Abstract
A major physiological role of insulin is the regulation of glucose uptake into skeletal and cardiac muscle and adipose tissue, mediated by an insulin-stimulated translocation of GLUT4 glucose transporters from an intracellular vesicular pool to the plasma membrane. This process is similar to the regulated docking and fusion of vesicles in neuroendocrine cells, a process that involves SNARE-complex proteins. Recently, several SNARE proteins were found in adipocytes: vesicle-associated membrane protein (VAMP-2), its related homologue cellubrevin, and syntaxin-4. In this report we show that treatment of permeabilized 3T3-L1 adipocytes with botulinum neurotoxin D, which selectively cleaves VAMP-2 and cellubrevin, inhibited the ability of insulin to stimulate translocation of GLUT4 vesicles to the plasma membrane. Furthermore, treatment of the permeabilized adipocytes with glutathione S-transferase fusion proteins encoding soluble forms of VAMP-2 or syntaxin-4 also effectively blocked insulin-regulated GLUT4 translocation. These results provide evidence of a functional role for SNARE-complex proteins in insulin-stimulated glucose uptake and suggest that adipocytes utilize a mechanism of regulating vesicle docking and fusion analogous to that found in neuroendocrine tissues.
One of the best characterized systems for analysis of function of SNARE-complex proteins is the transport, docking, and regulated fusion of synaptic vesicles with presynaptic membranes in neuroendocrine tissues (1–4). In these systems, the synaptic vesicle-associated SNARE (v-SNARE) proteins, such as the vesicle-associated membrane proteins (VAMPs) (also known as synaptobrevins) form high-affinity complexes with their cognate target membrane proteins (t-SNARE), such as syntaxins and SNAP-25 proteins (synaptosome-associated protein of 25 kDa), present on the presynaptic membrane (4).
Neurosecretory vesicle docking and fusion events share similarities with the process involved in insulin-stimulated glucose uptake, in which intracellular vesicles containing GLUT4 glucose transporters are translocated and fuse with the plasma membrane allowing increased glucose entry into the insulin-sensitive tissues: skeletal and cardiac muscle and adipose (5, 6). Recently, a number of protein components associated with immunopurified GLUT4-containing vesicles have been identified. These include a novel aminopeptidase (gp160), secretory carrier-associated membrane proteins (SCAMPs), low molecular weight GTP-binding proteins, and two members of the synaptobrevin family of proteins: VAMP-2 and cellubrevin (7–13). The function of all of these proteins in adipocytes remains unknown. VAMP-2 expression was initially thought to be restricted to neuroendocrine cells and associated with synaptic vesicles as described above. In contrast, the highly homologous protein, cellubrevin, shows a wide tissue distribution, is associated with endosomes, and is thought to be involved in constitutive recycling of vesicles (14, 15). The association of VAMP-2 and cellubrevin with immunopurified GLUT4 vesicles and their redistribution following stimulation with insulin (8) suggest that adipocytes may utilize similar molecular machinery and perhaps an analogous mechanism for GLUT4–vesicle docking and fusion as observed for synaptic vesicles in neuroendocrine cells. In addition, syntaxin-4 (a cognate t-SNARE for VAMP-2 in neuroendocrine cells) has also been reported to be expressed in 3T3-L1 adipocytes (16). The purpose of the present study was to determine whether these molecules play a functional role in insulin-stimulated GLUT4–vesicle translocation to the plasma membrane in 3T3-L1 adipocytes.
EXPERIMENTAL PROCEDURES
Cell Culture and Permeabilization with Streptolysin O (SLO).
3T3-L1 fibroblasts were maintained in DMEM containing 10% calf serum. Adipogenesis was induced with DMEM containing 10% fetal bovine serum, dexamethasone, isobutylmethylxanthine, and insulin as described (17). Fully differentiated 3T3-L1 adipocytes were permeabilized with SLO for 5 min at 37°C in intracellular buffer (20 mM Hepes, pH 7.4/140 mM K+-glutamate/5 mM MgCl2/5 mM EGTA/5 mM NaCl/1 mM DTT) (18). An ATP-regenerating system was included in this buffer and consisted of 0.1 mg/ml creatine kinase, 5 mM creatine phosphate, and 1 mM ATP. Based on guanosine 5′-[γ-thio]triphosphate-stimulated GLUT4 translocation, 80–90% of the cells were permeabilized by this technique.
