Abstract
Reduced growth and viability is a common phenotype of plants with constitutively activated pathogen defenses. One branch of the plant innate immunity system, effector-triggered immunity, is especially potent and requires tight control to enable normal plant development. While some facets of this control that directly regulate resistance protein abundance or activity have been documented, general control of effector-triggered signaling sensitivity is poorly understood. We recently identified SUPPRESSOR OF rps4-RLD 1 (SRFR1), a novel negative regulator of avrRps4-triggered immunity. Mutations in SRFR1 were previously shown not to induce constitutive high expression of the defense gene PR1, and to be fully susceptible to the virulent Pseudomonas syringae pv. tomato strain DC3000. SRFR1 encodes a tetratricopeptide repeat-containing protein with weak similarity to transcriptional repressors in other organisms. By transient expression in Nicotiana benthamiana, SRFR1 was localized to the nucleus. Here we investigate more carefully whether expression of defense genes is misregulated in srfr1 mutant plants. Consistent with the hypothesized function of SRFR1 as a negative transcriptional regulator, we find that mRNA levels of several defense genes are upregulated in srfr1 mutants.
Key words: Arabidopsis thaliana, Pseudomonas syringae, disease resistance, avrRps4, RPS4, transcriptional repressor
Effector-Triggered Immunity
Effector-triggered immunity (ETI), which relies on the direct or indirect detection of pathogen effector proteins by host resistance (R) proteins, can lead to localized programmed cell death called the hypersensitive response (HR).1–4 Even when ETI is not accompanied by HR, in Arabidopsis the detrimental effects of the resistance response can be evident as chlorosis.5 As a result, stunted growth and poor seed-set are common phenotypes associated with constitutive ETI responses. Consequently, the plant must fine-tune the response to pathogens by exerting tight positive and negative control.6
SRFR1 is a Novel Regulator of ETI
By performing a genetic suppressor screen, we previously identified mutants with specifically enhanced responses to the P. syringae effector protein AvrRps4. The Arabidopsis accession RLD is a natural mutant in the cognate R protein RPS4 and is fully susceptible to DC3000 expressing avrRps4.7,8 Mutations in RLD in a gene we called SUPPRESSOR OF rps4-RLD 1 (SRFR1) enhanced resistance to DC3000 (avrRps4), but not to virulent DC3000. The mutant srfr1 alleles were recessive, suggesting that genetically SRFR1 functions as a negative regulator of AvrRps4-triggered immunity.9
SRFR1 encodes a novel tetratricopeptide repeat-containing protein that is conserved between plants and animals. To date none of the proteins in other organisms have an assigned function. SRFR1 orthologs appear to be absent in Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster. While proteins that share amino acid sequence similarity over the whole length of SRFR1 are absent in these organisms, the SRFR1 TPR domain shares some sequence similarity with the TPR-containing transcriptional repressors ScSSN6 and CeOGT1. We therefore proposed that AtSRFR1 functions as a transcriptional repressor of plant defense genes that fine-tunes the Arabidopsis defense response.10
Altered Expression of Defense Genes in srfr1 Mutants
Initial characterization of srfr1 mutants by RNA gel blots showed that the defense gene PR1 is not constitutively upregulated, consistent with the absence of elevated resistance to virulent DC3000.9 After cloning of SRFR1, we quantified the expression of RPS4 in the mutants and determined that RPS4 mRNA levels, which are induced approximately 10-fold by avrRps4 in resistant wild-type plants,11 are approximately two-fold higher in uninduced srfr1 mutants compared to uninduced wild-type plants.10 This raised the possibility that other defense-related genes are also slightly upregulated in srfr1 mutants, albeit not highly enough to trigger constitutive activation of plant defenses. This would be consistent with our model that srfr1 mutants are closer to a threshold for defense activation.
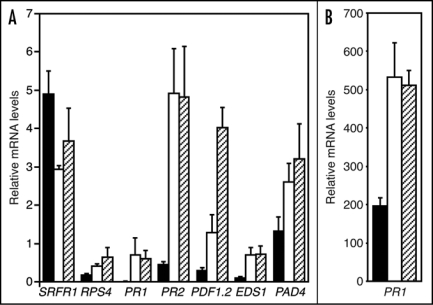
Several defense-related genes were indeed upregulated in uninduced srfr1-1 and srfr1-2 leaf tissue, including PR1 (Fig. 1A). PR1 expression has been reported to be induced over 1,000-fold in resistant plants upon pathogen inoculation,12 and the level of PR1 induction in uninduced srfr1 mutants was still considerably lower than in SA-induced tissue from a separate batch of wild-type or mutant plants (Fig. 1B). This perhaps explains why lower levels of PR1 induction in srfr1 mutants was previously not detected by RNA gel blot analysis.10
Figure 1.
