Summary
The ubiquitin-proteasome system (UPS) includes 3 enzymes that conjugate ubiquitin to intracellular proteins that are then recognized and degraded in the proteasome. The process participates in the regulation of cell metabolism. In the kidney, the UPS regulates the turnover of transporters and signaling proteins and its activity is down regulated in acidosis-induced proximal tubular cell hypertrophy. In chronic kidney disease (CKD), muscle wasting occurs because complications of CKD including acidosis, insulin resistance, inflammation, and increased angiotensin II levels stimulate the UPS to degrade muscle proteins. This response also includes caspase-3 and calpains which act to cleave muscle proteins to provide substrates for the UPS. For example, caspase-3 degrades actomyosin, leaving a 14kD fragment of actin in muscle. The 14 kD actin fragment is increased in muscle of patient with kidney disease, burn injury and surgery. In addition, acidosis, insulin resistance, inflammation and angiotensin II stimulate glucocorticoid production. Glucocorticoids are also required for the muscle wasting that occurs in CKD. Thus, the UPS is involved in regulating kidney function and participates in highly organized responses that degrade muscle protein in response to loss of kidney function.
Keywords: ubiquitin-proteasome system (UPS), muscle wasting, protein degradation, chronic kidney disease (CKD), 14kD actin fragment, caspase-3
All intracellular proteins and many extracellular proteins are continually “turning over”, being degraded to their constituent amino acids and replaced by synthesis of new proteins. Specific nuclear or cytosolic proteins, including proteins in the endoplasmic reticulum and mitochondria, are degraded at widely different rates; these rates vary from minutes for some regulatory enzymes, to days or weeks for proteins like actin and myosin in skeletal muscle, to months for hemoglobin in the red cell. Complex regulatory mechanisms ensure that proteolytic and synthetic processes are highly selective and precisely balanced because even a small decrease in synthesis or a small acceleration of degradation, if sustained, will result in marked loss of protein stores [36].
There are different proteolytic processes but in all tissues, the majority of intracellular proteins are degraded by the ubiquitin-proteasome system (UPS) [41]. However, extracellular proteins and some cell surface proteins are engulfed by endocytosis and degraded within lysosomes containing several acid-optimal proteases (e.g., cathepsins). Degradation of some cytosolic proteins in muscle are degraded in in lysosomes [61]. In mammalian cells, there also are calcium-activated, ATP-independent, cysteine proteases (calpains) and caspases. The latter cytosolic proteases cleave proteins at aspartic acid residues and are critical for the apoptotic process [43].
The ubiquitin-proteasome system
Initial steps in protein degradation by the UPS involve a series of 3 enzymes that link the cofactor, ubiquitin (Ub), onto proteins [26,36]. The enzymatic components that link chains of Ub onto proteins include the E1 (Ub-activating enzyme) and E2 proteins (Ub-carrier or conjugating proteins), which prepare Ub for conjugation. The third enzyme, E3 Ub-protein ligase, is a key factor in terms of the specificity of proteolysis because specific E3 enzymes will recognize a specific protein substrate. E3 enzymes catalyze the transfer of activated Ub to the substrate until a chain of 4-5 Ub's are attached. The conjugation reactions form Ub-conjugated proteins which can be recognized by the 26S proteasome. The proteasome removes Ub and degrades the substrate protein into small peptides [2]. In some cases, only a single ubiquitin is conjugated to a substrate protein (i.e. monoubiqutination). These substrate proteins are not degraded; instead, they are sorted into endocytic compartments and the Golgi network for routing to the plasma membrane or lysosomes [45]. The discovery of Ub and the biochemistry of its conjugation to substrate proteins led to the 2004 Nobel Prize in Chemistry awarded to Avram Hershko, Aaron Ciechanover and Irwin Rose (http://nobelprize.org/chemistry/laureates/2004/).
