Abstract
Human apurinic/apyrimidinic endonuclease (hApe1) encodes two important functional activities: an essential base excision repair (BER) activity and a redox activity that regulates expression of a number of genes through reduction of their transcription factors, AP-1, NFκB, HIF-1α, CREB, p53 and others. The BER function is highly conserved from prokaryotes (E. coli exonuclease III) to humans (hApe1). Here, we provide evidence supporting a redox function unique to mammalian Apes. An evolutionary analysis of Ape sequences reveals that, of the 7 Cys residues, Cys 93, 99, 208, 296, and 310 are conserved in both mammalian and non-mammalian vertebrate Apes, while Cys 65 is unique to mammalian Apes. In the zebrafish Ape (zApe), selected as the vertebrate sequence most distant from human, the residue equivalent to Cys 65 is Thr 58. The wild-type zApe enzyme was tested for redox activity in both in vitro EMSA and transactivation assays and found to be inactive, similar to C65A hApe1. Substitution of Thr 58 with Cys in zApe, however, resulted in a redox active enzyme, suggesting that a Cys residue in this position is indeed critical for redox function. In order to further probe differences between redox active and inactive enzymes, we have determined the crystal structures of vertebrate redox inactive enzymes, the C65A human Ape1 enzyme and the zApe enzyme at 1.9 Å and 2.3 Å, respectively. Our results provide new insights on the redox function and highlight a dramatic gain-of-function activity for Ape1 in mammals not found in non-mammalian vertebrates or lower organisms.
1.0 Introduction
Redox regulation has been shown to play an important role in modulating the DNA binding activity of a number of transcription factors including AP-1, NFκB, HIF-1α, HLF, CREB, PAX, p53 and others [1–11] (reviewed in [12]). As described for AP-1 (Fos/Jun), this regulation involves the reduction of a critical Cys residue(s) within c-Jun that allows it to bind DNA with much higher affinity than the oxidized c-Jun [13,14]. The factor responsible for reducing AP-1 was found to be a nuclear factor, termed redox effector factor 1 (Ref1) [15], and later identified as apurinic/apyrimidinic endonuclease (Ape1, also referred to as HAP1 or APEX1) [2]. Ape1 is an essential enzyme in the base excision repair pathway catalyzing the second step in the pathway, cleavage of the phosphodiester backbone 5’ of the abasic site leaving a 3’ hydroxyl and 5’-deoxyribose phosphate. Deletion of the APE1 gene results in very early embryonic lethality in mice [16], and knock-down of Ape through the use of a morpholino oligonucleotide injected in zebrafish embryos results in death at the midblastula transition [17]. In addition, Ape has been shown to be involved in heart and blood development in zebrafish [17].
The redox activity has been shown to require the N-terminal region of the protein (reviewed in [12]). The human Ape1 includes 61 residues not present in exonuclease III from E. coli, and while truncations of up to 40 residues do not affect the redox activity, removal of the 62 N-terminal residues from the human Ape1 renders this enzyme redox inactive [14,18]. It was hypothesized that Ape1’s redox activity might involve a Cys residue [14]. Accordingly, each of the seven Cys residues in Ape1 was substituted individually with Ala, and the resulting enzymes were tested for their ability to reduce AP-1 thereby allowing it to bind target DNA specifically in an electrophoretic mobility shift assay (EMSA) [18]. Of these single Cys mutants, all when reduced retained wild-type redox activity except C65A hApe1, which was redox inactive. When oxidized, C93A hApe1 was found to be redox active, while all of the other mutants were inactive. Thus, a redox mechanism involving a disulfide bond between Cys 65 and Cys 93 was proposed [18].
Subsequently, however, several crystal structures of Ape1 have been reported [19–21] providing three important observations with regard to the redox activity of Ape1. First, no disulfide bonds are present in any of the structures. Second, Cys 65 is a buried residue in the conformation of hApe1 observed in the crystal structures and therefore not accessible to the transcription factors that Ape1 reduces. Third, the structures of hApe1 that have been reported to date in different crystallographic lattices are remarkably similar. Even in the hApe1-DNA complex, there are no significant conformational changes in the enzyme [20]. Thus, the mechanism by which Cys 65 would participate in a potential redox reaction between Ape1 and a transcription factor requires further investigation. One possible explanation offered by Gorman et al. [19] was that substitution of Cys 65 with Ala may affect the stability and/or folding of Ape1 and result in subsequent loss of the redox activity. More recently, the role of Cys 65 in Ape1’s redox activity has been challenged through the creation of a Ref1C64A/C64A mouse (C64 is equivalent to the human C65 residue) that was shown to develop normally [22]; in this study the authors concluded that Cys 65 is not required for redox activity.
Ape vertebrate sequences are highly conserved with the most distantly related sequence, that of zebrafish Ape (zApe) being 66% identical to the human Ape1. We had initially selected zebrafish as a potential model system to study the role of Ape in development as mice knockouts were lethal. However, as reported here, the wild-type zApe lacks redox activity. Thus, we have not pursued the zebrafish development experiments. We report the results of biochemical studies including hApe1, zApe, and a zApe mutant engineered to acquire redox function, the first crystal structures of redox inactive enzymes from vertebrates, and a discussion of the mechanistic implications for the redox activity of Ape.
2.0 Materials and Methods
2.1 Evolutionary analysis of Ape sequences
Orthologous sequences for APE1 in human and other species except zebrafish were obtained from the 28 species alignment at the UCSC genome browser [23]. To obtain the zebrafish APE1 gene sequence, the BLAT web server at UCSC was used to search for matches to the human APE1 gene within the zebrafish genome. We next identified the 3-nucleotide codons corresponding to the 7 human Cys residues in the APE1 genes in all species and aligned the concatenated Cys codons using CLUSTALW [24]. The branch length for the tree was determined using PHYLIP [25], and the tree was drawn using MEGAN [26].
