Abstract
Perturbations of the chemical shifts of a small subset of residues in the catalytically active domain of Escherichia coli signal peptidase I (SPase I) upon binding signal peptide suggest the contact surface on the enzyme for the substrate. SPase I, an integral membrane protein, is vital to preprotein transport in prokaryotic and eukaryotic secretory systems; it binds and proteolyses the N-terminal signal peptide of the preprotein, permitting folding and localization of the mature protein. Employing isotopically labeled C-terminal E. coli SPase I Δ2–75 and an unlabeled soluble synthetic alkaline phosphatase signal peptide, SPase I Δ2–75 was titrated with the signal peptide and 2Δ 1H-15N hetero-nuclear single-quantum correlation nuclear magnetic resonance spectra revealed chemical shifts of specific enzyme residues sensitive to substrate binding. These residues were identified by 3D HNCACB, 3D CBCA(CO)NH, and 3D HN(CO) experiments. Residues Ile80, Glu82, Gln85, Ile86, Ser88, Gly89, Ser90, Met91, Leu95, Ile101, Gly109, Val132, Lys134, Asp142, Ile144, Lys145, and Thr234, alter conformation and are likely all in, or adjacent to, the substrate binding site. The remainder of the enzyme structure is unperturbed. Ramifications for conformational changes for substrate docking and catalysis are discussed.
Keywords: membrane protein, NMR, signal peptidase, signal peptide, substrate binding
Virtually all proteins destined for extracytoplasmic locations are synthesized as preproteins with a cleavable N-terminal extension sequence called the signal peptide. The signal peptide earmarks the preprotein for entry into the appropriate transport pathway and promotes the membrane translocation process via interactions with components of the transport machinery. In the penultimate step of transport, recognition by a signal peptidase is critical for proteolytic removal of the signal peptide and sets the stage for folding of the mature protein and its final localization.
The Escherichia coli signal peptidase I (SPase I) is an integral membrane protein of 37 kDa with two transmembrane segments (residues 4–28 and 58–76), a periplasmic N-terminus and C-terminal catalytic domain (residues 77–323) (1,2). The enzyme is thought to catalyze peptide bond hydrolysis via a catalytic dyad involving the hydroxyl of Ser90 for nucleophilic attack and the ε-amine of Lys145 as the general base to cleave the preprotein. Crystal structures of the soluble catalytic domain (SPase I Δ2–75) in the apo-form (3) and with bound inhibitors (4,5) revealed two shallow hydrophobic cavities adjacent to the Ser–Lys dyad that would be appropriate for docking residues −1 and −3 of the signal peptide.
Although members of the SPase I family of enzymes are associated with diverse transport pathways in prokaryotic and eukaryotic systems, the bacterial SPase I uniquely uses a Lys for catalysis; in most higher organisms a conserved His is used instead in a Ser–His dyad. The SPase I family is further set apart from most serine proteases that use a Ser–His–Asp catalytic triad mechanism. Moreover, the enzyme is essential for viability of the bacterium. These features make bacterial SPase I a potentially valuable target for antimicrobial agents and its exposure to the periplasm makes the enzyme accessible to drugs.
To pursue this possibility we must understand the mode of recognition of the signal peptide sequence of the preprotein by the enzyme including the topological alignment and the key contacts that lead to peptide proteolysis. To date, these interactions have been inferred from co-crystals of SPase I Δ2–75 with 5S, 6S β-lactam (penem) (4) or with the fatty acid derivative, Arylomycin A2 (5), both inhibitors of SPase I activity, and from molecular modeling using the β-lactamase signal peptide to predict its binding locale (5). However, no direct structural analysis of the enzyme with bound signal peptide or preprotein has been accomplished. This has proven a formidable problem, in part, because of the hydrophobic nature of the components involved. One of two β-sheet domains in SPase I Δ2–75 includes a large exposed hydrophobic surface thought to interact with the membrane surface in the cell. In addition, a key feature of functional signal peptides is the central 10–15 residue hydrophobic core region, making them highly susceptible to aggregation at the high concentration typically required for structural studies.
Here we report the first direct measurements on a molecular scale of SPase I-signal peptide interactions using a non-perturbing experimental approach, nuclear magnetic resonance (NMR) spectroscopy. We have expressed and purified doubly 13C, 15N- and singly 15N-isotopically labeled C-terminal E. coli SPase I Δ2–75 and an unlabeled soluble synthetic signal peptide corresponding to the wild-type alkaline phosphatase signal sequence plus four residues of the mature protein. 2D 1H-15N heteronuclear single-quantum correlation (HSQC) spectra were collected as SPase I Δ2–75 was titrated with signal peptide. The chemical shifts of a small subset of specific residues were sensitive to the binding of the signal peptide. The subset of perturbed residues includes: Ile80, Glu82, Gln85, Ile86, Ser88, Gly89, Ser90, Met91, Leu95, Ile101, Gly109, Val132, Lys134, Asp142, Ile144, Lys145, and Thr234. These residues mapped to a defined patch on the protein. This strategy provides the first direct experimental data that reveal which residues of the enzyme are impacted upon substrate binding.