The initial step in the conjugation of Ub onto proteins is the activation of Ub at its carboxy-terminus by the E1, Ub-activating enzyme. A single E1 enzyme uses ATP to generate a Ub thiolester which is subsequently transferred to a sulfhydryl group of one of 30-40 E2 carrier proteins [20]. The major determinant of the specificity of the Ub conjugating process, however, is the presence of more than a thousand E3's which can recognize specific proteins and transfer Ub to it. For example, in muscle wasting conditions, two E3 Ub-conjugating enzymes, Atrogin-1 (also known as MAFbx) and MuRF-1, are critical for the breakdown of muscle proteins [3]. In cultured muscle cells, the content of atrogin-1 mRNA correlates closely with rates of protein breakdown [42,44,49]. In models of muscle wasting conditions, their expression increases dramatically (8-20 fold) and this increased expression of Atrogin-1 and MuRF-1 occurs just when muscle atrophy is most rapid. Thus, the muscle content of these E3 mRNAs might prove useful as biomarkers of excessive proteolysis in muscle. The signals that activate these E3 Ub-conjugating enzymes have been extensively studied and at least two transcription factors regulating E3 enzyme expression have been identified. Forkhead transcription factors (FoxO) and the inflammatory transcription factor, NFκB, act on the promoters for Atrogin-1 and MuRF-1 respectively to stimulate their expression. Moreover, it has been shown that activation of FoxO or NFκB causes accelerated muscle wasting presumably via these E3s in conjunction with other atrogenes [6,28,44].
In the kidney and other organs, at least five major functions of the UPS can be identified: 1) he UPS permits cells to adapt to changes in physiological functions by rapidly removing proteins to terminate an enzymatic or regulatory process. 2) The UPS can change gene expression by degrading transcription factors or cofactors/inhibitors that regulate transcription. An example of this process is the phosphorylation that initiates IκB degradation, releasing the transcriptional activator NFκB. This factor acts to accelerate inflammatory responses. 3) The UPS eliminates abnormally folded or damaged proteins. Patients with cystic fibrosis have a mutant transmembrane conductance regulator protein (CFTR) which is selectively degraded and hence, does not reach the cell surface. The result is inadequate removal of secreted proteins, congesting a patient's airways [19]. 4) The UPS functions to present antigen on the major histocompatibility complex class I molecules [41]. 5) Finally, the UPS degrades cellular proteins (including muscle proteins) when calories are inadequate or in response to catabolic illnesses. These reactions provide amino acids that are used for gluconeogenesis and the synthesis of new proteins.
Protein metabolism and the UPS in the kidney
In the kidney, the UPS plays a critical role in the turnover of proteins that affect cellular function, including the regulation of ion channels. Malik et al., found that the UPS played a key role in regulating the turnover of the amiloride-sensitive epithelial sodium channel (ENac) in renal cells [29,30]. The physiologic relevance of changing sodium channel activity was demonstrated by inhibiting the UPS which led to greater activity of ENac, a condition reminiscent of Liddle's syndrome, characterized by hypertension. Evidence that the UPS is involved in functions of the kidney by regulating transcriptional responses was found in studies of hypoxia-Inducible Factor 1 or HIF-1 [17]. It was determined that blocking the UPS changes expression of HIF-1 and hence affects the kidney response to low oxygen supply. Another example of how the UPS affects the regulation of kidney cell function was found in studies of the responses to TGF-β. It was shown that Smurf-2 (Smad ubiquitination regulartory factor-2), an E3 ubiquitin ligase was upregulated in the kidney tubule cells of patients with obstructive kidney disease [50]. Smurf-2 can suppress Smad transcriptional corepressors, SnoN and Ski thereby altering TGF-β signaling transmission to promote fibrosis in the kidney.
The UPS is also critically involved in regulating the bulk of proteins in kidney cells in conditions associated with an increase in kidney cell mass. In response to accumulation of excess acid, proximal tubule epithelial cells hypertrophy and increase glutamine/glutamate metabolism to promote ammonia formation and the excretion of acid. The hypertrophic response involves a decrease in protein degradation along with a minor increase in protein synthesis [21]. Franch et al., examined mechanisms underlying the increase in proximal tubule cell mass by studying the response to EGF as a signal for cell hypertrophy [13]. EGF stimulated two hypertrophic responses, an increase in protein synthesis and suppression of protein breakdown. The mechanism involved both the UPS and lysosomes. The mechanisms underlying the hypertrophy of kidney cells in response to diabetes or unilateral nephrectomy has not been established.
