounds (4). Thus, while the finer details of the spatial arrangement of trimethylated lysines appear to be maintained in postmortem brain (28), as demonstrated by the preferred occupancy of H3K4me3 in nucleosomes downstream of transcription start sites (22, 39), other histone modifications may not be as well preserved. Why some histone modifications are more resilient under conditions of autolysis remains to be determined.
DNA methylation
In vertebrates, methylation of CpG dinucleotides within proximal gene promoters is frequently linked to transcriptional repression (40). For a recent review on DNA methylation and additional references, see (41). Recently, several studies have reported alterations in DNA methylation of some genes in cerebral cortex of subjects diagnosed with psychosis (42-46), although the reported changes were not entirely consistent across studies, in particular for COMT AND RELN (46-48). However, despite the potential importance of CpG methylation for the molecular pathology of schizophrenia and depression, it is important to point out that CpG dinucleotides of many gene promoters normally remain unmethylated (18), and thus epigenetic mechanisms operating at these loci are likely to depend on the local chromatin environment, including histone modifications. The complex interplay between DNA methylation and histone methylation is further illustrated by the finding that histone H3 residues that remain unmethylated at the lysine 4 position become a docking site for the DNA methyltransferases, DNMT3L and DNMT3A2, resulting in de novo DNA methylation at these sites (49). Notably, a recent study describes hypermethylation at the sites of ribosomal rRNA promoter sequences in the hippocampus of suicide victims (50). Therefore, in addition to promoters of single copy genes, examination of DNA methylation within repeat sequences may shed additional light on the molecular pathology of neuropsychiatric disease.
Methodological considerations and limitations
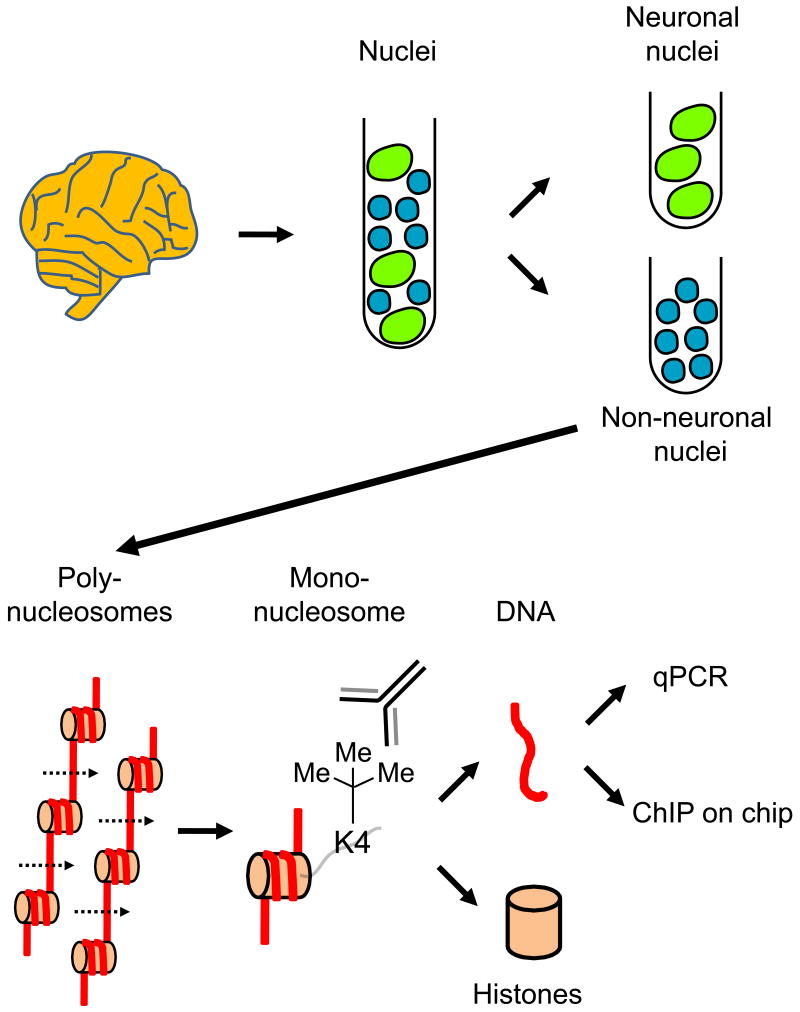
Histones are bound with very high affinity to genomic DNA (51) and so it is not surprising that bulk levels of nucleosome-bound DNA remain relatively unchanged, even in brain tissues subject to 30 hours of autolysis (38). However, in a living cell, the chromatin architecture of the interphase nucleus is complex. In addition to an array of subnuclear bodies (e.g., nucleosomes, PML bodies, Cajal bodies), highly organized 3-dimensional chromatin domains and transcription factories exist that could cluster together a number of individual loci on a given chromosome, even loci residing on different chromosomes (52). These higher-order chromatin structures may become disorganized or subject to random and spurious rearrangements after death, though specific information on this subject is lacking. These considerations should be taken into account when choosing between the two principle alternatives of chromatin preparation techniques: Cross-linking (typically via UV light or formaldehydes) followed by sonication versus unfixed (native) chromatin that is either sheared via sonication or digested with micrococcal nuclease, a DNAse that spares nucleosomal-bound DNA (53, 54). If crosslinking would indeed bind together chromosomal domains that came in close proximity only because of randomly drifting in the dying nucleus, then native chromatin preparations would be preferable. On the other hand, immunoprecipitation of native chromatin may only be feasible for the core histones and a very limited number of additional molecules bound with high affinity to the genomic DNA (53). In contrast, crosslinking is deemed to be important to assess the binding of transcription factors, RNA polymerases, enzymes and other proteins that are likely to be more loosely attached to genomic DNA. Finally, given the fact that many postmortem investigators work with a limited amount of tissue, commonly recommended steps to optimize