2.2 Expression and purification of proteins
GST-hApe1 and mutants
The wild-type and mutant hApe1 proteins were expressed as GST-fusions. A pGEX-4T hApe1 vector was constructed and confirmed by DNA sequencing. GST-C65A-hApe1 and GST-C93A-hApe1 mutants were introduced by site-directed mutagenesis into the pGEX-4T hApe1 vector using the Stratagene Quikchange kit and confirmed by DNA sequencing. The GST-hApe1 and mutants were transformed into Rosetta (DE3) E. coli (Novagen), grown in 1 L of LB media with 100 µg/mL ampicillin and 34µg/mL chloramphenicol until the OD at 600 nm reached 0.5, and then induced overnight with 1 mM IPTG at 22°C. The cells were harvested by centrifugation, resuspended in PBS buffer, and purified using the standard glutathione affinity purification protocol (Amersham Biosciences). Final concentrations of the proteins were calculated from absorbance measurements at 280 nm.
C65A hApe1
Site-directed mutagenesis of a pET28A vector encoding human Ape1 (40–318) in order to replace Cys 65 with Ala was performed using the Stratagene Quikchange kit and confirmed by DNA sequencing. Transformation, cell growth, and induction were performed as described for the GST-Ape1 proteins with the exception that 6 × 1L were grown and induced at 37°C for 4 hrs. The cultures were harvested by centrifugation at 4000 × g for 30 minutes, and the pellets were stored at −80°C. The cell pellets were each resuspended in 20 mL of 50 mM sodium phosphate buffer pH 7.8, 0.3 M NaCl, 10 mM imidazole, and then lysed by using a French press (SLM-AMINCO, Spectronic Instruments, Rochester, USA) at 1000 psi. The suspension was centrifuged at 35,000 rpm for 35 min, and the supernatant was then loaded on a Ni-NTA column at 4°C. The protein was eluted with a linear imidazole gradient (0.02– 0.5 M), and fractions containing C65A hApe1 were further purified on an S-Sepharose column using 50 mM MES pH 6.5, 1 mM DTT, and a linear NaCl gradient (0.05–1 M). The fractions were incubated overnight with 2 units/mg of thrombin to cleave the N-terminal hexa-His tag and then subjected to a final S-Sepharose chromatographic purification step. The protein was then concentrated to 54 mg/mL.
Zebrafish Ape
Expression vectors encoding the full-length zApe and zApe (33–310) proteins in pET15b were constructed using standard subcloning techniques and verified by DNA sequencing. The zApe vectors were transformed into Rosetta (DE3) E. coli; 6 × 1L cultures were grown in LB media with 100 µg/ml ampicillin until the OD at 600 nm reached approximately 0.6 and then induced with 1 mM IPTG at 37°C for 4–5 hrs. Harvesting of cell pellets, subsequent disruption, and initial Ni-NTA purification were done as described for the C65A hApe1. The Ni-NTA fractions containing zApe were combined and loaded on a pre-equilibrated MonoQ (16/10) column after dilution to a final NaCl concentration of 50 mM with 50 mM Tris, pH 8.0. The protein was eluted using a linear NaCl gradient (0.05– 1M), and fractions containing the zApe were combined, digested with thrombin overnight at 4°C, and then subjected to a final MonoQ (16/10) column, as described in the previous step. The purified proteins were concentrated to 50–75 mg/ml in 10 mM Hepes, pH 7.5.
Zebrafish T58C Ape
Site-directed mutagenesis was performed on the full-length zebrafish Ape pET15b vector using Stratagene’s Quikchange kit and verified by DNA sequencing. The T58C zApe vector was then transformed into Rosetta (DE3) E. coli. Cell growths and subsequent purification of the T58C zApe were performed as described for zApe above with the exception that a Q-sepharose column was used instead of the MonoQ column for the ion exchange chromatography.
2.2 Redox assays
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described [27] with the following modifications. 10 µg/µl purified Ape1 protein was reduced with 1.0 mM DTT at 37°C for 10 min and diluted in PBS buffer to yield final concentrations of 2 µg/µl protein and 0.2 mM DTT. Two µl of reduced Ape1 protein was added to EMSA reaction buffer, 10 mM Tris pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 5% (v/v) glycerol, with 6 µg of nuclear extract (treated with 0.01mM diamide for 10 min) from Hey-C2 cells as the source of AP-1 protein in a total volume of 18 µl and incubated for 30 min. at room temperature. Oxidized Ape1 samples were not reduced with DTT prior to addition to the reactions including Hey-C2 nuclear extracts. One µl poly(dI-dC) · poly(dI-dC) (1µg/ul) (Amersham Biosciences, Piscataway, NJ) was added for 5 min followed by one µl of the 5’ hexachloro-fluorescein phosphoramidite (HEX)-labeled double-stranded oligonucleotide DNA (The Midland Certified Reagent Company, Midland, TX) containing the AP-1 consensus sequence (5’CGCTTGATGACTCAGCCGGAA-3’) (0.1 pmol), and the mixture was further incubated for 30 min. at room temperature. The final concentration of DTT in the redox reactions was 0.02 mM. Samples were loaded on a 5% nondenaturing polyacrylamide gel and subjected to electrophoresis in 0.5X TBE buffer (200 V for 1 h at 4°C) and detected using the Hitachi FMBio II Fluorescence Imaging System (Hitachi Genetic Systems, South San Francisco, CA). The HEX fluorophore is excited by a solid-state laser at 532 nm (Perkin-Elmer) and emits a fluorescent light signal at 560 nm, which is then measured using a 585 nm filter.
Transactivation assay
The stable Skov-3X ovarian cancer cell line with NFkB- Luc gene (luciferase gene with the NFκB–responsive promoter) was established by transfecting Skov-3X cells with plasmid pNFκB-Luc (Stratagene, La Jolla, CA) using lipofectamine TM 2000 (Invitrogen Life Technologies, Carlsbad, CA) and screening the luciferase-positive colonies using a Luciferase Assay Kit ( Promega Corp., Madison, WI). The stable Skov-3X cells with the NFκB-Luc gene were cotransfected with plasmid pcDNA-wthApe1 or its mutants (pcDNA-C65A, pcDNA-C93A, pcDNA-C99A, pcDNA-C138A, pcDNA-C208A, pcDNA-C296A, pcDNA-C310A), or pcDNA- wtzApe or its mutant pcDNA-T58CzApe and a Renilla luciferase control reporter vector pRL-CMV (Promega Corp., Madison, WI) in a 1:10 ratio using lipofectamine TM 2000. After 24 h transfection, cells were lysed and the Firefly and Renilla luciferase activities were assayed using the Dual Luciferase Reporter Assay System ((Promega Corp., Madison, WI) with Renilla luciferase activity for normalization in a 96-well microtiter plate luminometer (Thermolabs systems, Franklin, MA). All of the transfection experiments were performed in triplicate and repeated at least three times in independent experiments. Student’s t-tests were performed in order to evaluate the statistical significance of the signals obtained in the assay.