Maintenance of protein stores in chronic kidney disease
Based on epidemiologic studies, chronic kidney disease afflicts millions of adults in the U.S. and the world and sharply increases their risk of cardiovascular disease [9]. In addition, CKD is associated with muscle wasting and loss of protein stores, conditions associated with excessive morbidity [25]. The loss of protein stores in CKD has been attributed to “malnutrition” (i.e., abnormalities caused by an insufficient or imbalanced diet). This is incorrect because changing the diet generally does not correct these abnormalities because they are due to complex metabolic adaptation [34]. With progressive loss of kidney function, the overall rates of muscle protein synthesis may decrease, but the more prominent response is an increase in rates of protein degradation [1,14-16,33,40]. Because protein turnover in humans is very high (3.5-4.5 g protein/kg/day) [36], even a small, persistent increase in proteolysis will cause marked protein depletion. Programmed activation of the UPS accounts for most of the accelerated muscle protein degradation CKD.
Mechanisms causing loss of muscle protein in CKD
Recent studies in rodent models of CKD have established that the accelerated muscle wasting induced by uremia involves cellular mechanisms that are similar to the mechanisms causing muscle wasting in other catabolic conditions, such as cancer cachexia, starvation, insulin deficiency/resistance, or sepsis [26,36]. The mechanism of muscle wasting in these catabolic states involves accelerated proteolysis via the UPS plus higher levels of mRNAs encoding certain components of the UPS. There are also changes (both increases or decreases) in the expression of about 100 atrophy-related genes called atrogenes [27]. The increase in muscle mRNA levels of atrophy-related genes in muscle wasting states occurs because of increased gene transcription [1,27,35,38]. In fact, there is a common transcriptional program that includes decreased expression of various growth-related genes in atrophying muscle leading to the conclusion that multiple transcriptional factors change coordinately, resulting in loss of muscle mass [27]. The strongest evidence for activation of the UPS in muscles of animals undergoing atrophy due to uremia (or other catabolic diseases) is that inhibitors of the proteasome provided in vitro will block the increase in protein degradation present in muscles isolated from rodents with various models of catabolic human diseases [1,38,51]. In humans as well, catabolic conditions stimulate activation of the UPS in muscle (i.e., an increase in mRNAs encoding Ub and proteasome subunits) [31,37,53,58].
Loss of kidney function is associated with many abnormalities, some of which have been identified as signals stimulating protein degradation in muscle. For example, the accumulation of acid from the reduced capacity to excrete acid stimulates protein degradation in muscle [33]. Secondly, insulin resistance can be present in CKD patients with only moderate renal insufficiency and it stimulates protein degradation in muscle [24,28,35,38,56].
Another common problem for CKD patients is hypertension and cardiovascular disease linked to an increase in angiotensin II levels. This response, like those related to acidosis or insulin resistance, causes muscle atrophy in rodents [5,47]. Finally, CKD patients frequently have high levels of inflammatory cytokines, raising the possibility that muscle protein degradation is accelerated because of inflammation [6,23].
Caspase-3 and the initial cleavage of myofibrillar proteins in CKD
Muscle atrophy in catabolic conditions specifically affects contractile proteins [8]. Since myofibrillar proteins comprise about 2/3 of the protein in muscle, loss of these proteins is largely responsible for the disability of patients who experience muscle wasting. Notably, degradation of myofibrillar proteins requires more than one proteolytic step because the UPS can readily degrade major components of the myofibril (actin, myosin, troponin or tropomyosin) but when these same proteins are present in complexes or in intact myofibrils, they are degraded very slowly by the UPS [46]. Therefore, other proteases must initially cleave proteins to break down the complex structure of muscle to produce substrates for the UPS. In exploring proteases that could initiate cleavage of myofibrillar proteins, we examined caspases because several catabolic states are characterized by high circulating levels of TNFα and insulin resistance, conditions that activate the caspase cascade [11,18,23]. We found that activated caspase-3 cleaves actomyosin in vitro and when stimulated in cultured muscle cells, there is cleavage of myofibrillar proteins that are rapidly degraded by the UPS.