chromatin preparations from a given sample (such as “titrating” the extent of shearing or enzyme-based digestion in multiple aliquots) appear less feasible under these circumstances. In our hands, a standardized protocol aimed to obtain mono-nucleosomal preparations by micrococcal nuclease-based digestion of homogenates from as little as 75 mg postmortem brain is a suitable option, at least for the purposes of immunoprecipitation with site-specific anti-methyl-histone antibodies (31, 38, 55) (Fig. 1). However, alternative methods involving shearing of chromatin by sonication with or without prior fixation, have been used to study histone H3-lysine 9 methylation at promoters in human striatum (37) and FOXP2, a transcription factor implicated in speech and language functions, in cerebral cortex (56).
Figure 1. Chromatin immunoprecipitation (ChIP) in postmortem brain.
Schematic overview of ChIP in postmortem brain, starting with tissue homogenization, collection and purification of nuclei, followed by the optional step of immunotagging and fluorescence-activated sorting of nuclei (into neuronal and non-neuronal, for example), followed by nuclei lysis and enzymatic digestion of chromatin fibers and polynucleosomes) into mono-nucleosomes, followed by immunoprecipitation with site- and modification-specific anti-histone antibodies (anti-tri-methyl-H3-lysine 4, for example), followed by separation of DNA and histones, and quantification of specific DNA sequences in the immunoprecipitate, relative to input. See (38, 55, 58) for details.
Importantly, most chromatin assays lack single cell resolution and instead require an “input” homogenate comprised of at least thousands of nuclei. This may be not a significant issue when working with simple eukaryotes such as yeast, or with a cell line, but it poses a considerable limitation when working with brain tissue comprised of an extremely heterogeneous set of neurons, glia, and other cells. Under some circumstances, methylation-sensitive PCR in situ hybridization may be a possible approach to examine cell-specific epigenetic modifications of the genomic DNA (57), but this approach is not applicable to histones. Furthermore, the large number of nuclei required for chromatin immunoprecipitation would translate into a considerable effort if one aspires to collect nuclei by laser capture and microdissection. However, nuclei, in contrast to cell somata, are fairly resilient to freeze-thaw and other types of tissue stress. Therefore, it is feasible to extract, purify and immunotag nuclei from frozen, unfixed human brain tissues for subsequent fluorescence-activated sorting of the different nuclei populations (Fig. 1); this approach was used to confirm that histone methylation at the BDNF (brain-derived neurotrophic factor) locus, and at other gene promoters, is differentially regulated in neurons, compared to the non-neuronal cells residing in human cerebral cortex (58).
Histone methylation studies in human postmortem brain
Gene expression in development and disease is associated with histone methylation changes at gene promoters human brain
Both the metabotropic (GRM1-7) and ionotropic (e.g., NMDA, AMPA, and kainate) glutamate receptor genes undergo dynamic, region- and cell-specific changes in expression during the course of brain development. In human brain, these developmental and region-specific changes of mRNA levels are accompanied by complementary alterations in H3K4 di- and tri-methylation at the sites of the corresponding promoters (31). Likewise, the progressive up-regulation of GABAergic mRNAs during development of human prefrontal cortex is reflected by parallel increases in H3K4 methylation at sites of the promoters (28) (Fig.2). Taken together, these studies confirm that H3K4 di- and tri- methylation defines actively expressed genes in the human brain, as has been previously shown for simple eukaryotes (yeast) and mouse and human cell lines (20, 30, 59).