2.3 Crystallographic studies
Crystal structure of C65A hApe1
The concentrated C65A hApe1 protein was diluted to 10 mg/mL in 10 mM Hepes pH 7.5 and then crystallized by hanging drop vapor diffusion using 10 mM MES pH 6.0, 7.5 mM Sm(OAc)3, 4% (v/v) Dioxane, 10–20% (w/v) PEG 8000 as the precipitating solution at 20°C. Crystals were soaked in stabilizing solutions including increasing concentrations of ethylene glycol to a final concentration of 20%, and data were collected at the Advanced Photon Source, Argonne, IL, beamline 19-ID (Table 1) at 100K and processed using HKL2000 [28] (see Table 1). The crystal structure was determined by molecular replacement with AMoRe [29] using 1E9N (PDB accession id) as the search model. The substitution of Cys 65 for Ala was verified in initial Fo-Fc maps in which a large negative peak was observed for the sulfhydryl of Cys 65, which was present in the initial model. Iterative cycles of manual rebuilding in O [30] and crystallographic refinement in CNS [31] were used to obtain the final structural model. The structure includes 2 Sm3+ atoms coordinated to E96 and D70 or E216 and E217 with the first site including two partially occupied sites for which occupancies were estimated by inspection of resulting Fo-Fc electron density maps. These sites are similar to two of the four Sm3+ sites found in the original hApe1 structure (PDB accession id 1BIX). The quality of the final model was assessed using PROCHECK [32].
Table 1.
Crystal structures of C65A hApe1 and zApe
| Crystallographic Data | C65A hApe1 | zApe |
|---|---|---|
| Cell dimensions (Å) | a=46.69 | a=54.74 |
| b=143.60 | b=117.89 | |
| c=45.39 | c=86.01 | |
| β=98.41 ° | ||
| Space group | P21212 | P21 |
| Resolution range (Å) | 33.0-1.90 | 49.15-2.30 |
| Total number of reflections | 118818 | 141633 |
| Number of unique reflections | 22948 | 46075 |
| Completeness (%) | 93.4 (92.1) | 96.5 (85.5) |
| aRmerge | 0.07 (0.16) | 0.059 (0.26) |
| I/σ | 33.1 (13.2) | 14.3 (3.4) |
| Refinement statistics | ||
| bRvalue | 0.177 | 0.199 |
| bRfree | 0.226 | 0.236 |
| Rmsd bonds Å | 0.0045 | 0.0058 |
| Rmsd angles ° | 1.37 | 1.38 |
| Average B-factor | 17.6 | 28.3 |
| No. of ions | 2 / 4 (Sm/OAc) | 3 (Pb) |
| No. of waters | 326 | 362 |
| Ramachandran plot statistics | ||
| Most favored | 87.3 | 86.9 |
| Addn. favored | 12.7 | 12.0 |
| Generously allowed | 1.1 | |
Highest resolution shells are (1.97-1.90) and (2.4- 2.30) for C65A hApe1 and zApe, respectively.
Rmerge= ΣΣi|Ii-<I>|/ Σ<I> where I is the integrated intensity of a reflection.
Rvalue=Σhkl∥Fobs − kFcalc|/Σhkl|Fobs|. 5% of all reflections were omitted from refinement, and Rfree is the same statistic calculated for these reflections.
Crystal structure of zApe
Crystals of zApe (33–310) (74 mg/ml in 10mM Hepes, pH 7.5 buffer) were grown by hanging drop vapor diffusion at 20°C using 200 mM Ammonium acetate, 100 mM Bis-Tris pH 5.5, 7% (w/v) PEG 3350, 2% (v/v) glycerol and 0.8 mM lead acetate as the precipitating solution. Microseeding was used to improve the size and quality of the crystals in the same precipitant with the exception that 4% glycerol was used. The crystals were cryocooled using saturated disodium malonate as the cryoprotectant. Data were collected at the Advanced Light Source, Berkeley, CA at beamline 4.2.2 and processed using d*trek [33] (see Table 1). The 2.3 Å resolution zebrafish Ape structure was solved by molecular replacement with MOLREP [34] using the human Ape1 (PDB accession id, 1BIX) as a search model. The asymmetric unit was found to contain 3 molecules. Crystallographic refinement was done using CNS [31]. Three partially occupied Pb2+ sites were included in the model; occupancies were estimated by inspection of Fo-Fc electron density maps following crystallographic refinement. Inclusion of non-crystallographic symmetry restraints for main chain atoms in residues 36–310 of each chain did not affect the final R-values with R=0.201 and Rfree = 0.237 using CNS [31]. The rms deviation for A vs. B was 0.55 Å and for A vs. C was 0.38 Å. Each refinement cycle was followed by manual model building in O [30]. The quality of the model was checked throughout refinement by using PROCHECK [32].
Figure 3, Figure 4, and Figure 5 were generated using MOLSCRIPT[35] and RASTER3D[36]. Figure 7 was generated using GRASP [37]. Coordinates have been deposited with the PDB: 203C and 203H, zApe and C65A hApe1 structures, respectively.
3.0 Results
3.1 Cell-based analysis of redox activity for hApe1 Cys mutants
The human Ape1 includes seven Cys residues, 65, 93, 99, 138, 208, 296, and 310. In a previous study [18], the redox activity of single Cys mutants was assessed using an in vitro EMSA redox assay in which reduction of AP-1 by hApe1 results in a shifted band, indicating the formation of an AP-1/DNA complex. Of the single Cys mutants tested, only C65A hApe1 was found to be redox inactive [18].
