Abstract
Tamoxifen is a powerful drug used to treat breast cancer patients, and more than 500,000 women in the U. S. are being treated with this drug. In our study, tamoxifen is found to be photomutagenic in Salmonella typhimurium TA102 at concentrations as low as 0.08 μM and reaches maximum photomutagenicity at 0.4 μM under a light dose equivalent to 20 min sunlight. These concentrations are comparable to the plasma tamoxifen concentration of 0.4 to 3 μM for patients undergoing tamoxifen therapy. The toxicity seems to be the result of DNA damage and/or lipid peroxidation caused by light irradiation of tamoxifen. The DNA damage caused by irradiation of ΦX174 DNA in the presence of tamoxifen appears to be formation of DNA-tamoxifen covalent adducts, not single strand/double strand cleavages, and there is no oxygen involvement. This is confirmed by EPR experiments that carbon-centerd radicals are formed by light irradiation of tamoxifen and there is no singlet oxygen formation. Although superoxide radical is formed, it is not involved in DNA damage.
Keywords: Tamoxifen, phototoxicity, HaCaT keratinocytes, Salmonella TA102, DNA damage, Lipid peroxidation
1. Introduction
Breast cancer is the most common cancer in women in the United States, with an estimated 215,000 cases diagnosed in 2004 or one new case in two minutes [1]. Endocrine therapy is the standard care for most women with hormone receptor-positive tumors in adjuvant and metastatic setting. Tamoxifen, trans-2-[4-(1,2-diphenyl-1-butenyl)phenoxy]-N,N-dimethylethylamine, or Nolvade by brand name, is a powerful drug used to treat breast cancer patients, and more then 500,000 women in the US are being treated with this drug. Tamoxifen is a member of the drug class called selective estrogen response modifiers, which compete with estrogen for binding to the estrogen receptor. Tamoxifen therefore acts by blocking (antagonizing) the proliferative effects of estrogen to stop or slow down tumor growth.
According to recent studies, long-term treatment with tamoxifen significantly increases women’s endometrial cancer [2–4]. DNA-tamoxifen adducts have been found in rat liver and in the endometrial tissue of certain women undergoing tamoxifen therapy [5–7]. It is not known if the induction of endometrial cancer in women is through DNA adducts or the estrogenic nature of the drug [8]. Gene mutation assays of tamoxifen in bacteria as well as assays for chromosome aberrations in mammalian cells are routinely performed [9]. The major class of mutation induced by tamoxifen in the cII gene was G:C→T:A transversion [10]. Mutation frequency at the lacI gene locus in the livers of dosed female lambda/lacI transgenic rats increased 4-fold when the rats were fed with food containing tamoxifen (20 mg/kg per day) for 6 weeks compared with control [11]. In contrast, tamoxifen or its derivatives are not mutagenic in Salmonella TA97a, TA98, TA100, and TA102 strains, and show protective effects at certain concentrations in either plate or pre-incubation tests [12].
Many light-absorbing chemicals are toxic to living systems when exposed to both light and chemical at the same time [13, 14]. Tamoxifen has a C=C double bond linking three benzene rings (Figure 1). It adsorbs light in the solar irradiation’s UVB region (280–320 nm) extending to short UVA region (320–400 nm). Upon absorption of sunlight, the higher energy state tamoxifen may undergo various reactions leading to toxicity. Possible photochemical reaction products of tamoxifen have been isolated and identified as isomerization to the cis-isomer, ring cyclization to form two isomeric substituted phenanthrenes, and oxidation products of the C=C double bonds [15–18]. The photoproducts are usually less toxic than the parent tamoxifen for aquatic organisms of breast cancer cell line MCF-7 [15, 18]. However, several recent publications report that various skin sensitivities including phototoxicity have been observed for patients undergoing tamoxifen therapy [14, 15, 19–21]. Since the photoproducts are less toxic than the parent tamoxifen, it is likely that reactive intermediates are generated during the phototransformation process of tamoxifen that causes toxicity. Therefore, we wish to report the study on phototoxicity including photomutagenicity of tamoxifen in Salmonella typhimurium TA102 and human skin HaCaT keratinocyte, a transformed human epidermal cell line, and provide mechanistic insights on light-induced generation of reactive intermediates.
Figure 1.
Tamoxifen, trans-2-[4-(1,2-diphenyl-1-butenyl)phenoxy]-N,N-dimethylethylamine.
