Abstract
For proper development of plants auxin levels need to be tightly controlled. For this, several routes have evolved and it is plausible that different organisms use these differently. To determine whether members of the family of GH3 proteins, which partially act as auxin conjugate synthetases in Arabidopsis thaliana, have similar roles in the moss Physcomitrella patens, we have investigated the in vitro activity of the two GH3 members in moss. We showed that both proteins can form amino acid conjugates with indole-3-acetic acid (IAA) but also with jasmonic acid (JA). Confirming these findings, single and double knockout-mutants showed lower levels of IAA conjugates than wild type. We discuss the results in light of the possible functions of IAA conjugate formation in lower land plants.
Key words: Arabidopsis thaliana, auxin metabolism, jasmonic acid, GH3 genes, moss, Physcomitrella patens
Auxins play diverse roles in many aspects of plant growth and development. Their activity is relying on the correct concentration in a given tissue and developmental stage.1 If higher levels of indole-3-acetic acid (IAA) are present, the hormone can also have an inhibitory effect on growth processes.2 Therefore, the tight control of IAA concentrations is absolutely necessary. To this end plants have evolved different mechanisms.3 First, biosynthesis is contributing to increasing IAA concentrations, mostly in young tissues such as meristems. Second, IAA can be transported in a polar way, depending on transport molecules, from cell to cell, away from the site of synthesis, thereby forming an auxin gradient along the plant axis. Third, IAA can be degraded, and fourth, IAA can be reversibly inactivated by conjugation to small molecules such as amino acids or sugars, but also be linked to larger molecules such as peptides or proteins.4 The inactive IAA conjugates can be hydrolyzed to yield free (i.e., active) IAA if needed. In higher plants the levels of free IAA constitutes between 5 and 20% depending on the tissue or age of the plant, whereas the conjugated form constitutes the major part.4 However, it is not yet clear in which way auxin homeostasis has evolved. The hypothesis that auxin has to be present during the evolution of a body plan has been tested by using different lower land plants which were compared in their mechanism to control auxin homeostasis. In algae, e.g., charophytes, the major metabolic way of controlling IAA is via biosynthesis. In bryophytes, the formation of IAA conjugates has been shown, although the amount was lower than for example in seed plants such as Arabidopsis thaliana.5,6 Since the molecular biology of auxin homeostasis in Arabidopsis is most advanced, we will use this model plant to compare the knowledge on seed plants with that in the moss Physcomitrella patens. The recent publication of the Physcomitrella genome7 gives the possibility to investigate components of the machinery controlling IAA levels in a lower land plant.
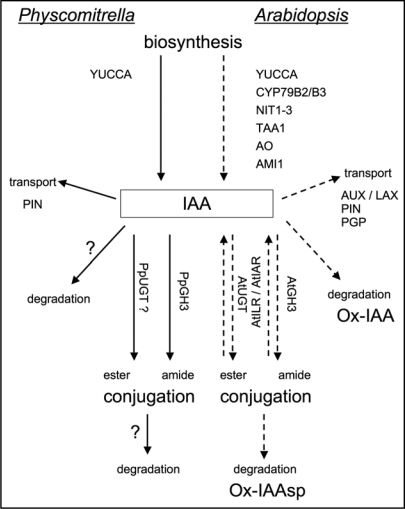
In general, there seem to be high levels of redundancy involved in the pathways leading to decrease or increase of IAA, respectively. In Figure 1 we compare the current knowledge about genes related to IAA concentrations in Physcomitrella with Arabidopsis. While in Arabidopsis many different biosynthetic routes leading to IAA were identified,8 in the Physcomitrella genome homologs of the YUCCA genes have been detected.7 The presence of auxin conjugate synthetases has been experimentally verified in the moss (see below) and additional evidence for ester conjugate synthesis comes from sequence homology to UDP-glucosyl transferases.7 There is also the possibility of degradation of either IAA or an amino acid conjugate with IAA9,10 as discussed below.
Figure 1.