In order to assess the redox activity of Ape1 and single Cys mutants within a cell providing data complementary to the in vitro EMSA data, we used a cell-based transactivation assay in which reduction of NFκB by hApe1 results in a quantifiable luciferase signal (See Materials and Methods). Although interpretation of in vitro analysis is more straight-forward, the level of confidence with regard to the biological significance of the results is increased by the inclusion of cell-based assays. In the transactivation assay, C65A hApe1 is redox inactive with a signal 1.2-fold that of the control, and C93A hApe1, shows a decrease in redox activity with a signal 2.0-fold that of the control as compared to wild-type, which is 2.8-fold that of the control (Fig. 1A). All other Cys mutants had wild-type redox activity in this assay. Statistical analysis was performed in order to determine that the signal for C65A hApe1 does not differ significantly from that of the control sample, while the signal for C93A hApe1 does differ significantly from wild-type hApe1. Thus, results obtained for the transactivation assays for reduction of NFκB by hApe1 are consistent with those obtained in previous in vitro EMSA assays for reduction of AP-1 [18], with the exception of the new finding that C93A hApe1 has intermediate redox activity. This finding was confirmed using the in vitro redox assay as shown in Figure 1B. Similar to the results from the transactivation assay, GST-C93A-hApe1 has intermediate redox activity.
Figure 1.
(A) Effect of human Ape1 and cysteine mutants on NFκB transactivation in a cell-based reporter assay system. SKOV-3X cells were transfected with a NFκB –Luc construct containing an NFκB –response promoter and driving the expression of a luciferase gene. The cells were cotransfected with plasmid pcDNA-wild-type zApe1, pcDNA-wild-type hApe1 or its mutants (pcDNA-C65A, pcDNA-C93A, pcDNA-C99A, pcDNA-C138A, pcDNA-C208A pcDNA-C296A, pcDNA-C310A) and a Renilla luciferase control reporter vector pRL-CMV. After a 24 h transfection period, cells were lysed and Firefly and Renilla luciferase activities were assayed using Renilla luciferase activity for normalization. All of the transfection experiments were performed in triplicates and repeated at least three times in independent experiments. Error bars denote standard deviation. Data are means± standard error from a representative experiment and the Student’s t-tests were performed. *Significant difference at the p< 0.01 level comparing the cysteine mutants and wt-hApe1. ‡ indicates a significant difference at the p< 0.01 level comparing the mutant with C65A hApe1, while ‡‡ indicates no significant difference (p> 0.05) compared with C65A. (B) Effect of wild-type zApe1, wild-type hApe1 and hApe1 C65A and C93A mutants on AP-1-DNA binding in a redox EMSA assay. The Ape proteins were reduced using 1mM DTT and then diluted to a final concentraton of 0.02mM DTT, which is used in the negative control lane 5. Reduced proteins with 0.02 mM DTT or oxidized (unreduced) proteins were preincubated with Hey-C2 nuclear extract, treated as described in Materials and Methods, for 30 min. Oxidized proteins (Lane 6–9) did not stimulate extract AP1-DNA binding. Reduced wild-type hApe1 (Lane 2) enhanced AP-1-DNA binding, reduced wild-type zApe1 and C65A-hApe1 (Lane 1 and 3) did not, and C93A-hApe1 (Lane 4) demonstrated an intermediate binding pattern. Specificity of AP-1 binding was determined using antibody to AP-1 (data not shown).
3.2 Evolutionary analysis of Ape sequences
In order to investigate the evolution of the redox function, we have analyzed the codons corresponding to the seven Cys residues in human Ape1 from vertebrates for which genome information is available including human, rhesus macaque, mouse, dog, horse, opossum, platypus, lizard (Anolis Carolinensis), frog (Xenopus tropicalis), and two fish, stickleback and zebrafish (See Material and Methods). We sought to identify divergence of Cys residues within vertebrates in order to correlate this with redox activity. An evolutionary tree depicting the relationship between the vertebrate sequences is shown in Fig. 2A. The reptilian and amphibian sequences represent a separate branch on the tree from the mammalian sequences. Excluding platypus in which the residue equivalent to Cys 138 is Arg, all Cys residues are conserved within the mammalian sequences as shown in Table 2. Within reptilian and amphibian sequences, Cys is not conserved in the position equivalent to Cys 65 or Cys 138. The residue equivalent to Cys 65 is either Ser or Thr and that equivalent to Cys 138 is Lys or Glu in non-mammalian sequences (Table 2).
Figure 2.
(A) An evolutionary tree is shown for vertebrate Ape sequences based on analysis of the codons in each sequence aligned with the 7 Cys codons in hApe1. Branch length is indicated as a number on each branch. Mammalian and non-mammalian vertebrate sequences are found in distinct branches with zebrafish representing the sequence most evolutionarily distant from the human Ape1. (B) PSI-BLAST sequence alignment of hApe1 and zApe sequences, top and bottom respectively. Identical residues are shown in the intervening row between the two sequences and conserved residues are indicated by a (+). The two sequences are 66% identical overall. Positions of Cys residues are denoted with an (*).
Table 2.
Comparison of Cys residues found in vertebrates
| 65 | 93 | 99 | 138 | 208 | 296 | 310 | |
|---|---|---|---|---|---|---|---|
| Human | C | C | C | C | C | C | C |
| Macaque | C | C | C | C | C | C | C |
| Mouse | C | C | C | C | C | C | C |
| Dog | C | C | C | C | C | C | C |
| Horse | C | C | C | C | C | C | C |
| Opossum | C | C | C | C | C | C | C |
| Platypus | C | C | C | R | C | C | C |
| Lizard | T | C | C | K | C | C | C |
| Frog | S | C | C | K | C | C | C |
| Stickleback | T | C | C | E | C | C | C |
| Zebrafish | T | C | C | E | C | C | C |
| Consensus | C/T/S | C | C | C/R/K/E | C | C | C |
See Materials and Methods for details on alignments of sequences and species. The line separating Platypus from Lizard within the table marks the division between mammalian species above the line and non-mammalian vertebrates below the line. The consensus sequence is indicated for each position on the bottom line.
Thus, based on this analysis of Cys conservation as well as biochemical characterization of substituted hApe1 enzymes, we predicted that zebrafish Ape (zApe), representing the non-mammalian vertebrate most distant from humans on the evolutionary tree shown in Fig. 2A, would be redox inactive. The zebrafish genome includes two copies of AP endonuclease, zfAPEX1a and zfAPEX1b, with identical coding sequences [17]. Therefore, at the protein level the two are indistinguishable from one another. The residue equivalent to the human Cys 65 in the zApe sequence is Thr 58 (Table 2, Fig. 2B). In order to test our hypothesis, we analyzed redox activity for the wild-type zApe in both in vitro and cell-based transactivation assays (Fig. 1). zApe was cloned, expressed, and purified as described in the Materials and Methods for use in the in vitro redox assays. As predicted, the zApe enzyme is redox inactive.