Treatment of Permeabilized Cells with Clostridial Botulinum Neurotoxins (BoNTs), IgA Protease, and Glutathione S-transferase (GST) Fusion Proteins.
BoNTs were obtained from Sigma. IgA protease was purchased from Boehringer Mannheim. Before incubation with permeabilized cells, the BoNTs were reduced with 20 mM DTT for 30 min at 37°C. Permeabilized cells were treated with buffer alone, 100 nM BoNT D, 100 nM BoNT C, or 940 nM IgA protease for 20–30 min at 37°C. In experiments using GST fusion proteins, the permeabilized cells were incubated for 10 min in the absence of GST protein or in the presence (10 μM) of GST alone, a soluble form of GST–VAMP-2 fusion protein (amino acids 1–94 of rat VAMP-2 sequence, which lacks the transmembrane domain), or a soluble form of GST–syntaxin-4, which lacks the C-terminal 25 amino acids encoding the transmembrane domain (kind gift of Richard H. Scheller, Stanford University Medical Center). The cells were then incubated for a further 10 min in the absence or presence of insulin (100 nM), and plasma membrane sheets or membrane proteins were prepared as described below.
Immunofluorescence of Plasma Membrane Sheets.
Translocation of GLUT4 to the cell surface was assessed using an immunofluorescence staining of plasma membrane sheets (19). 3T3-L1 cells were grown and differentiated on glass coverslips. Following the indicated treatments, the coverslips were washed in ice-cold buffer containing 50 mM Hepes (pH 7.4) and 100 mM NaCl. The cells were then subjected to sonication in buffer containing 20 mM Hepes (pH 7.4), 100 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 1 μg/ml leupeptin, 10 μg/ml aprotinin, and 2 mM phenylmethylsulfonyl fluoride (PMSF). The plasma membrane sheets were incubated with a rabbit antisera raised against a C-terminal GLUT4 peptide (East Acres Biologicals, Southbridge, MA) followed by a secondary incubation with a rhodamine-conjugated anti-rabbit IgG. Images were obtained using a Zeiss confocal microscope.
Preparation of Subcellular Membrane Fractions and Analysis of Proteins by Immunoprecipitation and Immunoblotting.
Subcellular membrane fractions were prepared as described (7). Briefly, 3T3-L1 cells were harvested in HES buffer (20 mM Hepes, pH 7.4/250 mM sucrose/1 mM EDTA/10 μg/ml aprotinin/1 μg/ml leupeptin/2 mM PMSF) and homogenized using a Teflon pestle in a glass homogenizer. The supernatant from a 19,000 × g centrifugation was centrifuged at 48,000 × g to obtain a high-density membrane pellet. The 48,000 × g supernatant was centrifuged at 210,000 × g to obtain the low density microsomal (LDM) pellet. Equal amounts of protein from LDM fractions were solubilized, separated by SDS/PAGE, transferred to nitrocellulose or poly(vinylidene difluoride), and immunoblotted with antisera against GLUT1, GLUT4 (East Acres Biologicals), SCAMP (kind gift from David Castle, Charlottesville, VA), VAMP-2 (8), or cellubrevin (kind gift from Pietro De Camilli, New Haven, CT). Immunoreactive material was detected by incubation with [125I]-labeled protein A or by incubation with a secondary IgG coupled to horseradish peroxidase followed by chemiluminescence detection. In vitro IgA protease reactions were carried out using 20 μg of LDM protein incubated in the absence or presence of IgA protease (240 nM) for 60 min at 37°C. The reactions were terminated by addition of SDS/PAGE sample buffer and subjected to immunoblot analysis as described above. For immunoprecipitation and analysis of syntaxin-4, membrane preparations from 3T3-L1 adipocytes were solubilized in buffer containing 20 mM Hepes (pH 7.8), 0.9% Triton X-100, 100 mM KCl, 1 mM DTT, 2 mM EDTA, and 1 mM PMSF. Samples containing equal amounts of protein were incubated with protein A-Sepharose beads coupled to an affinity-purified syntaxin-4 antibody (αSynt4) raised against a syntaxin-4 fusion protein encompassing amino acids 1–274 (20) or nonimmune serum. The immunoprecipitated proteins on the beads were washed and solubilized in SDS/PAGE sample buffer, separated by SDS/PAGE, and analyzed by immunoblotting.