Notably, caspase-3 activation produces a “footprint” of its activity, a 14kD C-terminal fragment of actin that is found in the insoluble fraction of muscle [12]. Localization of the fragment to the insoluble fraction presumably occurs because it is less susceptible to degradation by the UPS. We find accumulation of the 14 kD actin fragment in muscles of animals with accelerated protein degradation due to acidosis, diabetes, and angiotensin II-induced hypertension, all complications of CKD [12,28,56,56]. In patients with CKD or other causes of muscle wasting as well, we find that the level of the 14 kD actin fragment is increased in muscle. In addition, the level of the 14 kD actin fragment in muscle of CKD patients decreases in response to an exercise program directed at increasing the patient's endurance. In addition, we found that the level of the fragment was highly correlated (r = 0.78) with the measured rate of protein degradation in muscles of patients with muscle wasting from osteoarthritis. Finally, the 14 kD actin fragment in unburned muscle of patients who had suffered a major burn injury was sharply increased [60]. These results indicate that at least in these conditions, the level of the 14 kD fragment in muscle is closely related to the rate of protein degradation and is present in specific disorders characterized by muscle wasting [1,7]. Additional testing will be needed to determine if this method could serve as a biomarker of accelerated muscle protein degradation in other conditions causing muscle wasting.
Another family of proteases, the calpains, have also been suggested as the protease that initially cleaves myofibrillar proteins in conditions associated with muscle wasting. Calpains are calcium-dependent, cysteine proteases that could play a role in disorders that include muscular dystrophy or sepsis-induced muscle wasting [54,57]. At least in rodents with uremia or certain other types of atrophy the inhibition of calcium-activated proteases in muscle does not block the increase in protein degradation, the degradation of myofibrillar proteins, or the accumulation of the 14kD actin fragment in muscle cells [1,12]. Additional studies are needed to (a) determine whether different catabolic disorders stimulate caspase-3 and/or calpains and (b) explain the specific roles of these proteases in the breakdown of different muscle proteins.
Signals triggering muscle wasting in CKD or other catabolic states
CKD is associated with several complications that can trigger the UPS to degrade muscle protein. These include metabolic acidosis, decreased insulin action and angiotensin II levels, and/or inflammation [1,38,47,48]. There is evidence that these complications can function in concert to cause muscle wasting. For example, metabolic acidosis stimulates protein breakdown by the UPS in muscle by integrating abnormalities, including an increase in glucocorticoid production and the development of insulin resistance in muscle [28,32]. Notably, however, acidification alone does not stimulate muscle protein breakdown unless a physiological amount of glucocorticoids is also present [32,39]. Likewise, glucocorticoids are required to for the accelerated protein degradation documented to occur in models of diabetes, high levels of angiotensin II, and sepsis [35,47,52].
Inflammatory conditions are frequently present in CKD patients but it is not clear how they affect muscle protein degradation [22]. One possibility is that Inflammation suppresses insulin signaling and increases glucocorticoid production. These responses could explain why there is a requirement for glucocorticoids in the accelerated muscle protein degradation that occurs in response to sepsis [52].
Thirdly, decreased insulin/IGF-1 signaling could be the stimulus for accelerated muscle protein breakdown. One consequence of decreased insulin signaling would be a decrease in protein synthesis. The mechanism would involve decreased signaling through the phosphatidylinositol 3-kinase/Akt (PI3K/Akt) pathway [4,28]. Specifically, when insulin or IGF-1 signaling is low, PI3K activity falls, reducing the production of phosphadidylinositol-3,4,5 phosphate, the active product of PI3K. This results in a decrease in the phosphorylation and activity of the serine/threonine kinase, Akt. With a decrease in Akt activation, there would be decreased phosphorylation of downstream kinases, GSK1 and mTOR/S6kinase, suppressing protein synthesis. Similarly, decreased PI3K/Akt signaling is the key step in stimulating protein degradation in muscle. Decreased PI3K/Akt signaling will induce the expression of the E3's Ub conjugating enzymes, Atrogin-1 and MuRF-1, enhancing muscle protein degradation [28,44,49]. Expression of these E3 enzymes occurs because there is decreased phosphorylation of the forkhead family of transcription factors (FoxO1, 3, 4). When FoxO1, 3 and 4 are not phosphorylated, they can migrate into the nucleus to stimulate transcription of Atrogin-1 [28,44,49]. Insulin or IGF-1 can block this process by stimulating activity of the PI3K/Akt pathway to suppress the expression of Atrogin-1.
Besides stimulating the expression of specific E3 Ub ligases, the insulin/IGF-1-PI3K/Akt pathway regulates the activation of caspase-3 to initiate the breakdown of muscle proteins. For example, in insulin-deficient rats with accelerated muscle protein degradation, we found activation of the pro-apoptotic factor, Bax. This leads to the release of cytochrome C from mitochondria and activation of caspase-3 [28]. The result is increased production of the 14 kD actin fragment. This pathway was confirmed in cultured muscle cells with inhibited PI3K activity using genetic or pharmacological techniques. Together, these results provide evidence that muscle wasting in response to the complications of CKD is due to a common signaling pathway that alters key enzymes modulating protein synthesis and degradation.