3.3 Crystal structures of redox inactive Ape enzymes
In order to determine structural differences that may exist between vertebrate redox inactive and redox active Ape1 enzymes, we have determined the crystal structures of two vertebrate redox inactive enzymes, C65A hApe1 enzyme and zApe. N-terminally truncated C65A (40–318) hApe1 and zApe (33–310) enzymes were expressed and purified from E. coli (See Materials and Methods). The truncations were based on previously reported crystal structures of the wild-type hApe1 enzyme in which residues 1–42 are disordered [19–21]. The equivalent truncation in zApe begins with residue 33 as it lacks 7 residues found in the N-terminal region of the human enzyme as discussed below (Fig. 2B). C65A hApe1 and zApe samples used for structural studies were found to be redox inactive in redox EMSA assays (data not shown). Each of the redox inactive enzymes crystallized in a lattice distinct from those previously reported for wild-type hApe1 (See Table 1 for cell parameters) despite the use of similar crystallization condition containing Sm(OAc)3 for C65A hApe1 to that of Gorman et al.[19]. The crystal structures were determined by molecular replacement (See Materials and Methods for details).
Both zApe and C65A hApe1 are monomeric in solution as characterized by size exclusion chromatography (data not shown). The C65A hApe1 structure (PDB identifier 203H) includes one molecule in the asymmetric unit, 2 Sm3+ atoms, and 4 acetate molecules as shown in Fig. 3A. Within the repair active site, the Sm3+ atom occupies two sites each of partial occupancy, with one site coordinated by both Asp 70 and Glu 96 and the second coordinated only to Glu 96. The second Sm3+ atom is coordinated by Glu 216 and Glu 217 on the surface of the molecule. Two of the acetate molecules form hydrogen-bonding interactions with the backbone amides of residues Ser 129 and Ser 275; one is hydrogen-bonded to the N-terminus of the molecule, residue Met 37; and the fourth is hydrogen-bonded to the amine of Lys 78. These sites potentially represent general anionic binding sites on the surface of hApe1. In the zApe structure (PDB identifier 203C), there are three zApe molecules in the asymmetric unit along with 3 Pb2+ atoms, one per molecule, which are coordinated in the repair active site by Asp 63 in all sites and by Asp 63 and Glu 89, equivalent to Asp 70 and Glu 96, in two of the three sites (See Fig. 3B). The three zApe molecules are very similar to one another as shown in Fig. 3C with pairwise root mean square deviations (rmsds) of 0.48 to 0.86 Å for 276 Cα atoms. Metal ions bound in the structures of C65A hApe1 and zApe are bound in the “B” site, the putative post-cleavage metal ion binding site rather than the proposed catalytic “A” site [38].
Figure 3. Crystal structures of redox inactive Ape enzymes.
(A) The crystal structure of C65A hApe1 is shown as a ribbon rendering in red. Bound Sm3+ ions are shown as gold spheres and acetate ions in ball-and-stick representations in cyan. The two Sm3+ ions with overlapping positions occupy slightly different positions within the same binding site within the repair active site and have each been modeled with partial occupancy. (B) The crystal structure of wild-type zApe including three molecules in the asymmetric unit is shown. Each molecule is shown as a ribbon rendering with the A molecule in gold, B in blue, and C in purple. Pb2+ ions bound within the repair active site are shown as green spheres. (C) A comparison of the three zApe molecules is shown. The three zApe molecules are shown superimposed in the same ribbon rendering and same color scheme as in (B). The molecules are very similar to one another with differences limited to loop regions.
The crystal structures of both C65A hApe1 and zApe provide new structural information on the N-terminus of the protein. The C65A hApe1 structure includes 5 additional residues, 37 through 41 (37 – 39 are encoded by the vector), that have not been observed in previously reported Ape1 structures, even those in which the full-length Ape1 was used [21] (See Fig. 4C). The conformation of these N-terminal residues is partially stabilized by a hydrogen bonding interaction between the guanidium NH2 of Arg 221 and the carbonyl of Gly 41. No interactions between residues 37–40 and the rest of the molecule were observed. Each of the three zApe molecules also have an extended N-terminus as in the C65A hApe1 with one molecule (designated the B molecule) including a fully ordered N-terminus including residues 33–310 as well as 2 additional N-terminal residues encoded by the vector. The conformation of the N-terminal residues of zApe (B) differs from that seen in the C65A hApe1 structure and lacks the hydrogen-bonding interaction observed between Arg 221 and Gly 41; the equivalent residues in zApe are Lys 214 and Ala 34 as shown in Fig. 4A.
Figure 4. Comparison of wild-type hApe1 with redox inactive enzymes.
(A) Ribbon renderings of C65A hApe1(red) and zApe (blue) are shown highlighting the differences in the N-terminal region of the protein with structurally equivalent residues R221 and G41 in hApe1 vs. K214 and A34 in zApe shown in ball-and-stick models. (B) Ball-and-stick renderings are shown as a stereo pair for the superimposed C65A hApe1 (red) and the hApe1 (green) structures in the region of residue 65 highlighting the similarities in the two structures. Ala 65 is shown in yellow superimposed on the wild-type Cys 65, shown in navy. (C) A similar comparison of structural conservation in the vicinity of Cys 65 is shown as a stereo pair for wild-type zApe crystal structure (B-molecule in blue) with Thr 58 in red superimposed on the wild-type hApe1 structure (green) with Cys 65 in navy. Labeled residues farther from Thr 58 differ in the two enzymes; those preceded by a “z” indicate the zApe residue, otherwise from human Ape1.