2. Methods and Materials
2.1 Material
Tamoxifen, NaN3, 2-mercaptoethanol, Superoxide Dismutase (SOD), histidine, D2O (>99% D), KI, α-(4-Pyridyl-1-oxide)-N-tert-butylnitrone (POBN) for trapping carbon-centered radicals [22], and 2,2,6,6-Tetramethyl-piperidine (TEMP) for trapping singlet oxygen [23] were purchased from Sigma-Aldrich Chemical Company and used without further purification (St. Louis, MO). The nitrone spin trap for superoxide radicals, 5-tert-butoxycarbonyl-5-methyl-1-pyrroline N-oxide (BMPO), was a gift from Prof. B Kalyanaraman (Medical College of Wisconsin) [24]. Salmonella typhimurium TA102 was kindly provided by Dr. Bruce Ames (University of California, Berkeley). Rat liver S9, induced by Aroclor1254, was purchased from ICN Biochemical Company (Aurora, OH). HaCaT keratinocytes, a transformed human epidermal cell line, were obtained from Dr. Norbert Fusenig of the German Cancer Research Centre (Heidelberg, Germany).
2.2 Light sources
A 300-W Xe lamp from ORIEL Instruments (Stratford, CT) producing a simulated solar light, was used for photomutagenicity, photocytotoxicity in HaCaT cells, and DNA cleavage experiments. The average light intensity was 7.3 mW/cm2 for visible, 3.6 mW/cm2 for UVA, and 11.6 μW/cm2 for UVB. This intensity matches well with local sunlight intensity at 11:00 am on a clear day during February or July in Jackson, Mississippi, USA (11–15 mW/cm2 visible and 3.1–5.0 mW/cm2 UVA). A custom made UVA light box with a 4-lamp unit using UVA lamps (National Biologics) was used for peroxidation essays. The irradiance of light was determined using an Optronics OL754 Spectroradiometer (Optronics Laboratories, Orlando, FL), and the light dose was routinely measured using a Solar Light PMA-2110 UVA detector (Solar Light Inc., Philadelphia, PA). The maximum emission of the UVA is between 340 – 355 nm. The light intensities at wavelengths below 320 nm (UVB light) and above 400 nm (visible light) are about two orders of magnitude lower than the maximum at 340–355 nm.
2.3 Bacteria photomutagenicity and viability assays
Photomutagenicity test was conducted based on the bacteria mutagenicity test developed by Maron and Ames [25] with modification [26, 27]. Bacteria viability was a modified procedure described in previous publications [28, 29].
2.4 Phototoxicity test in human skin HaCaT keratinocyte cells
The cell culture method was reported in a previous publication [30, 31]. After the HaCaT cells grew to a concentration of no less than 1×105 cells/mL, the cells were harvested and centrifuged at 2000 rpm for 5 min. The supernatant was discarded and the pellet was washed twice with 1× PBS. Finally the pellet was resuspended in 1× PBS to reach a cell concentration of approximately 5×105 cells/mL. The cell suspension was placed in a 96-well plate with 100 μL in each well. Then 100 μL of a given TAM concentration in 1× PBS was added through a serial dilution of the freshly prepared 0.11 mM tamoxifen stock solution in DMSO. The final concentration of DMSO was 2.3%. Two sets of 96-well plates were used with one covered with aluminum foil as the dark control, while the other was irradiated for 20 min (UVA 4.6 J/cm2 and visible light 8.4 J/cm2). Among the nine wells at each tamoxifen concentration, the cells in three wells were taken out for Comet assay and the remaining six for cell viability assay.
2.5 Cell viability
The six wells for cell viability assay received 100 μL of fluoroscein diacetate (FDA, 10 ng/mL) in each well and incubated at 37°C for 35 min. The plates were then read immediately using a Fluoroscan Ascent FL (Thermo Labsystems) with filters set at an excitation wavelength of 485 nm and emission wavelength of 538 nm. All experiments were in triplicates.