Methods and Materials
Signal peptide synthesis and purification
The E. coli wild-type alkaline phosphatase signal peptide plus four residues from the mature region of the preprotein, three Lys at each termini, a cysteine and a C-terminal amide, KKKMKQSTIALA LLPLLFCPVTKARTPEKKK-NH2, was chemically synthesized and purified as described previously (6,7). Thr at position 19 was replaced with Cys to tether reporter groups for future studies. Cleavage of the peptide was confirmed by incubating the peptide with SPase I Δ2–75 at 37 °C in 1 M Tris, pH 8.0 and 1% Triton X-100 buffer. The sample was run on a 12% Tris Tricine gel and silver stained. Prior to the NMR experiments, 5 mg of lyophilized peptide was dissolved in 5 µL of dimethylsulfoxide. The peptide was then resuspended in 20 mm NaH2PO4, 10 mm EDTA, pH 6.3 to a stock concentration of 10 mm. This was either used immediately or frozen at −20 °C for short-term storage.
Construction, isotopic labeling, and purification of the catalytic domain of signal peptidase, SPase I Δ2–75
Plasmid pET23b coding for the full-length signal peptidase (generously provided by Ross Dalbey, The Ohio State University, Columbus, Ohio, USA) was used to generate a truncated form of the enzyme in which residues 2–75, corresponding to the transmembrane segments, are deleted. BL21(DE3) cells transfected with the plasmid coding for SPase I Δ2–75 were expressed in M9 Minimum Media labeled with 13C and 15N (11.1 mm 13C-labeled glucose, and 18.7 mm 15N-labeled ammonium chloride). The next day, the cells were resuspended in 10 mm EDTA, 20 mm Tris–HCl pH 7.4 (buffer A), lysed by two passages through a French Pressure cell at 1500 psi and purified as described by Paetzel et al. (8) with minor modifications. The inclusion bodies were pelleted from the cell lysate by centrifugation at 12,000 g for 5 min, and washed four times with buffer A plus 0.5% Triton X-100. The protein was solubilized in 6 m guanidine–HCl in 20 mm Tris–HCl pH 7.4 at room temperature for 1 h. Triton X-100 was eliminated by mixing the sample with BioBeads at 5 g/25 mL (Bio-Rad Laboratories, Hercules, CA, USA) for 2 h at room temperature. The sample was dialyzed against three 1L changes of buffer A. The isolated protein of >95% purity was concentrated with a 10K cutoff centricon, and buffer exchanged with 20 mm NaH2PO4 pH 6.3, 10 mm EDTA. D2O to 10% was added to the sample before the NMR analysis. Enzymatic activity was confirmed by cleavage of a GST-signal peptide fusion protein. Singly 15N-labeled SPase I Δ2–75 was also prepared using the same protocol except in the minimal media isotopically labeled 13C-labeled glucose was replaced with unlabeled glucose.
NMR experiments
All NMR experiments were performed on a Varian Inova spectrometer operating at a 1H resonance frequency of 600 MHz and a sample temperature of 25 °C. The 2D 1H-15N HSQC were acquired as 2048 and 128 complex points in the 1H and 15N dimensions, respectively, on a 0.30 mm uniformly 13C- and 15N-labeled sample of SPase I Δ2–75 in 90% H2O/10% D2O using 20 mm NaH2PO4, 10 mm EDTA, pH 6.3. To make the amide 1H and 15N as well as 13Cα, 13Cβ and 13CO sequence specific resonance assignments, a combination of sensitivity enhanced 3D CBCA(CO)NH, 3D HNCACB and 3D HN(CO) experiments were utilized on a 0.30 mm uniformly 13C- and 15N-labeled sample of SPase I Δ2–75 in 90% H2O/10% D2O using 20 mm NaH2PO4, 10 mm EDTA, pH 6.3. 3D CBCA(CO)NH were acquired with 1024 points in the 1H dimension, 64 points in the 15N dimension, and 16 points in the 13C dimension over 16 transients. 3D HNCACB data were acquired with 1024 complex points in the 1H dimension, 64 points in the 15N dimension, and 32 points in the 13C dimension, averaged over 8 transients. 3D HN(CO) data were acquired with 1024 points in the 1H dimension, 64 points in the 15N dimension, and 16 points in the 13C dimension over 16 transients.