The structure of C65A hApe1 is similar to the wild-type hApe1 structures with rmsds of 0.24–0.58 Å for superimpositioning of Cα coordinates. The zApe enzyme is also structurally similar to the human enzymes with rmsds of 0.71–0.76 Å for pairwise comparisons of Cα coordinates in the three zApe molecules as compared to the C65A or wild-type hApe1 structures. In particular, for the region of the structure in the vicinity of residue 65, not only the Cα atoms but all the atoms in the amino acids superimpose well including the Cβ atoms in Ala 65 and Cys 65 in the C65A and wild-type hApe1 structures, respectively, as shown in Fig. 4B. The chemical environment immediately surrounding Thr 58, the residue equivalent to Cys 65 in hApe1, is conserved in both zApe and wild-type hApe1 structures with identical amino acids in very similar conformations (Fig. 4C). Further away from Thr 58, the residues in zApe and hApe1 differ as shown in Fig. 4C. Other structural differences between the zApe and hApe1 enzymes include the loop regions, residues 114–122 and 121–129 in zApe and C65A hApe1, respectively, and residues 238–241 in zApe corresponding to a loop that is one residue longer in hApe1, residues 245–249. We also note that of the 7 Cys residues present in the wild-type hApe1, 5 are structurally conserved in the zApe enzyme, namely, 93, 99, 208, 296, and 310 as shown in Fig. 5.
Figure 5. Comparison of Cys residues in zApe and hApe1.
hApe1 is shown in a green ribbon rendering, zApe in blue. Cys residues are shown in a ball-and-stick model with conserved Cys residues shown in magenta, and those associated only with hApe1 in green and zApe in blue. Five of the Cys residues found in hApe1 are also found in zApe. Residue numbers with a “z” prefix indicate the zApe sequence.
3.4 The T58C zApe enzyme is redox active
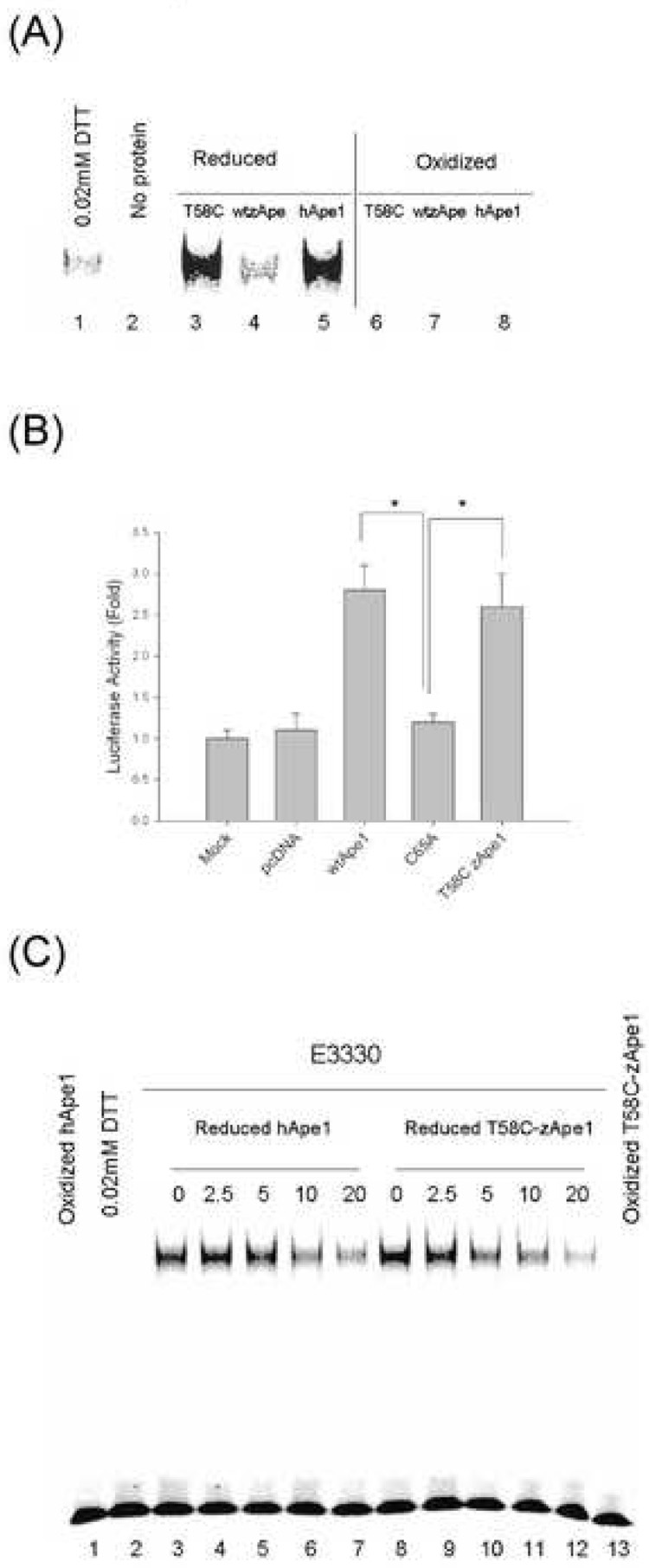
Based on our structural and evolutionary analysis, we predicted that substitution of Thr 58 with Cys would confer redox activity to zApe. The amino acid sequence of zebrafish Ape1 is 66% identical to that of the human Ape1 retaining all catalytic residues required for the endonuclease activity but lacking Cys residues equivalent to Cys 65 and Cys 138 in the hApe1. Other differences in sequence include a deletion of 7 residues in the N-terminus corresponding to residues 15–21 in the human enzyme and deletion of the residue corresponding to 248 (Fig. 2B). The T58C zApe was expressed and purified from E. coli (see Methods) and found to be redox active with near wild-type human Ape1 (hApe1) activity in the EMSA as shown in Fig. 6A. Similarly, T58C zApe has near wild-type human Ape1 redox activity in the transactivation assay (Fig. 6). Transactivation assays in which AP-1 is reduced resulting in an increased luciferase signal were also performed for hApe1, C65A hApe1, zApe, and T58C zApe, and results obtained were very similar to those obtained for the reduction of NFκB (data not shown).
Figure 6.