2.6 UV light–induced DNA damage by tamoxifen
Light-induced DNA damage by tamoxifen was examined with supercoiled plasmid DNA (Promega) in 10% methanol buffered with PBS at 7.1 [32]. Briefly, supercoiled ΦX174 phage DNA (27 μM in base pairs, Sigma-Aldrich, St. Louis) was mixed with TAM (0–200 μM) to make a total of 60 μL of solution. These solutions were placed in the wells of a 3×8 Titertek plate (ICN Biochemicals). The plate was irradiated for 20 min. After irradiation, each sample was mixed with 12.5 μL of a dye solution (bromophenol blue and xylene cyanole in 50% glycerol) and 13 μL of the mixture was loaded onto a preprepared 1% agarose gel. After gel-electrophoresis, the DNA was stained with ethidium bromide and visualized with a gel documentation system.
2.7 Lipid peroxidation
Experiments were conducted with a solution of 100 mM methyl linoleate and 0.2 mM of tamoxifen in methanol. Samples were placed in a UV-transparent cuvette and irradiated with 0, 14, 35, and 70 J/cm2 of UVA light. After irradiation, the methyl linoleate hydroperoxide products were separated by HPLC using a Prodigy 5 m ODS column (4.6 × 250 mm, Phenomenex, Torrance, CA) eluted isocratically with 10% water in methanol (v/v) at 1 mL/min. The extent of lipid peroxides formed by the photo-irradiation of methyl linoleate in the absence and presence of tamoxifen was determined by calculating the amount of methyl linoleate-hydroperoxides detected in resolved HPLC peak areas detected at 235 nm as previously described [33–35].
2.8 Detection of reactive intermediates using ESR
Superoxide radical generation by tamoxifen under UV irradiation (330 nm or 280 nm) was spin trapped with BMPO. Samples containing 25 mM BMPO in 50% ethanol in two 50 μL quartz capillaries were used. The UVA light was provided by a Schoeffel 1000 W Xenon lamp coupled with a Schoeffel grating monochromator. The excitation light had a maximum centered at 389 nm. All experiments were performed in duplicate. The data were obtained with error of less than 10%. Conventional ESR spectra were obtained with a Varian E-109 X-band spectrometer. ESR signals were recorded with 15 mW incident microwave and 100 kHz field modulation of 1G. The scan width was 100 G for TEMP experiments. All measurements were performed at room temperature.
2.9 Biostatistical Analysis
Differences between light exposure and control groups were performed by one-way analysis of variance (ANOVA; The SAS System for windows, version 8, SAS Institute, Gary, NC, USA). Means were separated by Tukey’s test. Differences at p< 0.05 are considered significant.
3. Results
3.1 Phototoxicity and photomutagenicity of tamoxifen in Salmonella TA102
Salmonella TA102, in the presence of tamoxifen with concentrations of 0, 0.5, 1.0, 1.5, 2.0 μM or 0, 0.2, 0.5, 1.0, 2.0, 5.0, and 25 μM, was irradiated for 5 or 20 min with the 300-W Xe lamp. Bacteria viability is shown in Figure 2. In the concentration range of 0–2 μM, there was a slight increase in bacteria viability. However, at 25 μM, 99% of bacteria death was observed for both light doses and 80% of bacteria death for that without light irradiation. These results indicate that tamoxifen is toxic to TA102 at concentrations >5 μM with or without light irradiation, but the toxicity is greater in the presence of light. At concentrations lower than 5 μM, tamoxifen is not phototoxic. Actually, there is a slight bacteria growth in this concentration range. This is also observed that low dose tamoxifen can stimulate the breast cell growth and high dose tamoxifen is toxic to the cells [36].
Figure 2.
Salmonella typhimurium TA102 survival in the presence of tamoxifen with or without light irradiation. Light dose were 1.2 J/cm2 UVA and 2.1 J/cm2 visible (5 min) and 4.6 J/cm2 UVA and 8.5 J/cm2 of visible light (20 min). Bacteria colonies were grown in Petri dishes with nutrient agar. All data points were the average reading from three Petri dishes.