Titrations with substrate were performed-using a 0.30 mm uniformly 15N-labeled sample of SPase I Δ2–75 in 90% H2O/10% D2O in 20 mm NaH2PO4, 10 mm EDTA, pH 6.3 buffer. Unlabeled signal peptide was added to the SPase I Δ2–75 sample to concentrations of 0.025, 0.050, 0.10, 0.3 and 1 mm. No changes in pH were detected over the course of the titration. After each addition of unlabeled signal peptide, a 1H–15N HSQC spectrum was collected using the same parameters as described above. All spectra were processed using nmrPipe/nmrDraw software (9) and analyzed using the program Sparky (http://www.cgl.ucsf.edu/home/sparky).
Modeling of a signal peptide bound to SPase I Δ2–75
A model of a wild-type alkaline phosphatase signal peptide cleavage region plus the C-terminal most core region Phe residue (Phe-Cys-Pro-Val-Thr-Lys-Ala) bound to the binding site of SPase I Δ2–75 was prepared using the atomic coordinates from the model of the SPase I Δ2–75 complex with the cleavage region of the beta-lactamase signal peptide deduced from molecular modeling and generously provided by Mark Paetzel (5). Backbone-dependent changes in the signal peptide sequence of the model were prepared using the PyMOL Viewer Software.
Results and Discussion
Peptide design
To examine the interaction between SPase I and signal peptide, we used a well-characterized synthetic signal sequence corresponding to the wild-type alkaline phosphatase signal peptide. The peptide includes the first four residues from the mature region of the preprotein and ends in a C-terminal amide to more closely approximate the presentation of substrate during preprotein translocation. A major obstacle in the biophysical analysis of the SPase I-signal peptide interaction centers on the low solubility of these components in aqueous solution. To surmount this problem, we strategized a peptide design that would retain solubility during complex formation. Toward this end, we used the hydrophobicity scale and TM finder developed by Deber and colleagues (10,11) for estimating the number of flanking Lys residues needed to enhance solubility of membrane interactive peptides (12,13). Using this approach, the aggregation was overcome with the addition of three Lys, each, to the N- and C-termini. Good solubility was retained with 300 µm SPase I Δ2–75 and peptide to millimolar levels. Deber has extensively characterized membrane interactive peptides with flanking lysine tags and found these do not alter secondary structure, extent of oligomerization, and membrane association (12,13). Thus, these solubility tags should not influence molecular recognition of soluble SPase I Δ2–75 and the alkaline phosphatase signal peptide. Indeed, we observe proteolysis of the peptide by SPase I Δ2–75 similar to that observed for the signal peptide without solubility tags (data not shown) and an affinity comparable with previous studies (14).
Identification of SPase I Δ2–75 residues that are sensitive to signal peptide binding
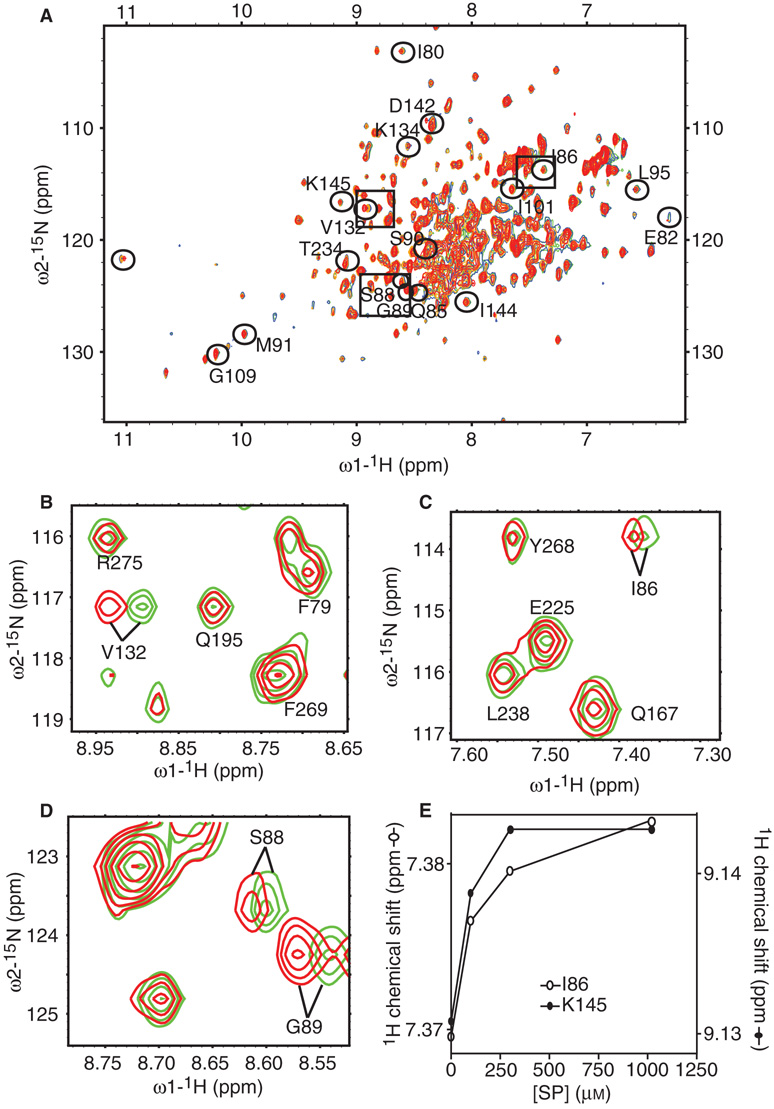