Defense gene mRNA levels are higher in srfr1 mutants than in the wild-type. Total RNA was isolated from three biological replicates. mRNA levels were quantified by real-time quantitative reverse transcription PCR and were normalized using SAND gene (At2g28390) mRNA levels as an internal standard.10 Error bars denote standard error. (A) Defense gene mRNA levels in uninduced tissue of RLD wild-type (closed bars), srfr1-1 (open bars) and srfr1-2 (hatched bars). (B) PR1 mRNA levels 24 h after induction by spraying leaf tissue of RLD (closed bars), srfr1-1 (open bars) and srfr1-2 (hatched bars) with 1.5 mM SA. Note difference in scale. Primers used were: 5′-AAC TCT ATG CAG CAT TTG ATC CAC T-3′ and 5′-TGA TTG CAT ATC TTT ATC GCC ATC-3′ for SAND; 5′-CTG GAT ATG CCT CAC TAG AAG-3′ and 5′-CAC TGG GTC ACA AGG CTC TG-3′ for SRFR1 (At4g37460); 5′-CCT AAC ATT ATG GGC ATC ATC A-3′ and 5′-CCG CCT TCA CAA TTT CAT TGA-3′ for RPS4 (At5g45250); 5′-GCA ATG GAG TTT GTG GTC AC-3′ and 5′-GTT CAC ATA ATT CCC ACG AGG-3′ for PR1 (At2g14610); 5′-ATC TCC CTT GCT CGT GAA TC-3′ and 5′-GGA TCG TTA TCA ACA GTG GAC-3′ for PR2 (At3g57260); 5′-AAG TTG TGC GAG AAG CCA AG-3′ and 5′-CCA TGT TTG GCT CCT TCA AG-3′ for PDF1.2 (At5g44420); 5′-GAC GGG GAA GTA GAT GAG AAG-3′ and 5′-TCA TCC ATC ATA CGC TCA CG-3′ for EDS1 (At3g48080); and 5′-GAG GAG ATC TTT GTT ACG GG-3′ and 5′-TCG CCT CCC ACA CAC TAT AA-3′ for PAD4 (At3g52430).
In summary, we have found additional support for our model that SRFR1 function determines a set-point in the plant innate immune response system by negatively regulating defense gene expression levels. In this model, weak recognition of AvrRps4 in the natural RPS4 mutant RLD is not sufficient to trigger resistance because of the suppressive function of SRFR1. In srfr1-1 and srfr1-2 plants on the other hand, weak recognition of AvrRps4 is sufficient to exceed a threshold for resistance activation. Whether SRFR1 directly downregulates defense gene expression, or whether this regulation is indirect, awaits further study.
Acknowledgements
This work was supported by National Science Foundation grant IOS-0715926 to W.G., and in part by the Missouri Agricultural Experiment Station (project No. MO-PSSL0603).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7682
References
- 1.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Goodman RN, Novacky AJ. The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon. St. Paul, MN: APS Press; 1994. [Google Scholar]
- 4.Greenberg JT, Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 5.Gassmann W. Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol Plant-Microbe Interact. 2005;18:1054–1060. doi: 10.1094/MPMI-18-1054. [DOI] [PubMed] [Google Scholar]
- 6.McDowell JM, Simon SA. Recent insights into R gene evolution. Mol Plant Pathol. 2006;7:437–448. doi: 10.1111/j.1364-3703.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- 7.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 8.Hinsch M, Staskawicz BJ. Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol Plant-Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 9.Kwon SI, Koczan JM, Gassmann W. Two Arabidopsis srfr (suppressor of rps4-RLD) mutants exhibit avrRps4-specific disease resistance independent of RPS4. Plant J. 2004;40:366–375. doi: 10.1111/j.1365-313X.2004.02213.x. [DOI] [PubMed] [Google Scholar]
- 10.Kwon SI, Kim SH, Bhattacharjee S, Noh JJ, Gassmann W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 2009;57:109–119. doi: 10.1111/j.1365-313X.2008.03669.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X-C, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007;145:1577–1587. doi: 10.1104/pp.107.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao SY, Calis O, Patrick E, Zhang GG, Charoenwattana P, Muskett P, Parker JE, Turner JG. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005;42:95–110. doi: 10.1111/j.1365-313X.2005.02356.x. [DOI] [PubMed] [Google Scholar]