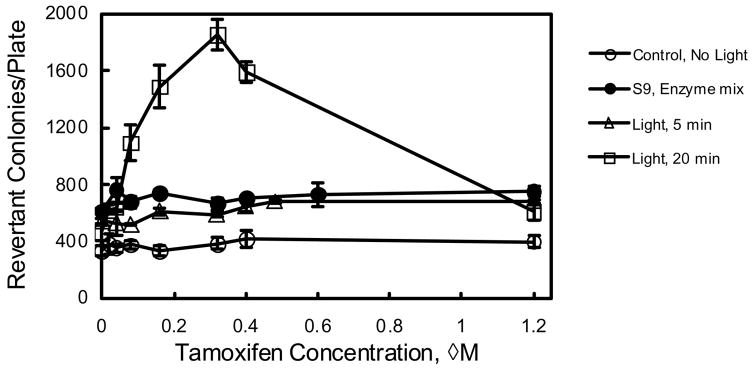
Thus, the photomutagenicity of tamoxifen in the concentration range of 0 to 1.2 μM, where it is not phototoxic, was examined. These data are presented in Table 1 and Figure 3. Without light irradiation, tamoxifen is not mutagenic (lowest line in Figure 3). With 5 min irradiation or with the S9 mix (2 mg/plate), the number of revertant colonies caused by these treatments were about twice as much as the negative control caused by spontaneous mutation (two lines in the middle of Figure 3). This indicates that TAM is mutagenic with the S9 mix and photomutagenic with 5 min light irradiation. With 20 min light irradiation, the number of revertant colonies due to exposure to 0.32 μM tamoxifen is 5.6 time of the negative control and 4 time of the light irradiation control (first point of the top line in Figure 3). This mutagenic effect is seen at concentrations as low as 0.08 μM, where there is still 2.3-time the number of revertant colonies than the control with light irradiation. The 5-fold increase in the number of revertant colonies indicates that tamoxifen is strongly photomutagenic in TA 102 with 20-min irradiation at concentrations greater than 0.08 μM. At concentrations greater than 0.32 μM, the number of revertant colonies decreased with the increase of tamoxifen concentration, indicating that some of the revertant bacteria could not survive the toxicity of > 0.32 μM tamoxifen and 20 min light.
Table 1.
Number of revertant colonies per plate for Salmonella TA 102 underwent various treatments*
| Tamoxifen (μM) | Control | With S9 | 3.3 J/cm2 (5 min) | 13.1 J/cm2 (20 min) |
|---|---|---|---|---|
| 0 | 303 ± 14 | 486 ± 16 | 545 ± 21 | 463 ± 56 |
| 0.04 | 403 ± 48 | 665 ± 8 | 527 ± 92 | 623 ± 29 |
| 0.08 | 399 ± 73 | 619 ± 14 | 519 ± 11 | 637 ± 29 |
| 0.16 | 426 ± 50 | 710 ± 21 | 610 ± 26 | 1095 ± 128 |
| 0.32 | 413 ± 28 | 720 ± 35 | 591 ± 17 | 1490 ± 148 |
| 0.4 | 417 ± 44 | 715 ± 14 | 648 ± 37 | 1856 ± 109 |
| 0.6 | 422 ± 40 | 697 ± 40 | 682 ± 16 | 1590 ± 76 |
| 1.2 | 413 ± 14 | 770 ± 28 | 682 ± 16 | 605± 50 |
Experiment was in duplicate
Figure 3.
Photomutagenicity of tamoxifen with Salmonella typhimurium TA102 with 1.2 J/cm2 UVA and 2.1 J/cm2 visible (5 min) and 4.6 J/cm2 UVA and 8.5 J/cm2 of visible light (20 min) irradiation or in the presence of S9 mix. The points are the average reading of three parallel experiments.
3.2 Photocytotoxicity and photogenotoxicity of Tamoxifen in human skin keratinocytes
The viability of HaCaT cells was examined by measuring the fluorescence intensity of FDA taken into the cells. Human HaCaT cells were treated with tamoxifen at concentrations of 0, 0.04, 0.08, 0.16, 0.32, 0.4, 0.6, and 1.2 μM under both light doses, 1.2 J/cm2 UVA and 2.1 J/cm2 visible (5 min) and 4.6 J/cm2 UVA and 8.5 J/cm2 of visible light (20 min). There is no observable photocytotoxicity under these test conditions (data not shown).