Comparison of possibilities to regulate auxin homeostasis in Physcomitrella (solid lines) and Arabidopsis (dotted lines). Biosynthesis—AO, aldehyde oxidase; AMI1, amidase; CYP, cytochrome P450; NIT, nitrilase; TAA1, tryptophan aminotransferase; YUCCA, flavin monooxygenase; transport—AUX/LAX, auxin influx facilitator family; PIN, auxin efflux carrier family; PGP, ABC transporter type auxin efflux carrier family; conjugation/hydrolysis—UGT, UDP-glucosyl transferase; GH3, auxin conjugate synthetase family; ILR/IAR, auxin conjugate hydrolase family; Ox-IAA, oxindole-3-acetic acid; Ox-IAAsp, oxindole-3-aspartic acid.
So far our work has focussed on the characterization of two members of the so called GH3 family, of which several from Arabidopsis can form conjugates of IAA with a variety of amino acids.11 While 19 members of this family have been described in Arabidopsis, only two are present in Physcomitrella.12 The Arabidopsis family clusters in three groups: group I containing the jasmonic acid conjugate synthetase JAR1 and a few others with as yet unkown function, group II the auxin conjugate synthetases, and group III with mostly as yet uncharacterized members.11,13 Sequence similarity of the GH3 genes from Physcomitrella showed that both cluster within the JAR1 group.12 Therefore, we analyzed the enzymatic activity of the two Physcomitrella GH3 proteins (PpGH3-1 and PpGH3-2) in vitro14 and found that both were active on jasmonic acid and a variety of different amino acids, whereas PpGH3-2 was active mostly with IAA. PpGH3-1 showed only weak activity with IAA and only two amino acids. For this reason, it could be assumed that the two Physcomitrella genes evolved by gene duplication, from which the initial activities would be for IAA and jasmonic acid. One of these genes might have evolved into a jasmonate conjugate synthetase (maybe AtJAR1),13 thereby loosing its activity on IAA. The second may have given rise to the auxin conjugate synthetase family in Arabidopsis,11 but the conjugate synthetases of Physcomitrella have still activity with both hormones. Interestingly, there is no evidence as yet that jasmonic acid itself has a role during Physcomitrella development, although a possible function of JA-conjugates has not been closely investigated. Since in Arabidopsis the JA conjugate with isoleucine is the active compound to be recognized by the COI1 receptor protein,15 it could be the case that JA itself has no effect in Physcomitrella. However, in our growth experiments a small growth promoting effect of JA, independently on the presence of GH3 genes was found. Similar observations were made with gibberellins in Physcomitrella.16
Further characterization of single and double KO mutants in each of the PpGH3 genes has led to the hypothesis that GH3 proteins are indeed involved in regulating the auxin homeostasis in Physcomitrella.14 Both single KO mutants were more sensitive to increasing IAA concentrations in the medium than the wild type. Furthermore, the levels of free IAA were higher and the levels of conjugated IAA concomitantly dropped. A double KO mutant had almost no IAA conjugates when compared to the wild type. However, this mutant was still able to synthesize ester conjugates with IAA. Interestingly, the role of GH3 proteins in auxin conjugation seemed to be only important in the gametophore stage, whereas protonema cultures of GH3 KO mutants did not show any changes in auxin homeostasis. Therefore, we hypothesize that the role of GH3 proteins is dependent on a certain developmental stage of the moss. Additionally, we propose other detoxification mechanisms for example, export or degradation, in protonema.
In higher plants the ester conjugate formation of IAA has been shown to be dependent on UDP-glucosyl transferases (AtUGT84B1 for Arabidopsis17 and ZmIAGLU for maize18). In the genome of Physcomitrella we could detect candidate sequence(s) for these genes, indicating that Physcomitrella has indeed the potential to synthesise the ester conjugates found in the gametophores in addition to amide conjugates. However, in the Physcomitrella genome, no homolog for an auxin conjugate hydrolase was found. In higher plants, auxin conjugate hydrolysis is thought to contribute to free IAA and depending on the plant species, large gene families with overlapping but distinct substrate preferences for individual amino acid conjugates with IAA are present.19,20 Since this is not the case for Physcomitrella, one has to ask the question whether the conjugation of auxin is a one-way road for inactivation of excess auxin and whether auxin conjugate hydrolysis has evolved later during plant evolution.