The amide 1H–15N HSQC resonances were monitored for a uniformly 15N-labeled 0.30 mm sample of SPase I Δ2–75 before and after the addition of unlabeled signal peptide to final concentrations of 0.025, 0.05, 0.10, 0.30, and 1 mm (Figure 1A). Chemical shift changes were observed for a limited number of residues after the addition of signal peptide, directly identifying residues of SPase I Δ2–75 impacted by signal peptide binding (15,16). An overlay of the 2D 1H-15N HSQC spectra of SPase I Δ2–75 in association with signal peptide at different concentrations over the spectrum of SPase I Δ2–75 identified which resonances shifted in the presence of the substrate. Examples of peaks corresponding to residues that shifted with signal peptide addition are shown in Figure 1B, C, and D, respectively. The extent of the chemical shift changes of some peaks were dose-dependent and saturable, indicating that the binding was specific (e.g. Figure 1E).
Figure 1.
Assignment of peaks corresponding to residues in the SPase I Δ2–75 amino acid sequence that change in backbone conformation upon signal peptide binding. (A) 2D 1H-15N HSQC spectra of SPase I Δ2–75 with increasing concentration of signal peptide ([SPase I Δ2–75] = 0.3 mm, no signal peptide (green), [signal peptide] = 0.100 mm (blue), [signal peptide] = 0.300 mm (yellow), [signal peptide] = 1 mm (red)). Peaks that shifted due to peptide binding are circled and marked with the corresponding residue identification where applicable. (B) Representative region of 2D 1H-15N HSQC spectra of SPase I Δ2–75, shows the superposition of the spectra of free SPase I Δ2–75 (green) and SPase I Δ2–75 in complex with 1 mm signal peptide (red) as in (A). Shown is a peak corresponding to residue Val132 that shifted with signal peptide addition. Other peaks that did not shift are also identified. (C) Representative region of 2D 1H-15N HSQC spectra of SPase I Δ2–75 as in (B). Shown is a peak corresponding to residue Ile86 that shifted with signal peptide addition. Other peaks that did not shift are also identified. (D) Representative region of 2D 1H-15N HSQC spectra of SPase I Δ2–75 as in (B). Shown are peaks corresponding to residues Ser88 and Gly89 that shifted with signal peptide addition. (E) Example titration curves showing saturation of chemical shift changes corresponding to residues Ile86 and Lys145 of SPase I Δ2–75 upon increasing signal peptide concentration.
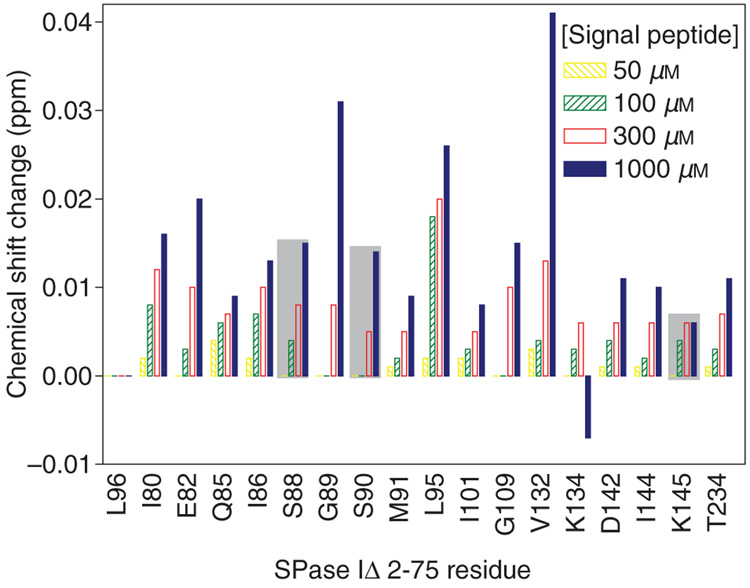
Using this strategy we found changes in chemical shifts of a small subset of amide proton CαH correlations, i.e. chemical shifts in some backbone resonances, corresponding to a likely change in backbone conformation of 18 residues of SPase I Δ2–75. Seventeen of these residues were identified: Ile80, Glu82, Gln85, Ile86, Ser88, Gly89, Ser90, Met91, Leu95, Ile101, Gly109, Val132, Lys134, Asp142, Ile144, Lys145, and Thr234. One peak in the HSQC whose chemical shift changed upon signal peptide binding could not be assigned because key resonances were missing in the HNCACB, the CBCA(CO)NH, and the HN(CO) data sets, perhaps due to rapid relaxation. Peaks that shifted with signal peptide binding are identified on the 2D 1H-15N HSQC spectrum (Figure 1A). As shown in the spectrum and in the graph summarizing the extent of the changes in chemical shifts of resonances (Figure 1A and Figure 2), peaks corresponding to residues Val132 and Gly89 change most dramatically. These chemical shift differences in amino acids between the presence and absence of signal peptide permit the elucidation of the enzyme conformational changes upon substrate binding.