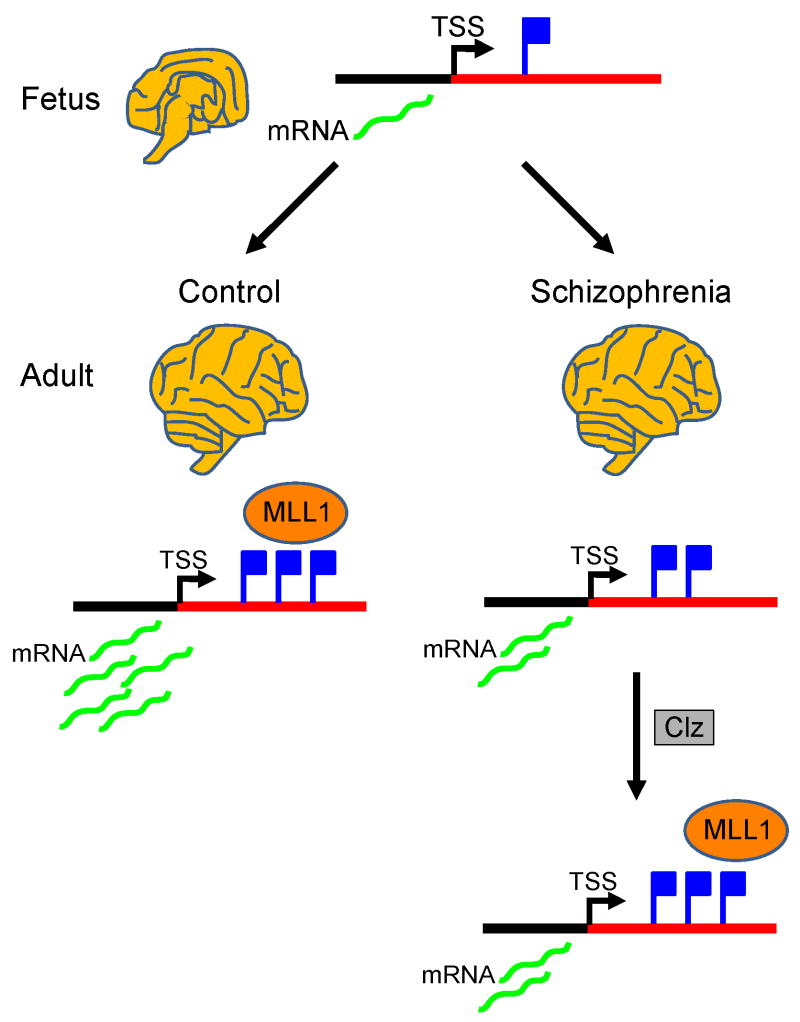
Figure 2. Epigenetic determinants of dysregulated GABAergic gene expression in schizophrenia.
A subset of GABAergic gene promoters, including GAD1 encoding GAD67, show a progressive upregulation in mRNA levels and open chromatin-associated histone H3-lysine (K) 4 trimethylation (blue tags) during the course of prefrontal development and maturation. Chromatin remodeling at the GAD1 locus is likely to involve MLL1 methyltransferase. A subset of subjects diagnosed with schizophrenia shows a deficit both in GAD1 mRNA levels and trimethylated H3K4. Treatment with the atypical antipsychotic, clozapine, leads to increased MLL1 recruitment, and H3K4 methylation at the GAD1 locus. Of note, these drug-induced methylation changes may not necessarily be accompanied by increased GAD1 mRNA. See (28) for details.
Given the above, it is possible that disease-related changes in mRNA levels could be explained by alterations of H3K4 trimethylation or other histone modifications. Indeed, a recent study reported a deficit in the H3K4me3 mark at the promoter of the GABA synthesis gene, glutamic acid decarboxylase, 67kDa (GAD1/GAD67) in postmortem schizophrenic brain, in conjunction with a deficit in GAD1 transcript and increased levels of the repressive mark, H3K27me3 (28) (Fig. 2). Another example is the already mentioned FXN gene in Friedreich's Ataxia; here, an abnormal GAA triplet repeat expansion in intron 1 is associated with decreased FXN mRNA levels in brain, and this occurs in conjunction with decreased histone acetylation, but increased tri-methylation of H3K9, a repressive chromatin mark (37).