A key endocrine factor that participates in the regulation of muscle protein turnover is glucocorticoids. Pharmacologic doses of glucocorticoids have been used as a mechanism to activate the UPS to degrade muscle proteins and to study the activation of Atrogin-1 [10,42,55]. However, the response to pharmacologic doses of glucocorticoids differs from the physiologic responses. In vivo, catabolic conditions do not stimulate muscle wasting unless a physiologic level of glucocorticoids is present. Evidence for this difference in the responses is present in studies of adrenalectomized rodents: when they were starved or treated with NH4Cl to induce metabolic acidosis or made insulin-deficient with streptozotocin, muscle protein degradation did not increase unless the animals are also given a physiological dose of glucocorticoids [35,39,59]. Similarly, the increase in muscle wasting induced by angiotensin II or sepsis can be blocked by inhibiting the glucocorticoid receptor [47]. Thus, glucocorticoids exert a permissive role because the same dose of glucocorticoids does not stimulate muscle wasting in adrenalectomized animals. These results as well as the analysis of the complex transcriptional responses that are present in catabolic conditions indicate that muscle wasting results from activation of a regulated pathway that is of protein degradation that is integrated with changes in protein synthesis.
In this brief review of the UPS in kidney disease, we have emphasized that activity of the UPS in kidney cells is important for controlling levels of regulatory proteins. The UPS also participates in the turnover of the bulk of proteins in kidney epithelial cells. The responses of kidney and muscle cells to certain conditions such as acidosis, however, occur in opposite directions, and it is not clear how such opposing responses are regulated. Finally, specific complications of kidney disease have been shown to coordinate the activity of proteolytic systems (i.e., caspase-3 and the UPS) to degrade muscle proteins. These responses appear to apply to other catabolic conditions associated with muscle wasting and involve defects in insulin/IGF-1 signaling pathways and decreased PI3K/Akt signaling. Understanding these regulatory mechanisms could blunt the muscle wasting that occurs in different catabolic conditions.
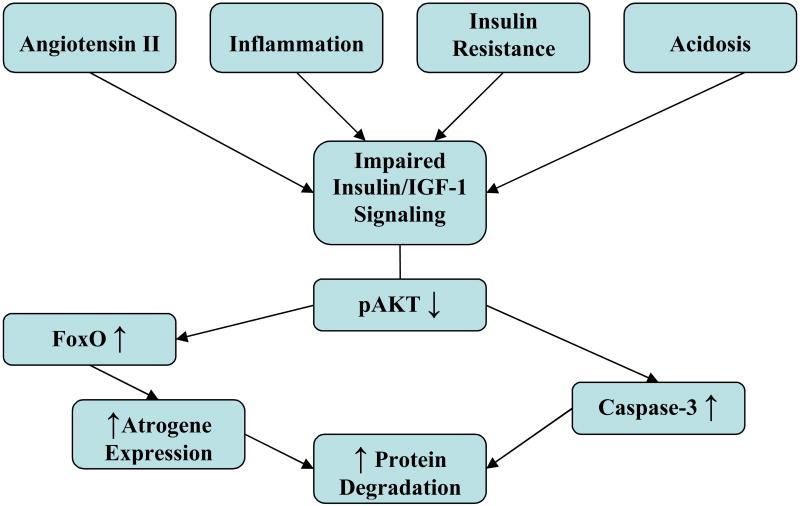
Fig. 1.
Complications of CKD including increased levels of angiotensin II, the presence of inflammation, insulin resistance and/or acidosis, impair insulin/IGF-1 signaling. The impairment decreases Akt phosphorylation, resulting in an increase in caspase-3 activity which disrupts the complex structure of muscle proteins and creates substrates for the UPS. The UPS activity is stimulated by Forkhead transcription factor (FoxO) which promotes expression of atrogenes including components of the UPS. The result is stimulation of muscle protein degradation.