Figure 2.
Residues identified that exhibit chemical shift changes as monitored in Figure 1A with increasing signal peptide concentration (50–1000 µm) as indicated in the legend. Major changes were observed only in the proton dimension as shown. The shift of a peak at each signal peptide concentration was compared to the peak measured without addition of the signal peptide. Changes of residues proposed to be critical to the proteolytic mechanism are shaded by a gray box. For comparison, Leu96 that did not shift is also shown.
Localization of assigned residues on the structure of SPase I Δ2–75
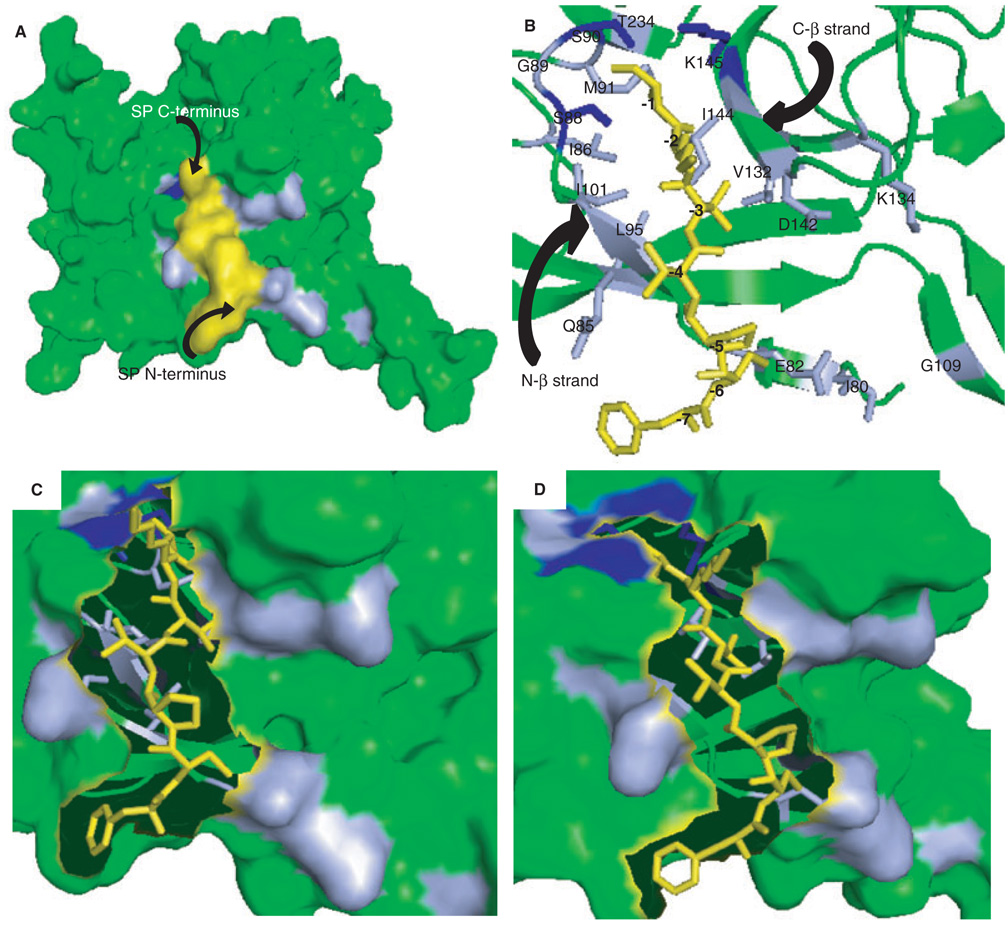
The residues with altered chemical shifts were mapped onto the crystal structure of the SPase I Δ2–75 (Figure 3) (5). Virtually all of the perturbed residues are located in one groove on the surface of the protein (Figure 3A). Since 93% of the residues of SPase I Δ2–75 showed no perturbation in the NMR spectra upon binding of substrate (only 18 out of 264 resonances were observed to have chemical shift changes that were signal peptide dependent), most of the protein did not change structure substantially upon substrate binding. This demonstrates that the signal peptide binding is specific and involves only local changes in SPase I Δ2–75 structure. These changes reflect flexibility in the enzyme active site that may be important for optimizing the binding surface for interacting with a wide range of signal peptides. Induced fit is a classic concept characterizing the conformational changes in binding partners that can accompany macromolecular interactions to enhance affinity and specificity. The observations of chemical shift changes arising from the backbone of SPase I Δ2–75 are consistent with induced fit of enzyme and signal peptide binding partners (although we cannot rule out the possibility that proximity to the peptide binding site might also lead to structural alteration of some enzyme residues).