(A) Effect of wild-type hApe1, wild-type zApe1, and its mutant protein on AP-1-DNA binding in a redox EMSA assay. The Ape1 proteins; wild-type hApe1, wild-type zApe1 and zebrafish Ape1 mutated at position 58 from a Thr to a Cys (T58C) were reduced by 1 mM DTT and then diluted as described in Materials and Methods and in Figure 1. Reduced proteins with 0.02mM DTT or oxidized (unreduced) proteins were preincubated with oxidized nuclear extract and assayed using EMSA as previously described. Oxidized (unreduced) proteins did not stimulate AP-1-DNA binding (Lane 6–8). Reduced wild-type hApe1, zApe1 mutant (T58C) zApe (Lane 3, and 5) enhanced AP-1-DNA binding and wild-type zApe1 (Lane 4) did not. (B) Effect of zApe1 mutant (T58C zApe1), human wild-type Ape1 and its mutant C65A (redox deficient/repair active Ape1) on NFκB transactivation. The stable Skov-3X cells with NFκB – Luc were cotransfected with plasmid pcDANA-T58C zApe1, pcDNA-wtApe1 or pcDNA-C65A-Ape1 and a Renilla luciferase control reporter vector pRL-CMV. After 24 h transfection, cells were lysed and Firefly and Renilla luciferase activities were assayed with Renilla luciferase activity for normalization. All of the transfection experiments were performed in triplicates and repeated at least three times in independent experiments. Error bars denote standard deviation. Data are means± standard error from a representative experiment and Student’s t-tests were performed. * indicates a significant difference, with p< 0.01 for C65A hApe1 compared with wthApe1 and for T58C zApe vs. C65A hApe1. There is no statistically significant difference between wthApe1 and T58C zApe.
Of particular interest is the fact that T58C zApe effectively reduces human transcription factors including AP-1 and NFκB in these experiments. The calculated pI of zApe is 5.8 while that of hApe1 is 8.3, and examination of the electrostatic potential surfaces mapped to the molecular surface of the two enzymes reveals that in the vicinity of the redox active residue, the electrostatic potential surface is substantially different in the two enzymes. Notably, zApe is more negatively charged as represented by red regions in Fig. 7; blue regions indicate positively charged areas. Cys 65 is located just behind Glu 87; similarly, Thr 58 is just behind Glu 80 (see Fig. 7). Comparison of the molecular surfaces calculated for wild-type zApe and hApe1 (1BIX) further highlight that although residues surrounding the redox active Cys (or Thr in zApe) are conserved, residues on the surface of the molecules in this same region of the structure are not conserved, and thus the properties of the molecular surfaces are not conserved. In contrast, the electrostatic potential surfaces of C65A hApe1 and hApe1 are very similar, and with the exception of the N-terminal residues 37–42 found only in the C65A hApe1 structure, the molecular surfaces are also quite similar. Thus, the surface properties of the redox active T58C zApe would be expected to be similar to those of the wild-type zApe. We further note that this “redox” surface of the molecule is relatively flat with no obvious binding pockets deep enough to accommodate a small aromatic molecule.
Figure 7. Comparison of electrostatic potential surfaces.
(A) The electrostatic potential surface of wild-type hApe1 is shown for the face of the molecule expected to be involved in its redox activity. Red regions are negatively charged, blue, positively charged. As a point of reference in each panel, either Glu 87 or Glu 80 is labeled along with R301 or R293. Cys 65, Ala 65, or Thr 58 in panels A, B, and C, respectively, is a buried residue located behind Glu 87 or Glu 80. Although it is not solvent accessible, a conformational change in the structure would likely expose Cys 65 on this face of the molecule. (B) The electrostatic potential surface of the C65A hApe1 is shown for the same face as that shown in (A). C65A hApe1 and hApe1 have similar patterns of charged regions and surface features. The C65A hApe1 differs from that of the wild-type structure in the vicinity of the N-terminus, denoted as N in all images, as there are more ordered residues present in this structure. (C) The electrostatic potential surface of zApe is shown for the same face of the molecule as in (A) and (B) and is distinctly different from that of the hApe1 enzymes. Most notably, this surface in zApe is more negatively charged, consistent with its calculated acidic pI. The molecular surface is also distinct in part due to the structure of the N-terminal portion of the protein.
4.0 Discussion
The results presented here impact our current understanding of the mechanism of Ape1’s redox activity. First, our evolutionary sequence analysis suggests that the redox function is found only in mammals and results from the acquisition of Cys 65. As shown in Table 2, the equivalent residue is either Ser or Thr in non-mammalian vertebrate Ape sequences. While Cys 138 is also conserved in most mammals and not in non-mammalian vertebrates, the platypus has a Ser residue in the equivalent position, and substitution of Cys 138 has no effect on redox activity in in vitro EMSA[18] or transactivation assays. The functional importance of Cys 65 has been demonstrated by loss of redox function resulting from substitution of this residue with Ala in transactivation assays, consistent with a previous report for EMSA redox analysis of Ala mutants [18]. A potential redox role for Cys 93 is suggested by transactivation assays and EMSA redox analysis in which C93A hApe1 has less redox activity than the wild-type enzyme albeit significantly more activity than the C65A hApe1. However, we note that Cys 93 is conserved in non-mammalian vertebrate sequences including zApe and that this enzyme is not redox active.
In assessing the results of the transactivation assays, it is important to consider the role of the base excision repair activity of hApe1 as well as its redox activity. As previously reported and in data not shown here, we have assayed all of the single Cys mutants and found that they retain wild-type repair activity [39]. Thus, it is unlikely that the reduction in redox activity observed for C65A and C93A hApe1 results from a reduction in repair activity. In addition, the results obtained for the in vitro and cell-based transactivation assays are entirely consistent suggesting that each is in fact monitoring the same activity of hApe1, namely the redox activity.
The gain of redox function observed for the T58C zApe provides compelling evidence for the role of Cys 65 in the redox activity of Ape1. Further, our results directly contradict the conclusion that Cys 65 is not critical for the redox activity of Ape1 as has recently been suggested in the C64A Ape1 mouse knock-in experiments [22]. Of particular relevance is the fact that the T58C zApe enzyme reduces human AP-1 and NFκB in our assays. Through comparative structural analysis, the crystal structures of the redox inactive C65A hApe1 and wild-type zApe were shown overall to be quite similar to the redox active wild-type hApe1 structure. Thus, it is unlikely that substitution of Ala for Cys 65 in the hApe1 results in a conformational change accounting for its loss of redox activity. One possible conclusion from our structural analysis is that the previously reported structures of the wild-type hApe1 enzyme do not represent this molecule in a redox active state. This would account for the overall structural similarity between the redox inactive and redox active enzymes, which extends to the repair active sites in which metal ions are bound in all of the structures. The analysis of the N-terminal regions of the zApe and C65A hApe1 supports previous results on deletion analysis of hApe1 in which the enzyme lacking 36 amino acid residues from the N-terminus retained redox activity. Our present analysis further suggests that while there is a requirement for N-terminal residues, sequence conservation of the N-terminal residues is not required for the redox function. This is supported by the fact that the N-terminal residues are poorly conserved between zApe and hApe1 (Fig. 2B).