Histone modifying enzymes and neuropsychiatric disease
As outlined above, histone lysine methylation signals at gene promoters could be viewed as a markers that differentiate between sites of active and silenced gene expression. In addition, there is increasing evidence that some components of the histone methylation machinery are critical for normal brain function and development. For example, the H3K9-specific histone methyl-transferase, SET domain bifurcated 1 (SETDB1) (60), also known Erg-associated protein with SET domain (ESET) (61), demonstrates increased expression in striatum in Huntington's disease, resulting in global H3K9 hyper-trimethylation and, probably, neuronal dysfunction (62). Therefore, the delicate balance of different histone methylation marks in brain chromatin appears to be essential for orderly brain development and function. This point is further underscored by the recent discovery that the minimal critical region of the 9q subtelomeric deletion syndrome associated with severe mental retardation encompasses the H3K9-specific histone methyltransferase, Euchromatin Histone Methyltransferase 1 gene (EHMT1; also termed GLP) (63). In addition, mutations in the X-linked gene Jumonji AT-rich interactive domain 1C (JARID1C; also termed SMCX), which encodes a H3K4me3-specific demethylase, results in mental retardation and autism (64, 65). Furthermore, iBRAF, a High Mobility Group (HMG) containing protein, is thought to promote neuronal differentiation by recruiting a H3K4-specific methyltransferase, mixed-lineage leukemia (Mll1), to sites of neuronal gene promoters, thus activating neuronal gene expression (66). Mice lacking both Mll1 alleles die during early intrauterine development, while mice carrying only one intact allele show a defect in synaptic plasticity in the hippocampus (67). Interestingly, there is evidence that the atypical antipsychotic, clozapine, upregulates Mll1-occupancy, and H3K4 methylation, at selected gene promoters (28) (Fig. 2). While the specific signaling pathways targeted by clozapine to induce these effects remain to be determined, intact brain circuitry does appear necessary because the drug does not elicit histone methylation changes in cultured cortical neurons that lack monoaminergic and other subcortical input (27). Therefore, several key regulators of histone H3K4 and H3K9 methylation, including MLL1, JARID1C/SMCX, EHMT1, and SETDB1, are essential for orderly brain development and could provide potential targets for new drug development.
Undoubtedly, the study of histone methylation and other types of epigenetic modifications bears great promise to enhance knowledge about the mechanisms of psychiatric disease. Recent technological advances, combining chromatin immunoprecipitation with single DNA molecule sequencing, now make it possible to study histone modifications and DNA methylation in a genome-wide and highly quantitative fashion (68, 69). These approaches will also shed light on how the epigenomic landscape of the human brain is shaped during development and altered in psychiatric disease.
Acknowledgments
The authors thank Caroline Connor for helpful comments on the manuscript. Supported by grants from the US National Institutes of Health (grant numbers MH071476 and HD048489).
Footnotes
Financial disclosures: The authors reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ginsberg SD, Mirnics K. Functional genomic methodologies. Prog Brain Res. 2006;158:15–40. doi: 10.1016/S0079-6123(06)58002-1. [DOI] [PubMed] [Google Scholar]
- 2.Hasenkamp W, Hemby SE. Functional genomics and psychiatric illness. Prog Brain Res. 2002;138:375–93. doi: 10.1016/S0079-6123(02)38087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61(3):300–8. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 4.Akbarian S, Ruehl MG, Bliven E, Luiz LA, Peranelli AC, Baker SP, Roberts RC, Bunney WE, Jr, Conley RC, Jones EG, Tamminga CA, Guo Y. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2005;62(8):829–40. doi: 10.1001/archpsyc.62.8.829. [DOI] [PubMed] [Google Scholar]
- 5.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22(7):2718–29. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, Mirnics K, Lewis DA. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13(2):147–61. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006;52(2):293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Knable MB, Barci BM, Webster MJ, Meador-Woodruff J, Torrey EF. Molecular abnormalities of the hippocampus in severe psychiatric illness: postmortem findings from the Stanley Neuropathology Consortium. Mol Psychiatry. 2004;9(6):609–20. 544. doi: 10.1038/sj.mp.4001471. [DOI] [PubMed] [Google Scholar]
- 9.Meador-Woodruff JH, Clinton SM, Beneyto M, McCullumsmith RE. Molecular abnormalities of the glutamate synapse in the thalamus in schizophrenia. Ann N Y Acad Sci. 2003;1003:75–93. doi: 10.1196/annals.1300.005. [DOI] [PubMed] [Google Scholar]
- 10.Karoutzou G, Emrich HM, Dietrich DE. The myelin-pathogenesis puzzle in schizophrenia: a literature review. Mol Psychiatry. 2008;13(3):245–60. doi: 10.1038/sj.mp.4002096. [DOI] [PubMed] [Google Scholar]
- 11.Sokolov BP. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? Int J Neuropsychopharmacol. 2007;10(4):547–55. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- 12.Haroutunian V, Katsel P, Dracheva S, Stewart DG, Davis KL. Variations in oligodendrocyte-related gene expression across multiple cortical regions: implications for the pathophysiology of schizophrenia. Int J Neuropsychopharmacol. 2007;10(4):565–73. doi: 10.1017/S1461145706007310. [DOI] [PubMed] [Google Scholar]