Figure 3.
Models of the signal peptide-SPase I Δ2–75 complex. (A) Residues identified in 2D 1H-15N HSQC spectra that changed in backbone conformation upon signal peptide binding (Figure 1A) are marked on the surface representation of SPase I Δ2–75 in complex with a model of the wild-type alkaline phosphatase signal peptide. Amino acids in light blue showed shifts in resonances, those proposed to be critical to the proteolytic mechanism and showed shifts are depicted in dark blue. The signal peptide model is shown in yellow. (B) Magnified view of the alkaline phosphatase signal peptide-SPase I Δ2–75 complex, represented by a stick and ribbon diagram, respectively. Residues of SPase I Δ2–75 involved in signal peptide binding as found by NMR analysis are indicated and color coded as in (A). The signal peptide model is shown in yellow and residues are numbered relative to the cleavage site: (−1) Ala, (−2) Lys, (−3) Thr, (−4) Val, (−5) Pro, (−6) Cys, (−7) Phe. (C) Surface representation of the magnified view of SPase I Δ2–75 and signal peptide (represented as sticks), showing the residues on the N-terminal beta strand (residues 85–90) of the signal peptide binding cleft. (D) Surface representation of a magnified view of SPase I Δ2–75 and signal peptide represented as sticks, showing the residues on the C-terminal beta strand (residues 142–145) of the signal peptide binding cleft. All figures were prepared using PyMOL Viewer Software and the coordinates from the model of the wild-type SPase I Δ2–75 complex with the cleavage region of the beta-lactamase signal peptide (5) as described in Materials and Methods.
The NMR data show that residues Gln85 and Asp142 are among those that change conformation upon substrate binding. Examination of the structure of the apo-enzyme shows that if these two residues move appropriately, the substrate gains access to a substantial hydrophobic groove. Hydrophobic residues of the signal peptide cleavage region, Val(−4) and Pro(−5), and of the C-terminal end of the core region, Phe(−7) and Leu(−8), can interact with this portion of the enzyme and thus the movement of Gln85 and Asp142 will significantly enhance the binding energy of the substrate docked with enzyme.
Interestingly, residues Ser90 and Lys145, considered to constitute the catalytic dyad (17), were identified from these data to be among the set of residues whose conformation changes upon signal peptide binding (though through-space polarization of electron density by binding of the substrate cannot be ruled out). This indicates that these two residues in the apo-enzyme are not in the proper conformation for enzyme catalysis. Rather, upon substrate binding, the peptide backbone at Ser90 and Lys145 changes conformation, presumably allowing the orientation necessary for attack of the scissile bond and consistent with the role of Ser90 as the nucleophile and Lys145 as the base catalyst in the cleavage reaction (17–19). This is in good agreement with the hydrogen bonding contact observed for these residues and Arylomycin A2 in enzyme-inhibitor co-crystals (5). A third residue, Ser88, whose role in catalysis was suggested by mutational analysis (18), also shows perturbation in the NMR spectra. The hydroxyl of this residue was found within hydrogen bonding distance of O45 of Arylomycin A2 (5) but not a beta-lactam inhibitor (4) in structures deduced from co-crystals. While other explanations are possible, our data support the role of this residue in contributing a stabilizing hydrogen bond with the oxyanion intermediate during catalysis.
When residues with altered resonances in the HSQC are mapped upon the structure of the protein, they define a small volume on the protein and cluster closely around the sites previously suggested to represent the S1 and S3 pockets of the enzyme. In particular, Ile86, Ser88, Gly89, Ser90, Met91, Ile144, and Lys145 help define the S1 pocket and Ile86, Leu95, Val132, Asp142, and Ile144 help define the S3 pocket. Since Pro residues do not have NH-CαH correlations, Pro87 was not identified on the NMR spectra although it is proposed to contribute to S1 (5). The S1 pocket is narrower in the structure of the apo-enzyme than in the acylenzyme (3) consistent with our findings that conformational change in this region is required with signal peptide binding. Collectively, these data suggest the reorientation of the residues of S1 and S3 to achieve close complementation with the docked −1 and −3 signal peptide residues, consistent with an induced fit hypothesis.