We propose that the hApe1 enzyme undergoes a conformational change in order to reduce transcription factors. Although we cannot at this time detail that conformational change, we propose that it ultimately involves exposure of Cys 65. This putative mechanism is consistent with the fact that Ape1 reduces a large number of structurally unrelated transcription factors including bZip family members AP-1 and HLF, immunoglobulin-like factors NFkB and p53, the homeodomain-like factor PAX, and the bHLH-PAS domain factor HIF-1α and presumes a thiol exchange type of mechanism, which is consistent with the requirement for Cys 65. As revealed in a comparison of the molecular surfaces of zApe and hApe1, there is little conservation both in the detailed surface features as well as the electrostatic potential surface that would be encountered by a transcription factor. Clearly the surfaces of the molecules prior to a conformational change are not conserved and therefore do not provide a conserved “binding site”. Therefore, we propose that in order to reduce the many structurally unrelated transcription factors, Ape changes conformation resulting in a binding site that will accommodate these different transcription factors and in which Cys 65 is now accessible. As noted above, within the immediate vicinity of Cys 65 or its equivalent, the chemical environment is highly conserved. While it is possible that DNA plays a role in the interactions of hApe1 and the transcription factors that it reduces, hApe1 was previously reported to cross-link to the DNA-binding domain of c-Jun in the absence of DNA[14]. In conclusion, our results provide new insights regarding the evolution of the redox function and a structural basis for beginning to elucidate the mechanism by which Ape1 reduces transcription factors.
Acknowledgements
We thank Steve Ginell, Marianne Cuff and Andrzej Joachimiak from the Structural Biology Center Collaborative Access Team at the Advanced Photon Source (APS), Jay Nix from the Molecular Biology Consortium at the Advanced Light Source (ALS), members of the Georgiadis and Kelley laboratories, and Tom Hurley for helpful discussions. Data were collected at beamline 19-ID in the facilities of the SBC-CAT at APS and at beamline 4.2.2 at the ALS. Results shown in this report are derived from work performed at Argonne National Laboratory, SBC at the APS. Argonne is operated by U Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Financial support for this work was provided by the National Institutes of Health (CA114571 to M.M.G., CA94025 and CA106298 to M.R.K.), the IU Cancer Center (internal seed funding to M.M.G. and M.R.K.), and the Riley Children’s Foundation (M.R.K).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: The authors declare that there are no conflicts of interest.
References
- 1.Lando D, Pongratz I, Poellinger L, Whitelaw ML. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J. Biol. Chem. 2000;275:4618–4627. doi: 10.1074/jbc.275.7.4618. [DOI] [PubMed] [Google Scholar]
- 2.Xanthoudakis S, Curran T. Identification and characerization of Ref-1, a nuclear protein that facilitates AP-1 DNA binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao KS, Xanthoudakis S, Curran T, O'Dwyer PJ. Activation of AP-1 and of a nuclear redox factor, Ref-1, in the response of HT29 colon cancer cells to hypoxia. Mol Cell Biol. 1994;14:5997–6003. doi: 10.1128/mcb.14.9.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 6.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 8.Hanson S, Kim E, Deppert W. Redox factor 1 (Ref-1) enhances specific DNA binding of p53 by promoting p53 tetramerization. Oncogene. 2005;24:1641–1647. doi: 10.1038/sj.onc.1208351. [DOI] [PubMed] [Google Scholar]
- 9.Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akamatsu Y, Ohno T, Hirota K, Kagoshima H, Yodoi J, Shigesada K. Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J. Biol. Chem. 1997;272:14497–14500. doi: 10.1074/jbc.272.23.14497. [DOI] [PubMed] [Google Scholar]
- 11.Huang RP, Adamson ED. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- 12.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 13.Abate C, Patel L, Rauscher FJI, Curran T. Redox regulation of Fos and Jun DNA-binding activity in vitro. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 14.Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc. Natl. Acad. Sci, USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abate C, Luk D, Curran T. A ubiquitous nuclear protein stimulates the DNA-binding activity of fos and jun indirectly. Cell Growth Differ. 1990;1:455–462. [PubMed] [Google Scholar]
- 16.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci, USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Shupenko CC, Melo LF, Strauss PR. DNA repair protein involved in heart and blood development. Mol Cell Biol. 2006;26:9083–9093. doi: 10.1128/MCB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell. Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorman MA, Morera s, Rothwell DG, La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structure and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature. 2000;403 doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 21.Beernink PT, Segelke BW, hadi MZ, Erzberger JP, Wilson DMI, Rupp B. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implicatin for the catalytic mechanism. J. Mol. Biol. 2001;307:1023–1034. doi: 10.1006/jmbi.2001.4529. [DOI] [PubMed] [Google Scholar]
- 22.Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol. Cell. Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felsenstein J. PHYLIP (Phylogeny Inference Package) 2005. version 3.6. [Google Scholar]
- 26.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gius D, Cao XM, Rauscher FJ, 3rd, Cohen DR, Curran T, Sukhatme VP. Transcriptional activation and repression by Fos are independent functions: the C terminus represses immediate-early gene expression via CArG elements. Mol Cell Biol. 1990;10:4243–4255. doi: 10.1128/mcb.10.8.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Navaza J. AMoRe: an Automated Package for Molecular Replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Brunger AT, Adams PA, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–290. [Google Scholar]
- 33.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 34.Vagin AA, Isupov MN. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr. 2001;57:1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 35.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 36.Merritt EA, Bacon DJ. Raster3d: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 37.Nicholls A, Sharp K, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct. Func. Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 38.Oezguen N, Schein CH, Peddi SR, Power TD, Izumi T, Braun W. A "moving metal mechanism" for substrate cleavage by the DNA repair endonuclease APE-1. Proteins. 2007;68:313–323. doi: 10.1002/prot.21397. [DOI] [PubMed] [Google Scholar]
- 39.Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid. Redox Signal. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]