RESULTS AND DISCUSSION
The various serotypes of BoNTs are Zn-dependent endopeptidases that have a unique substrate specificity. BoNT B, D, and F cleave VAMP-2 and cellubrevin; BoNT A and E cleave SNAP-25; and BoNT C cleaves certain syntaxin isoforms (21–23). These BoNT proteins have been powerful tools in defining functional roles for several of the proteins involved in the docking and/or fusion of synaptic vesicles with presynaptic membranes (4). To test the hypothesis that VAMP-2 and cellubrevin are involved in regulated GLUT4–vesicle translocation, we determined the effect of BoNT cleavage of the known substrate proteins present in adipocytes and examined their potential functional role in insulin-regulated translocation of GLUT4.
To introduce the BoNTs into 3T3-L1 adipocytes we established a permeabilized adipocyte system using SLO. The SLO-permeabilized adipocytes maintain the cellular components required for insulin-regulated GLUT4 translocation (18). Thus, treatment of permeabilized 3T3-L1 adipocytes with insulin resulted in a dramatic increase in the amount of GLUT4 associated with the plasma membrane as determined by immunofluorescence of plasma membrane sheets (Fig. 1A). Pretreatment of the permeabilized cells with 100 nM BoNT D completely inhibited this insulin-stimulated GLUT4 translocation (Fig. 1A). In contrast, pretreatment with BoNT C (which cleaves syntaxins-1A/1B, -2, and -3) did not alter GLUT4 translocation.
Figure 1.
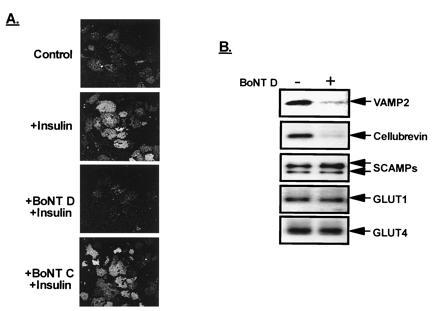
Treatment of 3T3-L1 adipocytes with BoNT D blocks insulin-stimulated GLUT4–vesicle translocation to the plasma membrane. (A) SLO-permeabilized 3T3-L1 adipocytes were pretreated for 20 min with buffer alone (control) or buffer containing 100 nM BoNT D or BoNT C, followed by an additional 10-min incubation with insulin. Plasma membrane sheets were then prepared and GLUT4 was detected by immunofluorescence. (B) Following treatment of permeabilized cells with buffer alone (−) or BoNT D (+), an LDM subcellular membrane fraction was prepared and LDM-resident proteins were examined by immunoblot analysis using antisera against the proteins indicated.
To determine the specificity and extent of cleavage of the BoNT D target proteins (VAMP-2 and cellubrevin) present in adipocytes, we prepared a LDM subcellular fraction that contains the intracellular GLUT4 vesicles and examined several resident protein components by immunoblot analysis. BoNT D treatment of permeabilized 3T3-L1 adipocytes resulted in an 80% decrease in immunodetectable levels of both VAMP-2 and cellubrevin (Fig. 1B), indicative of the specific cleavage by BoNT D. As expected, other LDM-resident proteins, such as GLUT1, GLUT4, and SCAMPs, were unaffected by BoNT D. In addition, the Coomassie blue staining pattern of proteins from control and BoNT D-treated samples was identical (data not shown), suggesting the absence of any global effect by the toxin.