3.3 Light-induced damage of plasmid DNA by tamoxifen
Irradiation of ΦX174 DNA solution in the presence of tamoxifen causes damage to the DNA. As can be seen in Figure 4 (left panel), ΦX174 DNA has two bands, and the intensity ratio of the lower band (supercoiled) versus the upper band (open circular) is about 9:1. As can be seen, there is no DNA damage with up to 50 μM tamoxifen. At higher concentrations, the intensity of both bands decreases and the DNA begins to spread out over the lane, but mostly moving slower than the original DNA. At the two highest concentrations, some of the DNA accumulates at the gel origin (lanes 9 and 10 left panel). Longer irradiation up to 40–60 min (lanes 6–8 right panel), DNA accumulated at the origin also spreads out. This indicates that there appears no single strand DNA cleavage which supposes to convert the supercoiled form to the open circular form as shown before [37]. It seems like that the damaged DNA has greater molecular weight or less negative charge. Both of which can be achieved by formation of DNA-tamoxifen covalent adducts. DNA-tamoxifen adducts have greater molecular weight and at the same time, less negative charge since the dimethylamino group in tamoxifen is positively charged in the test solution. Another possible damage would be DNA-DNA crosslinks. The fact that at the highest concentration and longest irradiation time, the DNA accumulated near the gel origin also disappears, indicating DNA fragmentation occurred.
Figure 4.
Damage of ΦX-174 supercoiled plasmid DNA (27 μM in base pairs) caused by light-irradiation of tamoxifen (TAM) in solution. Left: concentration dependence experiment; Right: irradiation time dependence experiment. Light dose was 4.6 J/cm2 of UVA and 8.5 J/cm2 of visible light.
3.4 Effect of scavengers on DNA damage induced by tamoxifen and light irradiation
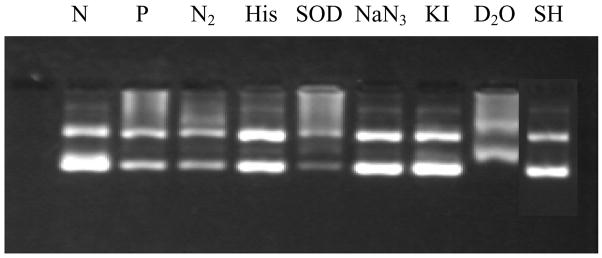
To find out which reactive species is involved in causing the light-induced DNA damage by tamoxifen, these experiments were carried out in the presence of several scavengers (Figure 5). The effects by scavengers on DNA damage are listed in Table 2. Each lane is compared with the positive control (P). As can be seen in Figure 5, the presence of histidine, NaN3, KI or 2-mercaptoethanol caused significant decrease in DNA damage. In the absence of oxygen (purged with nitrogen), in the presence of SOD, or in D2O, there is almost no effect. These results indicate that oxygen is not likely involved in DNA photodamage by tamoxifen.
Figure 5.
Effect of scavengers on light induced DNA damage by tamoxifen (200 μM). All experiments were tested with 27 μM ΦX174 DNA and irradiated for 20 min by 300 W Xe lamp except the negative control. Lane N: negative controls in the dark; lane P: positive control with tamoxifen and irradiation; lanes 3, 4, 5, 6, 7, and 8 are in the presence of nitrogen (N2), 50 mM histidine (His), 200 units SOD, 50 mM NaN3, 50 mM KI, 100% D2O, and 1.0 M mercaptoethanol (SH).
Table 2.
Effect of scavengers on DNA photodamage by tamoxifen
| Experiment | Reactive Species Affected | Effect on DNA damage |
|---|---|---|
| Nitrogen Purged | O2 | no effect |
| Histidine, 50 mM | OH•/1O2 | decrease |
| SOD, 200 units/mL | O2•− | no effect |
| NaN3, 50 mM | 1O2 | decrease |
| KI, 50 mM | excited singlet state | decrease |
| D2O, 100% | enhance 1O2 lifetime | no effect |
| 2-Mercaptoethanol, 1.0 M | carbon centered radical | decrease |
3.5 Induction of lipid peroxidation
With UVA light irradiation of tamoxifen solution in the presence of methyl linoleate, lipid perxidation is studied with light doses of 14, 35, and 70 J/cm2 (Table 3). At a low light dose of 14 J/cm2, the presence of tamoxifen caused 2.7 times more peroxides. However, at the highest dose of 70 J/cm2, the amount of lipid peroxide is about the same with or without of tamoxifen.
Table 3.