Even with conformational alterations, however, this region is too small to accommodate large side chains. Not surprisingly, signal peptides share very little sequence identity except in the C-terminal cleavage region with small and neutral residues found at –1 and – 3 positions (Ala and Thr in the alkaline phosphatase signal peptide, respectively), relative to the cleavage site. The adjacent long and positively charged side chain of Lys(−2) on the alkaline phosphatase signal peptide must point away from the groove due to its size and the lack of a neighboring hydrophilic residue to complement it.
Using recent molecular models of the SPase I-signal peptide complex, we have analyzed the predicted interactions in the context of our experimental data. Based on the crystal structure of SPase I Δ2–75 with Arylomycin A2, a model of bound beta-lactamase signal peptide suggested residues Phe84, Gln85, Ile86, Pro87, Ser88, Gly89, Ser90, Met91, Leu95, Val132, Asp142, Tyr143, Ile144, and Lys145 form the signal peptide binding site (5). Of these, we found Gln85, Ile86, Ser88, Gly89, Ser90, Met91, Leu95, Val132, Asp142, Ile144, and Lys145 in addition to Ile80, Glu82, Ile101, Gly109, and Lys134, to be responsive to signal peptide binding from the HSQC spectra. It is not surprising that Ile101 and Lys134 are not implicated in interactions with the peptide in earlier models because these residues may interact with the signal peptide only after a conformational change in the active site induced by substrate binding. In addition, Ile80, Glu82, and Gly109 are located away from the S1 and the S3 pocket, but are ideally oriented for interaction with the C-terminus of the hydrophobic core region of the peptide (Figure 3B, C, and D). Our data are the first to identify involvement of these residues in the signal peptide binding, indicating that at least the C-terminus of the core region of the peptide may be interacting with the enzyme. The crystal structure of the acyl-enzyme did not implicate Gln85 in the signal peptide binding (4) site but consistent with our findings, hydrogen bonding of the Arylomycin A2 inhibitor to the main chain carbonyl and amide of Gln85 suggested it may play some role in the signal peptide interaction. From molecular models the involvement of Ile101 has been unclear (5) although a SPase I Ile86Thr/Ile144Cys mutant revealed its likely role (5). The SPase I Ile86Thr/Ile144Cys mutant-signal peptide complex model indicates the side chains of Ile101 and Val132 require significant adjustment to accommodate the peptide consistent with the NMR data presented here that show conformational changes of Ile101 and Val132 upon peptide binding. Our data also point to the involvement of Ile144 previously inferred from the presence of a water hydrogen bonded to its main chain in the apo-enzyme (3) but displaced in the structure with the Arylomycin A2 inhibitor (5) and from the consequences of mutants at this position on fidelity of signal peptide cleavage (5,19). Leu95, located in the S3 pocket, was also identified in previous models (5). Since there is one unidentified resonance in the HSQC that changed because of peptide binding, we cannot exclude Phe84 or Tyr143, highlighted in models (5), from participating in the signal peptide binding site, nor can a role for Pro87 be excluded since our experiments are not sensitive to proline.
The dimensions of the region defined by the perturbed residues detected by NMR are appropriate for accommodation of the signal peptide cleavage region but little of the hydrophobic core of the signal peptide. This leaves open the possibility that the remainder of the hydrophobic core of the peptide is still within the translocon when the enzyme interacts with the cleavage region in vivo. This is consistent with the notion that during preprotein translocation, the periplasmic portion of the enzyme pivots relative to its transmembrane domains so that its large hydrophobic surface makes contact with the lipid bilayer for substrate interaction. The major phosphor-lipid in E. coli membranes is phosphatidylethanolamine, and since bilayers made of this phospholipid have a relatively hydrophobic surface (20), a favorable interaction could occur between the hydrophobic surface of SPase I and the bacterial membrane. Positioning the SPase I active site near the translocon would permit interaction with the cleavage region without release of the signal peptide from, for example, SecYEG. This also suggests a mechanism by which SPase I can be used by multiple transport pathways; pathway specificity may be imparted through recognition of the features of the signal peptide amino terminal and core regions by components of the translocation machinery unique to each pathway while the cleavage region and the C-terminus of the core region, only, contact SPase I.
Conclusions and Future Directions
NMR provides a powerful approach for the study of protein structure and dynamics. Here we report on the first NMR examination of soluble SPase I and on the structural analysis of its association with a signal peptide substrate. This work suggests that substrate binding to SPase I proceeds consistent with induced-fit recognition. This conformational adaptability may be ideally suited for the accommodation of signal peptide cleavage regions with a spectrum of different sequences. The conformational alterations we observe are largely confined to the enzyme active site, and include the catalytic dyad. These data further provide direct evaluation of predictions for substrate binding and catalysis based on molecular modeling and crystallography. That we were able to obtain high resolution NMR spectra using isotopically labeled enzyme, and in the presence of substrate by overcoming solubility issues, offers promise for future studies on this membrane protein including the development of an NMR-based approach to high throughput screening of potential SPase I inhibitors.