VAMP-2 and cellubrevin share extensive sequence homology; however, VAMP-2 contains an IgA protease recognition motif (PPXP, where X is alanine, threonine, or serine), located within its N terminus, that is not present in cellubrevin (24). To assess the relative contribution of VAMP-2 vs. cellubrevin in GLUT4–vesicle translocation, we pretreated permeabilized 3T3-L1 adipocytes with IgA protease. As indicated in Fig. 2A, IgA protease blocked the majority of insulin-stimulated translocation of GLUT4 to the plasma membrane. As predicted, analysis of LDM-resident proteins following an in vitro IgA protease reaction revealed that VAMP-2 was cleaved to a lower (≈14 kDa) form, while cellubrevin, GLUT4, and the 37-kDa SCAMP isoform were unaffected (Fig. 2B). IgA protease did cleave the 39-kDa form of SCAMP protein, and this protein was more sensitive than VAMP-2 to cleavage by IgA protease. However, at lower concentrations of IgA protease, when the 39-kDa SCAMP protein was cleaved and no VAMP-2 cleavage was observed, there was no effect on insulin-stimulated GLUT4 translocation to the plasma membrane (not shown), suggesting that this form of SCAMP is not required for insulin-regulated GLUT4–vesicle translocation or that in its cleaved form its function is not affected by proteolysis.
Figure 2.
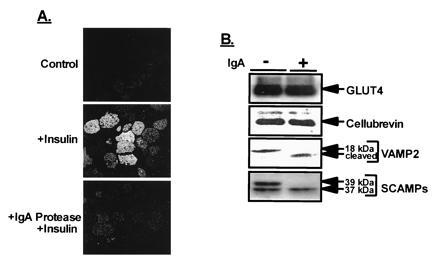
IgA protease blocks insulin-stimulated GLUT4–vesicle translocation to the plasma membrane. (A) SLO-permeabilized 3T3-L1 adipocytes were incubated in the absence (control) or presence of 940 nM IgA protease (Boehringer Mannheim) for 30 min at 37°C. The cells were then incubated a further 10 min with insulin (100 nM). Preparation of plasma membrane sheets and GLUT4 immunofluorescence were carried out as described above. (B) In vitro IgA protease reactions. LDM protein (20 μg) was incubated in the absence (control) or presence of 275 nM IgA protease for 60 min at 37°C. The reactions were terminated by addition of SDS/PAGE sample buffer, and the reaction products were examined by immunoblot analysis as described in Fig. 1. Full-length VAMP-2 is 18 kDa; its IgA fragment (cleaved) is indicated.
In neurosecretory cells, the VAMP proteins present on synaptic vesicles can form complexes with syntaxins and SNAP-25 present on presynaptic membranes (4, 25). Syntaxins-1A/1B and the neuronal form of SNAP-25 are not detected in 3T3-L1 adipocytes; however, they do contain syntaxin-4 (16). BoNT C can block exocytosis by cleavage of syntaxins-1A/1B, -2, and -3 (2, 22); however, syntaxin-4 is resistant to cleavage by BoNT C (26), perhaps explaining the lack of effect of BoNT C on GLUT4–vesicle translocation (Fig. 1A). To show whether, in 3T3-L1 adipocytes, VAMP-2 has the potential SNARE–protein–protein interactions with syntaxin-4, we used an antibody against syntaxin-4 and observed coprecipitation of VAMP-2 with syntaxin-4 in detergent extracts prepared from intact cells (Fig. 3A). These results indicate that the endogenous complements of these proteins are able to form stable complexes in these cells. In an attempt to further define a functional role for both VAMP-2 and syntaxin-4 in GLUT4 translocation, we pretreated permeabilized adipocytes for 10 min with soluble forms (lacking the transmembrane region) of GST-syntaxin-4 or GST-VAMP-2 (Fig. 3B). Both the GST–VAMP-2 protein and the GST–syntaxin-4 protein inhibited the insulin-stimulated translocation of GLUT4. In contrast, GST alone had no effect. Thus, by interacting with their cognate t-SNARE or v-SNARE, GST–VAMP-2 and GST–syntaxin-4, respectively, interfere with the GLUT4–vesicle translocation to the plasma membrane. Each protein, in this way, attests the involvement of the other and further supports the hypothesis that SNARE-complex proteins are necessary components in insulin-regulated GLUT4 translocation in adipocytes.