Lipid peroxidation induced by UVA light irradiation of tamoxifen (200 μM)
| 0 J/cm2 | 14 J/cm2 | 35 J/cm2 | 70 J/cm2 | |
|---|---|---|---|---|
| Methyl linoleate (ML) | 178, 192 | 282, 291 | 496, 514 | 1061, 1109 |
| ML + Tamoxifen | 193, 207 | 721, 784 | 827, 881 | 1093, 1118 |
3.6 Light-induced formation of reactive intermediates detected by ESR
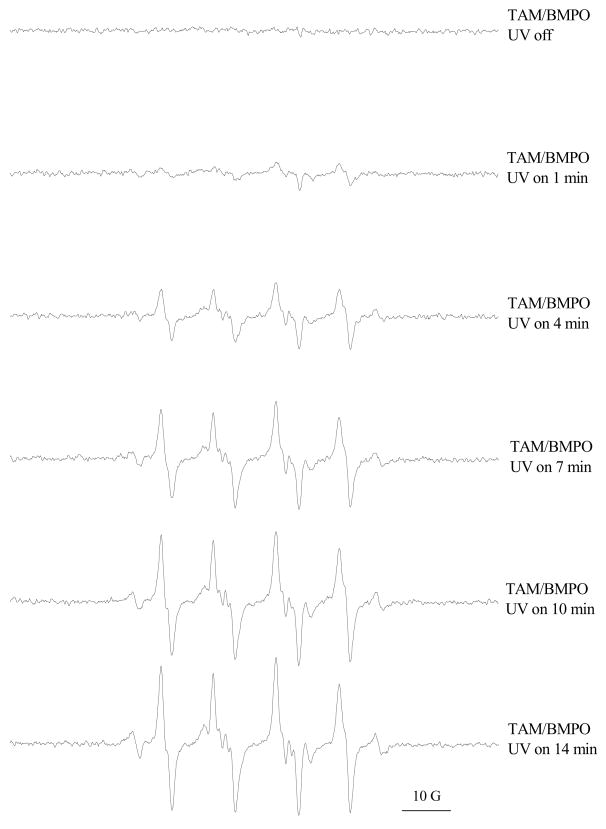
We tested the possible formation of three reactive intermediates with ESR: superoxide, singlet oxygen, and carbon-based radicals. Based on the results obtained, superoxide and carbon-based radicals are both formed, while singlet oxygen is not detected. (1) Superoxide: Irradiation with 330 nm light on the tamoxifen solution generates superoxide that can be trapped by BMPO (Figure 6). The amount of superoxide generated increases with the increasing concentration of tamoxifen in the range of 0–5 mM. The presence of 300 units/mL of SOD efficiently removed all the signals, indicating superoxide is indeed formed. Using 280 nm light seems to generate less superoxide compared with the 330 nm light. Figure 7 shows the irradiation time dependence for the formation of superoxide. As irradiation time (light dose) increases from 1 min to 14 min, the amount of superoxide generated increases linearly. (2) Singlet oxygen: TEMP is a specific probe for trapping singlet oxygen. The results indicated that photoirradiation of tamoxifen by UVA (330 nm) or UVB (280 nm) did not generate singlet oxygen (data not shown). (3) Carbon-centered radicals: POBN was used to trap carbon-centered radicals. As seen in Figure 8, UV irradiation of tamoxifen solution produces carbon-centered radicals. The amount of carbon-centered radicals formed is about the same whether irradiated for 10, 20, or 30 min.
Figure 6.
Superoxide radical generation by tamoxifen (TAM) by UV (330 nm or 280 nm) irradiation with spin trap BMPO. Samples contained 25 mM BMPO in 50% ethanol.
Figure 7.
Time dependence of superoxide generation by tamoxifen (TAM) under UV (330 nm) irradiation. Samples contained 10 mM Tamoxifen and 20 mM BMPO in 50% ethanol at room temperature.
Figure 8.
Time-dependent formation of carbon-centered radicals induced by UV (330 nm) irradiation of tamoxifen (TAM, 5 mM) in solution. The radicals were trapped with PBN.