- 13.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8(5):355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 14.Duman RS, Newton SS. Epigenetic marking and neuronal plasticity. Biol Psychiatry. 2007;62(1):1–3. doi: 10.1016/j.biopsych.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Buckley NJ. Analysis of transcription, chromatin dynamics and epigenetic changes in neural genes. Prog Neurobiol. 2007;83(4):195–210. doi: 10.1016/j.pneurobio.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8(1):57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Curr Opin Genet Dev. 2001;11(2):124–9. doi: 10.1016/s0959-437x(00)00168-4. [DOI] [PubMed] [Google Scholar]
- 18.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 19.Biron VL, McManus KJ, Hu N, Hendzel MJ, Underhill DA. Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev Biol. 2004;276(2):337–51. doi: 10.1016/j.ydbio.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 24.Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr Opin Struct Biol. 2006;16(6):753–60. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Rando OJ. Global patterns of histone modifications. Curr Opin Genet Dev. 2007;17(2):94–9. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27(42):11254–62. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):465–466. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 30.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Stadler F, Kolb G, Rubusch L, Baker SP, Jones EG, Akbarian S. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem. 2005;94(2):324–36. doi: 10.1111/j.1471-4159.2005.03190.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14(5):637–46. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 34.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 35.Horn PJ, Peterson CL. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14(1):83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- 36.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15(5):490–5. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17(5):735–46. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 38.Huang HS, Matevossian A, Jiang Y, Akbarian S. Chromatin immunoprecipitation in postmortem brain. J Neurosci Methods. 2006;156(12):284–92. doi: 10.1016/j.jneumeth.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 42.Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–45. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134(1):60–6. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 44.Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102(26):9341–6. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, Kato T. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25(22):5376–81. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy BC, O'Reilly RL, Singh SM. Site-specific cytosine methylation in S-COMT promoter in 31 brain regions with implications for studies involving schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2005;133(1):37–42. doi: 10.1002/ajmg.b.30134. [DOI] [PubMed] [Google Scholar]
- 48.Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, Kato T. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63(5):530–3. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Ooi SK, Q C, Bernstein E, L K, J D, Y Z, Erdjument-Bromage H, Tempst P, L SP, Allis CD, C X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS ONE. 2008;3(5):e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Travers A, Drew H. DNA recognition and nucleosome organization. Biopolymers. 1997;44(4):423–33. doi: 10.1002/(SICI)1097-0282(1997)44:4<423::AID-BIP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 52.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21(23):3027–43. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 53.O'Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31(1):76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 54.Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–46. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- 55.Matevossian A, Akbarian S. A Chromatin Assay for Human Brain Tissue. Journal of Visualized Experiments. 2008;13 doi: 10.3791/717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81(6):1144–57. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nuovo GJ. Methylation-specific PCR in situ hybridization. Methods Mol Biol. 2004;287:261–72. doi: 10.1385/1-59259-828-5:261. [DOI] [PubMed] [Google Scholar]
- 58.Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 60.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–32. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21(1):148–52. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- 62.Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA, Smith KM, Ferrante RJ. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci U S A. 2006;103(50):19176–81. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Genevieve D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79(2):370–7. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A(4):505–11. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- 65.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128(6):1077–88. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 66.Wynder C, Hakimi MA, Epstein JA, Shilatifard A, Shiekhattar R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat Cell Biol. 2005;7(11):1113–7. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- 67.Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J Biol Chem. 2007;282(13):9962–72. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- 68.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008 doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]