Figure 3.
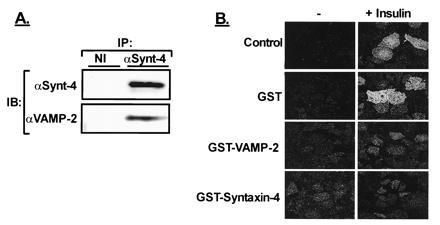
(A) Syntaxin-4 is present in 3T3-L1 adipocytes and associates with VAMP-2. Detergent extracts of 3T3-L1 adipocytes were subjected to immunoprecipitation (IP) with a nonimmune antiserum (NI) or anti-syntaxin-4 (αSynt4). The immunoprecipitated proteins were separated by SDS/PAGE and immunoblotted (IB) with either αSynt-4 or αVAMP-2. (B) Effect of various GST-fusion proteins on insulin-regulated GLUT4 translocation. Permeabilized adipocytes were incubated in buffer alone or in the presence of 10 μM GST alone, GST–VAMP-2, or GST–syntaxin-4 for 10 min. The cells were then stimulated with insulin for an additional 10 min followed by analysis of GLUT4 translocation by immunofluorescence of plasma membrane sheets.
The above data provide compelling evidence for a functional role for VAMP-family proteins in insulin-regulated GLUT4–vesicle translocation. BoNT D cleaves both VAMP-2 and cellubrevin and blocks regulated GLUT4–vesicle translocation. While IgA protease is not as selective as the BoNTs, its proteolysis of VAMP-2 and not cellubrevin also leads to a significant, although not complete, inhibition of GLUT4–vesicle translocation. Furthermore, GST–VAMP-2 effectively inhibits insulin’s ability to stimulate translocation of GLUT4, presumably by competing for formation of functional SNARE complexes. From these results, it appears that VAMP proteins are required functional components for insulin-regulated GLUT4 docking and/or fusion with the plasma membrane.
The exact functional distinction between VAMP-2 and cellubrevin is unclear. In fibroblasts, cellubrevin is localized to endosome-derived vesicles and cleavage of cellubrevin prevents constitutive recycling of these vesicles (14, 15). In addition, cellubrevin-containing vesicles immunopurified from adipocytes contain GLUT4 but do not contain detectable levels of VAMP-2 (8). Furthermore, Martin et al. (27) have recently shown that ablation of endosomes in 3T3-L1 adipocytes results in the loss of 90% of the cellubrevin found in intracellular vesicles but only a 10% loss in VAMP-2 and a 40% loss in GLUT4. Taken together these data suggest that VAMP-2 and cellubrevin may be localized to distinct pools of GLUT4 vesicles (Fig. 4), consistent with the hypothesis that one pool of GLUT4 vesicles originates from a distinct exocytotic compartment and the other is endosomal derived (6, 8, 27, 28). Our data are consistent with this notion since cleavage of VAMP-2 but not cellubrevin by IgA protease leads to a loss of insulin-stimulated translocation of GLUT4, suggesting that cellubrevin-associated vesicles may not be involved in the acutely regulated translocation of GLUT4. However, further studies are necessary to clearly define the contribution of each of these pools to insulin-regulated glucose uptake.
Figure 4.
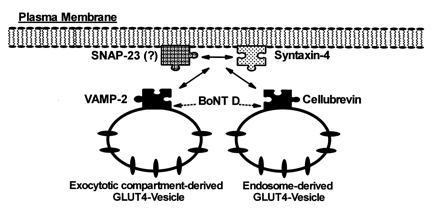
A potential model for the interaction of GLUT4-containing vesicles with the docking/fusion machinery in adipocytes.