Acknowledgments
Supported by NIH R01 grants DK37175 and P50 DK64233.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bailey JL, Wang X, England BK, Price SR, Ding X, Mitch WE. The acidosis of chronic renal failure activates muscle proteolysis in rats by augmenting transcription of genes encoding proteins of the ATP-dependent, ubiquitin-proteasome pathway. J Clin Invest. 1996;97:1447. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-comparmentalizing protease. Cell. 1998;92:367. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 3.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clark BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuel DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Sci. 2001;294:1704. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 4.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature Cell Biology. 2001;3:1014. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 5.Brink M, Price SR, Chrast J, Bailey JL, Anwar A, Mitch WE, Delafontaine P. Angiotensin II induces skeletal muscle wasting through enhanced protein degradation and down regulation of autocrine insulin-like growth factor-1. Endocrin. 2001;142:1489. doi: 10.1210/endo.142.4.8082. [DOI] [PubMed] [Google Scholar]
- 6.Cai D, Frantz JD, Tawa NE, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 7.Clark AS, Kelly RA, Mitch WE. Systemic response to thermal injury in rats: Increased protein degradation and altered glucose utilization in muscle. J Clin Invest. 1984;74:888. doi: 10.1172/JCI111506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6:376. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Coresh J, Astor BC, Greene T, Eknoyan G, Levey AS. Prevalence of chronic kidney disease and decreased kidney function in the adult US population: Third National Health and Nutrition Examination Survey. Am J Kid Dis. 2003;41:1. doi: 10.1053/ajkd.2003.50007. [DOI] [PubMed] [Google Scholar]
- 10.Dardevet DD, Sornet C, Taillandier D, Savary I, Attaix D, Grizard J. Sensitivity and protein turnover response to glucocorticoids are different in skeletal muscle from adult and old rats: Lack of regulation of the ubiquitin-proteasome proteolytic pathway in aging. J Clin Invest. 1995;96:2113. doi: 10.1172/JCI118264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeFronzo RA, Beckles AD. Glucose intolerance following chronic metabolic acidosis in man. Am J Physiol. 1979;236:E328–E334. doi: 10.1152/ajpendo.1979.236.4.E328. [DOI] [PubMed] [Google Scholar]
- 12.Du J, Wang X, Meireles CL, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE. Activation of caspase 3 is an initial step triggering muscle proteolysis in catabolic conditions. J Clin Invest. 2004;113:115. doi: 10.1172/JCI200418330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franch HA, Curtis PV, Mitch WE. Mechanisms of renal tubular cell hypertrophy: Mitogen-induced suppression of proteolysis. Am J Physiol. 1997;273:C843–C851. doi: 10.1152/ajpcell.1997.273.3.C843. [DOI] [PubMed] [Google Scholar]
- 14.Goodship THJ, Mitch WE, Hoerr RA, Wagner DA, Steinman TI, Young VR. Adaptation to low-protein diets in renal failure: Leucine turnover and nitrogen balance. J Am Soc Nephrol. 1990;1:66. doi: 10.1681/ASN.V1166. [DOI] [PubMed] [Google Scholar]
- 15.Graham KA, Reaich D, Channon SM, Downie S, Gilmour E, Passlick-Deetjen J, Goodship THJ. Correction of acidosis in CAPD decreases whole body protein degradation. Kidney Int. 1996;49:1396. doi: 10.1038/ki.1996.196. [DOI] [PubMed] [Google Scholar]
- 16.Graham KA, Reaich D, Channon SM, Downie S, Goodship THJ. Correction of acidosis in hemodialysis decreases whole-body protein degradation. J Am Soc Nephrol. 1997;8:632. doi: 10.1681/ASN.V84632. [DOI] [PubMed] [Google Scholar]
- 17.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98:1583. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spielgelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-a- and obesity-induced insulin resistance. Sci. 1996;271:665. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 19.Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome contribute CFTR processing. Cell. 1995;83:129. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 20.Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 21.Jurkovitz C, England BK, Ebb RG, Mitch WE. Influence of ammonia and pH on protein and amino acid metabolism in LLC-PK1 cells. Kidney Int. 1992;42:595. doi: 10.1038/ki.1992.323. [DOI] [PubMed] [Google Scholar]