In the Selaginella moellendorffii genome (http://genome.jgi-psf.org/Selmo1/Selmo1.home.html), an auxin conjugate hydrolase sequence related to higher plant ones, has been found based on homology searches, but the completion of the genome has to be awaited to draw final conclusions. Likewise, it is not clear, if this effect is specific for Physcomitrella, or found in bryophytes in general. Therefore, additional sequenced bryophyte genomes are needed.21
Since in Arabidopsis the degradation of the IAA-Aspartate conjugate to Ox-IAA-Asp (see Fig. 1) has been described,9,10 a similar scenario could be suggested to occur in Physcomitrella with the amino acid conjugates formed. Alternatively, the hydrolysis of IAA conjugates by members of the M20 dipeptidase family can be envisioned. However, this would need the activity of enzymes with very low sequence conservation to auxin conjugate hydrolases. These questions will be addressed in future research by studying the metabolism of IAA and IAA conjugates of Physcomitrella in more detail.
Acknowledgements
We acknowledge financial support from the Deutsche Forschungsgemeinschaft within SPP 1067 (Molecular analysis of phytohormone action).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7536
References
- 1.Davies PJ, editor. Plant hormones: biosynthesis, signal transduction, action! Dordrecht, the Netherlands: Kluwer Academic Publishers; 2004. [Google Scholar]
- 2.Bandurski RS, Cohen JD, Slovin JP, Reinecke DM. Auxin biosynthesis and metabolism. In: Davies P, editor. Plant hormones: physiology, biochemistry and molecular biology. 2nd edn. Boston, MA, USA: Kluwer Academic Publishers; 1995. pp. 39–65. [Google Scholar]
- 3.Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol. 2002;49:249–272. [PubMed] [Google Scholar]
- 4.Seidel C, Walz A, Park S, Cohen JD, Ludwig-Müller J. Indole-3-acetic acid protein conjugates: novel players in auxin homeostasis. Plant Biol. 2006;8:340–345. doi: 10.1055/s-2006-923802. [DOI] [PubMed] [Google Scholar]
- 5.Sztein AE, Cohen JD, Garcia de la Fuente I, Cooke TJ. Auxin metabolism in mosses and liverworts. Am J Bot. 1999;86:1544–1555. [PubMed] [Google Scholar]
- 6.Sztein AE, Cohen JD, Cooke TJ. Evolutionary patterns in the auxin metabolism of green plants. Int J Plant Sci. 2000;161:849–859. [Google Scholar]
- 7.Rensing SA, Lang D, Zimmer A, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist E, Kamisugi Y, et al. The Physcomitrella genome reveals insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 8.Woodward AW, Bartel B. Auxin: regulation, action and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Östin A, Moritz T, Sandberg G. Liquid chromatography/mass spectrometry of conjugates and oxidative metabolites of indole-3-acetic acid. Biol Mass Spectrom. 1992;21:292–298. [Google Scholar]
- 10.Östin A, Kowalyczk M, Bhalerao RP, Sandberg G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998;118:285–296. doi: 10.1104/pp.118.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierfreund NM, Tintelnot S, Reski R, Decker EL. Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J Plant Physiol. 2004;161:823–836. doi: 10.1016/j.jplph.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig-Müller J, Jülke S, Bierfreund NM, Decker EL, Reski R. Moss (Physcomitrella patens) GH3 proteins are involved in auxin homeostasis. New Phytol. 2009;181:323–338. doi: 10.1111/j.1469-8137.2008.02677.x. [DOI] [PubMed] [Google Scholar]
- 15.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar M, Liu G, Nomura K, He SJ, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 16.Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Van Der Straeten D. Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol. 2007;7:65. doi: 10.1186/1471-2229-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson RG, Lim E-K, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem. 2001;276:4350–4356. doi: 10.1074/jbc.M006185200. [DOI] [PubMed] [Google Scholar]
- 18.Szerszen JD, Szczyglowski K, Bandurski RS. iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science. 1994;265:1699–1701. doi: 10.1126/science.8085154. [DOI] [PubMed] [Google Scholar]
- 19.LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem. 2002;277:20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- 20.Campanella JJ, Smith SM, Leibu D, Wexler S, Ludwig-Müller J. The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J Plant Growth Regul. 2008;27:26–38. [Google Scholar]
- 21.Lang D, Zimmer AD, Rensing SA, Reski R. Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 2008;13:542–549. doi: 10.1016/j.tplants.2008.07.002. [DOI] [PubMed] [Google Scholar]