The expression of syntaxin-4 in adipocytes, its association with VAMP-2, the functional requirement of VAMP-2, and the observation that a GST–syntaxin-4 protein can block insulin-stimulated GLUT4 translocation provide strong evidence that syntaxin-4 can serve as a cognate t-SNARE molecule for VAMP-2 in 3T3-L1 adipocytes (Fig. 4). Thus, the requirement of syntaxin-4 and VAMP-2 (and perhaps cellubrevin) for insulin-stimulated translocation of GLUT4-containing vesicles to the plasma membrane in adipocytes suggests that this process utilizes proteins that are functionally comparable to those involved in regulated vesicular traffic in neuroendocrine systems. Furthermore, in neuroendocrine cells the formation of the heterotrimeric SNARE complex involves the binding of the vesicle-associated VAMP with syntaxins and SNAP-25 present on the plasma membrane (1–4). SNAP-25 is not detected in adipocytes using either a monoclonal antibody or a polyclonal serum raised against the C-terminal 12 amino acids of SNAP-25 (16, 29). However, an antiserum raised against amino acids 33–206 of SNAP-25 detected a protein in adipocytes, suggesting the existence of an adipocyte homologue (29). Indeed, SNAP-23 (a homologue of SNAP-25) was recently cloned from human B lymphocytes (30). Using an antibody to SNAP-23 we have detected this protein in membrane preparations from 3T3-L1 adipocytes (P. Wong, W. Trimble, A.K., M. Wilson, P. Roche, and B.C., unpublished results). This newly identified protein’s role in the mediation of SNARE–protein–protein interactions, as well as its possible role in GLUT4–vesicle translocation in adipocytes, has not yet been established.
In conclusion, although it has been traditionally thought that regulated secretory pathways were confined to neuroendocrine cells, we find functional elements of these pathways in adipocytes. While these different cell systems share similar endo- and exocytotic machinery, the signaling pathways leading to the regulation of these events appear to be quite different. In addition, the data presented here indicate that SNARE-complex proteins are necessary components in insulin-regulated GLUT4–vesicle translocation in adipocytes. The potential role of SNARE-complex proteins in the regulated secretion of other proteins found in adipocytes, such as adipsin (31) and the ob gene product, leptin (32), can now be determined.
Acknowledgments
This study was supported by Pilot and Feasibility Funding through National Institutes of Health Diabetes Endocrinology Research Care Grant 5 P30 DK-36836 (B.C.), National Institutes of Health Grants DK-33201 (C.R.K.) and DK-47919 (C.J.R.), and Medical Research Council Grant MT-7307 (A.K.).
Footnotes
Abbreviations: VAMP-2, vesicle-associated membrane protein; SCAMPs, secretory carrier-associated membrane proteins; BoNT, Clostridial botulinum neurotoxin; SLO, streptolysin O; GST, glutathione S-transferase; LDM, low density microsomal; PMSF, phenylmethylsulfonyl fluoride.