- 22.Kaysen GA, Dubin JA, Muller HG, Rosales L, Levin NW, Mitch WE. Inflammation and reduced albumin synthesis associated with stable decline in serum albumin in hemodialysis patients. Kidney Int. 2004;65:1408. doi: 10.1111/j.1523-1755.2004.00520.x. [DOI] [PubMed] [Google Scholar]
- 23.Kimmel PL, Phillips TM, Simmens SJ, Peterson RA, Weihs KL, Alleyne S, Cruz I, Yanovski JA, Veis JH. Immunologic function and survival in hemodialysis patients. Kidney Int. 1998;54:236. doi: 10.1046/j.1523-1755.1998.00981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi S, Maesato K, Moriya H, Ohtake T, Ikeda T. Insulin resistance in patients with chronic kidney disease. Am J Kid Dis. 2005;45:275. doi: 10.1053/j.ajkd.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 25.Kopple JD. National Kidney Foundation K/DOQI clinical practice guidelines for nutrition in chronic renal failure. Am J Kid Dis. 2001;37:S66–S70. doi: 10.1053/ajkd.2001.20748. [DOI] [PubMed] [Google Scholar]
- 26.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 27.Lecker SH, Jagoe RT, Gomes M, Baracos V, Bailey JL, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 28.Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol. 2004;15:1537. doi: 10.1097/01.asn.0000127211.86206.e1. [DOI] [PubMed] [Google Scholar]
- 29.Malik B, Schlanger L, Al-Khalili O, Bao HF, Yue G, Price SR, Mitch WE, Eaton DC. ENaC degradation in A6 cells by the ubiquitin-proteasome proteolytic pathway. J Biol Chem. 2001;276:12903. doi: 10.1074/jbc.M010626200. [DOI] [PubMed] [Google Scholar]
- 30.Malik B, Yue Q, Yue G, Chen XJ, Price SR, Mitch WE, Eaton DC. Role of Nedd4-2 and polyubiquitination in epithelial sodium channel degradation in untransfected renal A6 cells expressing endogenous ENaC subunits. Am J Physiol Renal Physiol. 2005;289:F107–F116. doi: 10.1152/ajprenal.00179.2002. [DOI] [PubMed] [Google Scholar]
- 31.Mansoor O, Beaufrere Y, Boirie Y, Ralliere C, Taillandier D, Aurousseau E, Schoeffler P, Arnal M, Attaix D. Increased mRNA levels for components of the lysosomal, Ca++-activated and ATP-ubiquitin-dependent proteolytic pathways in skeletal muscle from head trauma patients. Proc Natl Acad Sci USA. 1996;93:2714. doi: 10.1073/pnas.93.7.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.May RC, Kelly RA, Mitch WE. Metabolic acidosis stimulates protein degradation in rat muscle by a glucocorticoid-dependent mechanism. J Clin Invest. 1986;77:614. doi: 10.1172/JCI112344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May RC, Kelly RA, Mitch WE. Mechanisms for defects in muscle protein metabolism in rats with chronic uremia: The influence of metabolic acidosis. J Clin Invest. 1987;79:1099. doi: 10.1172/JCI112924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitch WE. Malnutrition: a frequent misdiagnosis for hemodialysis patients. J Clin Invest. 2002;110:437. doi: 10.1172/JCI16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitch WE, Bailey JL, Wang X, Jurkovitz C, Newby D, Price SR. Evaluation of signals activating ubiquitin-proteasome proteolysis in a model of muscle wasting. Amer J Physiol. 1999;276:C1132–C1138. doi: 10.1152/ajpcell.1999.276.5.C1132. [DOI] [PubMed] [Google Scholar]
- 36.Mitch WE, Goldberg AL. Mechanisms of muscle wasting: The role of the ubiquitin-proteasome system. N Engl J Med. 1996;335:1897. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 37.Pickering WP, Price SR, Bircher G, Marinovic AC, Mitch WE, Walls J. Nutrition in CAPD: Serum bicarbonate and the ubiquitin-proteasome system in muscle. Kidney Int. 2002;61:1286. doi: 10.1046/j.1523-1755.2002.00276.x. [DOI] [PubMed] [Google Scholar]
- 38.Price SR, Bailey JL, Wang X, Jurkovitz C, England BK, Ding X, Phillips LS, Mitch WE. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome pathway by a mechanism including gene transcription. J Clin Invest. 1996;98:1703. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price SR, England BK, Bailey JL, Van Vreede K, Mitch WE. Acidosis and glucocorticoids concomitantly increase ubiquitin and proteasome subunit mRNAs in rat muscle. Am J Physiol. 1994;267:C955–C960. doi: 10.1152/ajpcell.1994.267.4.C955. [DOI] [PubMed] [Google Scholar]