Acknowledgments
We thank Gregory Choi for help with initial NMR data collection and analysis. This work was supported in part by National Institutes of Health Grant GM37639 (to D.A.K.).
Abbreviations
- HSQC
heteronuclear single-quantum correlation
- NMR
nuclear magnetic resonance
- SPase I
type I signal peptidase
- SPase I Δ2–75
catalytic domain of signal peptidase I
- TAT
twin-arginine translocation
- TOM/TIM
translocase in the outer mitochondrial membrane/translocase in the inner mitochondrial membrane.
References
- 1.Wolfe PB, Wickner W, Goodman JM. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J Biol Chem. 1983;258:12073–12080. [PubMed] [Google Scholar]
- 2.Whitley P, Nilsson L, von Heijne G. Three-dimensional model for the membrane domain of Escherichia coli leader peptidase based on disulfide mapping. Biochemistry. 1993;32:8534–8539. doi: 10.1021/bi00084a020. [DOI] [PubMed] [Google Scholar]
- 3.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser-Lys dyad mechanism. J Biol Chem. 2002;277:9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- 4.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 5.Ekici OD, Karla A, Paetzel M, Lively MO, Pei D, Dalbey RE. Altered −3 substrate specificity of Escherichia coli signal peptidase 1 mutants as revealed by screening a combinatorial peptide library. J Biol Chem. 2007;282:417–425. doi: 10.1074/jbc.M608779200. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Miller A, Kendall DA. Signal peptide determinants of SecA binding and stimulation of ATPase activity. J Biol Chem. 2000;275:10154–10159. doi: 10.1074/jbc.275.14.10154. [DOI] [PubMed] [Google Scholar]
- 7.Izard JW, Doughty MB, Kendall DA. Physical and conformational properties of synthetic idealized signal sequences parallel their biological function. Biochemistry. 1995;34:9904–9912. doi: 10.1021/bi00031a012. [DOI] [PubMed] [Google Scholar]
- 8.Paetzel M, Chernaia M, Strynadka N, Tschantz W, Cao G, Dalbey RE, James MNR. Crystallization of a soluble, catalytically active form of Escherichia coli leader peptidase. Proteins. 1995;23:122–125. doi: 10.1002/prot.340230115. [DOI] [PubMed] [Google Scholar]
- 9.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 10.Deber CM, Wang C, Liu LP, Prior AS, Agrawal S, Muskat BL, Cuticchia AJ. TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 2001;10:212–219. doi: 10.1110/ps.30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu LP, Deber CM. Guidelines for membrane protein engineering derived from de novo designed model peptides. Biopolymers. 1998;47:41–62. doi: 10.1002/(SICI)1097-0282(1998)47:1<41::AID-BIP6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Melnyk RA, Partridge AW, Yip J, Wu Y, Goto NK, Deber CM. Polar residue tagging of transmembrane peptides. Biopolymers. 2003;71:675–685. doi: 10.1002/bip.10595. [DOI] [PubMed] [Google Scholar]
- 13.Tang YC, Deber CM. Aqueous solubility and membrane interactions of hydrophobic peptides with peptoid tags. Biopolymers. 2004;76:110–118. doi: 10.1002/bip.10566. [DOI] [PubMed] [Google Scholar]
- 14.Tschantz WR, Paetzel M, Cao G, Suciu D, Inouye M, Dalbey RE. Characterization of a soluble, catalytically active form of Escherichia coli leader peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry. 1995;34:3935–3941. doi: 10.1021/bi00012a010. [DOI] [PubMed] [Google Scholar]
- 15.Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins. Science. 1997;278:497–499. doi: 10.1126/science.278.5337.497. [DOI] [PubMed] [Google Scholar]
- 16.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 17.Tschantz WR, Sung M, Delgado-Partin VM, Dalbey RE. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 18.Carlos JL, Klenotic PA, Paetzel M, Strynadka NC, Dalbey RE. Mutational evidence of transition state stabilization by serine 88 in Escherichia coli type I signal peptidase. Biochemistry. 2000;39:7276–7283. doi: 10.1021/bi000301l. [DOI] [PubMed] [Google Scholar]
- 19.Karla A, Lively MO, Paetzel M, Dalbey R. The identification of residues that control signal peptidase cleavage fidelity and substrate specificity. J Biol Chem. 2005;280:6731–6741. doi: 10.1074/jbc.M413019200. [DOI] [PubMed] [Google Scholar]
- 20.Yeagle PL, Sen A. Hydration and the lamellar to hexagonal II phase transition of phosphatidylethanolamine. Biochemistry. 1986;25:7518–7522. doi: 10.1021/bi00371a039. [DOI] [PubMed] [Google Scholar]