References
- 1.Sollner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof T C, Niemann H. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sollner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1995;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 4.Sudhof T C, De Camilli P, Niemann H, Jahn R. Cell. 1993;75:1–4. [PubMed] [Google Scholar]
- 5.Cushman S W, Wardzala L J. J Biol Chem. 1980;255:4758–4762. [PubMed] [Google Scholar]
- 6.Birnbaum M J. Int Rev Cytol. 1992;137:239–297. [PubMed] [Google Scholar]
- 7.Volchuk A, Mitsumoto Y, He L, Liu Z, Habermann E, Trimble W, Klip A. Biochem J. 1994;304:139–145. doi: 10.1042/bj3040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volchuk A, Sargeant R, Sumitani S, Liu Z, He L, Klip A. J Biol Chem. 1995;270:8233–8240. doi: 10.1074/jbc.270.14.8233. [DOI] [PubMed] [Google Scholar]
- 9.Kandror K V, Pilch P F. J Biochem (Tokyo) 1994;269:138–142. [PubMed] [Google Scholar]
- 10.Cain C C, Trimble W S, Lienhard G E. J Biol Chem. 1992;267:11681–11684. [PubMed] [Google Scholar]
- 11.Laurie S M, Cain C C, Lienhard G E, Castle J D. J Biochem (Tokyo) 1993;268:19110–19117. [PubMed] [Google Scholar]
- 12.Jetton T L, Liang Y, Pettepher C C, Zimmerman E C, Cox F G, Horvath K, Matschinsky F M, Magnuson M A. J Biol Chem. 1994;269:3641–3654. [PubMed] [Google Scholar]
- 13.Cormont M, Tanti J F, Gremeaux T, Van Obberghen E, Le Marchand-Brustel Y. Endocrinology. 1991;126:3343–3350. doi: 10.1210/endo-129-6-3343. [DOI] [PubMed] [Google Scholar]
- 14.McMahon H T, Ushkaryov Y A, Edelmann L, Link E, Binz T, Neimann H, Jahn R, Sudhof T C. Nature (London) 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- 15.Galli T, Chilcote T, Mundigl O, Binz T, Niemann H, De Camilli P. J Cell Biol. 1994;125:1015–1024. doi: 10.1083/jcb.125.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volchuk A, Wang Q, Ewart H S, Liu Z, He L, Bennett M K, Klip A. Mol Biol Cell. 1996;7:1075–1082. doi: 10.1091/mbc.7.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Herreros A G, Birnbaum M J. J Biol Chem. 1989;264:19994–19999. [PubMed] [Google Scholar]
- 18.Robinson L J, Pang S, Harris D S, Heuser J, James D E. J Cell Biol. 1992;117:1181–1196. doi: 10.1083/jcb.117.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fingar D C, Hausdorff S F, Blenis J, Birnbaum M J. J Biol Chem. 1993;268:3005–3008. [PubMed] [Google Scholar]
- 20.Sumitani S, Ramlal T, Liu Z, Klip A. Biochem Biophys Res Commun. 1995;213:462–468. doi: 10.1006/bbrc.1995.2154. [DOI] [PubMed] [Google Scholar]
- 21.Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Sudhof T C, Jahn R, Niemann H. J Biol Chem. 1994;269:1617–1620. [PubMed] [Google Scholar]
- 22.Blasi J, Chapman E R, Yamasaki S, Binz T, Niemann H, Jahn R. EMBO J. 1993;12:4821–4828. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamasaki S, Baumeister A, Binz T, Blasi J, Link E, Cornille F, Roques B, Fykse E M, Sudhof T C, Jahn R, Niemann H. J Biol Chem. 1994;269:12764–12772. [PubMed] [Google Scholar]
- 24.Binscheck T, Bartels F, Bergel H, Bigalke H, Yamasaki S, Hayashi T, Niemann H, Pohlner J. J Biol Chem. 1995;270:1770–1774. doi: 10.1074/jbc.270.4.1770. [DOI] [PubMed] [Google Scholar]
- 25.Calakos N, Bennett M K, Peterson K E, Scheller R H. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- 26.Schiavo G, Shone C C, Bennett M K, Scheller R H, Montecucco C. J Biol Chem. 1995;270:10566–10570. doi: 10.1074/jbc.270.18.10566. [DOI] [PubMed] [Google Scholar]
- 27.Martin S, Tellam J, Livingstone C, Slot J W, Gould G W, James D E. J Cell Biol. 1996;134:625–635. doi: 10.1083/jcb.134.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holman G D, Leggio L L, Cushman S W. J Biol Chem. 1994;269:17516–17524. [PubMed] [Google Scholar]
- 29.Wong P P C, Wilson M C, Klip A. FASEB J. 1996;10:A536. (abstr.). [Google Scholar]
- 30.Ravichandran V, Chawla A, Roche P A. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa K, Rosen B S, Spiegelman B M, Lienhard G E, Tanner L I. Biochim Biophys Acta. 1989;1014:83–89. doi: 10.1016/0167-4889(89)90244-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman J M. Nature (London) 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]