- 40.Reaich D, Channon SM, Scrimgeour CM, Daley SE, Wilkinson R, Goodship THJ. Correction of acidosis in humans with CRF decreases protein degradation and amino acid oxidation. Am J Physiol. 1993;265:E230–E235. doi: 10.1152/ajpendo.1993.265.2.E230. [DOI] [PubMed] [Google Scholar]
- 41.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class 1 molecules. Cell. 1994;78:761. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 42.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-1 stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol. 2004;287:E591–E601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 43.Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 44.Sandri M, Sandri C, Gilbert A, Skuck C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003;278:35857. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 46.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271:26690. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 47.Song YH, Li Y, Du J, Mitch WE, Rosenthal N, Delafontaine P. Muscle-specific expression of insulin-like growth factor-1 blocks angiotensin II-induced skeletal muscle wasting. J Clin Invest. 2005;115:451. doi: 10.1172/JCI22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenvinkel P, Heimburger O, Paultre F, Diczfalusy U, Wang T, Berglund L, Jogestrand T. Strong association between malnutrition, inflammation and atherosclerosis in chronic kidney failure. Kidney Int. 1999;55:1899. doi: 10.1046/j.1523-1755.1999.00422.x. [DOI] [PubMed] [Google Scholar]
- 49.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Klinenber JR, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 50.Tan R, He W, Lin X, Kiss LP, Liu Y. Smad ubiquitination regulatory factor-2 in the fibrotic kidney: regulation, target specificity, and functional implication. Am J Physiol Renal Physiol. 2008;294:F1076–F1083. doi: 10.1152/ajprenal.00323.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tawa NE, Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J Clin Invest. 1997;100:197. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiao G, Fagan J, Roegner V, Lieberman M, Wang JJ, Fischer JE, Hasselgren PO. Energy-ubiquitin-dependent muscle proteolysis during sepsis in rats is regulated by glucocorticoids. J Clin Invest. 1996;97:339. doi: 10.1172/JCI118421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiao G, Hobler S, Wang JJ, Meyer TA, Luchette FA, Fischer JE, Hasselgren PO. Sepsis is associated with increased mRNAs of the ubiquitin-proteasome proteolytic pathway in human skeletal muscle. J Clin Invest. 1997;99:163. doi: 10.1172/JCI119143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tidball JG, Spencer MJ. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J Physiol. 2002;545:819. doi: 10.1113/jphysiol.2002.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran H, Brunet A, Griffith EC, Greenberg ME. The many forks in FOXO's road. Sci STKE. 2003;2003:RE5. doi: 10.1126/stke.2003.172.re5. [DOI] [PubMed] [Google Scholar]
- 56.Wang XH, Hu Z, Hu JP, Du J, Mitch WE. Insulin resistance accelerates muscle protein degradation: activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrin. 2006;147:4160. doi: 10.1210/en.2006-0251. [DOI] [PubMed] [Google Scholar]
- 57.Wei W, Fareed MU, Evenson A, Menconi MJ, Yang H, Petkova V, Hasselgren PO. Sepsis stimulates calpain activity in skeletal muscle by decreasing calpastatin activity but does not activate caspase-3. Am J Physiol Regul Integr Comp Physiol. 2005;288:R580–R590. doi: 10.1152/ajpregu.00341.2004. [DOI] [PubMed] [Google Scholar]
- 58.Williams AB, Sun X, Fischer JE, Hasselgren PO. The expression of genes in the ubiquitin-proteasome proteolytic pathway is increased in skeletal muscle from patients with cancer. Surgery. 1999;126:744. [PubMed] [Google Scholar]
- 59.Wing SS, Goldberg AL. Glucocorticoids activate the ATP-ubiquitin-dependent proteolytic system in skeletal muscle during fasting. Am J Physiol. 1993;264:E668–E676. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- 60.Workeneh B, Rondon-Berrios H, Zhang L, Hu Z, Ayehu G, Ferrando A, Kopple JD, Wang H, Storer TW, Fournier M, Lee SW, Du J, Mitch WE. Development of a diagnostic method for detecting increased muscle protein degradation in patients with catabolic conditions. J Am Soc Nephrol. 2006;17:3233. doi: 10.1681/ASN.2006020131. [DOI] [PubMed] [Google Scholar]
- 61.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]