4. Discussions
The light-induced toxicity results presented here clearly show that tamoxifen is photomutagenic in Salmonella typhimurium TA102 at doses as low as 0.08 μM (Figure 3 and Table 1), but is not acutely phototoxic to the human skin cells at concentrations below 5 μM. Light irradiation of tamoxifen generates superoxide and carbon-centered radicals, but not singlet oxygen (Figures 6–8). The observation of superoxide, but not singlet oxygen, through photoirradiation of tamoxifen agrees with the result reported by Onoue and Tsuda in their test of 26 pharmaceuticals [14]. The reactive intermediates cause damage to plasmid DNA and produce lipid peroxidation. The DNA damages are likely some forms of DNA-tamoxifen covalent adduct since the damaged DNA have greater molecular weight and move slower in the gel than the original DNA (Figure 4). Degassing with nitrogen, carrying out the experiment in D2O (lengthening 1O2 lifetime), or in the presence of SOD, all do not affect the damage, indicating oxygen is not involved (Figure 5). Although the EPR study indicates that superoxide radical is formed when tamoxifen is irradiated, superoxide is not involved in producing DNA damages, but may be involved in lipid peroxidation (Table 3). The scavenging effect by KI (excited singlet state) indicates that the DNA damage is a result of the excited state reaction of tamoxifen. Normally, histidine and NaN3 are used as singlet oxygen scavengers [37, 38]; however, their scavenging of the DNA damage shown here is probably not due to reaction with singlet oxygen, but may be due to the scavenging effect on a carbon-centered radical of tamoxifen. Histidine and NaN3 can also react with free radicals in addition to singlet oxygen [39–41]. EPR study clearly indicates the formation of carbon-centered radicals upon light irradiation of tamoxifen (Figures 6–8). A further proof of the involvement of carbon-centered radicals in DNA damage is the scavenging effect by 2-mercaptoethanol (Figure 5 lane SH), a widely used carbon-centered radical scavenger [42, 43].
Due to the involvement of carbon-centered radicals for DNA damage, it is likely some forms of DNA-tamoxifen covalent adduct is formed, shown by the smeared and slower moving bands of the DNA-tamoxifen adducts (Figure 4). The more tamoxifen molecules covalent linked to DNA, the greater the molecular weight and the slower the DNA-tamoxifen moves in the gel. Another possibility for the slower moving bands may be due to lower negative charge for DNA-tamoxifen adduct. Tamoxifen is positively charged on the dimethylamino group at neutral pHs.
The positively charged tamoxifen will cancel out the negative charge on the phosphate backbone of DNA, and thus causing the DNA-tamoxifen to move slower. It is known that tamoxifen can bind DNA non-covalently [44, 45]; and the DNA-tamoxifen non-covalent complex can facilitate the formation of DNA-tamoxifen covalent adducts upon light irradiation. However, the non-covalently bound DNA-tamoxifen complex seems to be very weak since they were not observed during electrophoresis as shown in lanes 3–7 in Figure 4 (left).
Formation of DNA-tamoxifen covalent adducts in cells or patients treated with tamoxifen have been widely studied [2–8, 36, 46–49]. The pathway of the formation of DNA-tamoxifen adduct is through the metabolism of tamoxifen to α-hydroxytamoxifen intermediate. The major DNA-tamoxifen adduct is therefore α-(N2-deoxyguanosyl)tamoxifen and it is believed to be responsible for the carcinogenicity of tamoxifen. In general, it was demonstrated metabolic activation and photolysis produce different DNA adducts as it was the case for DNA adducts formed with 7,12-dimethylbenz[a]anthracene through metabolism and photolysis [50]. Therefore, determination of light-induced DNA-tamoxifen adducts will possibly require a different set of standards and it is beyond the scope of the present paper.
The tamoxifen concentration for photomutagenicity is within the range of plasma tamoxifen concentration for patients treated with this drug. Plasma concentrations of tamoxifen and its metabolites are 0.40 to 0.65 μM in breast cancer patients with standard oral dose of 20 mg/day [51, 52] or 2.96 ± 1.32 μM in prostate cancer patients with orally administered 160 mg/m2/day [53]. The half-life of tamoxifen in human body is 7 days [54]. In another test, patients given tamoxifen tablets (20 mg/day) had a maximum plasma tamoxifen level of 95 μM and the metabolite desmethyltamoxifen at about 30 μM [55]. Tamoxifen had a half life of about 5 days while desmethyltamoxifen had a much longer half life of 22 days. This means that after a patient is administered with tamoxifen and goes in the sunlight outdoors, the patient may be at risk of light-induced mutagenic effects of tamoxifen.
Acknowledgments
This research was in part supported by the National Institutes of Health: NIH SCORE S06 GM08047. We thank NIH-RCMI for Core Molecular and Cellular Biology Facility and Analytical Chemistry Facility established at JSU.
Footnotes
This article is not an official U.S. Food and Drug Administration guidance or policy statement. No official support or endorsement by the U.S. Food and Drug Administration is intended or should be inferred.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harwood KV. Advances in endocrine therapy for breast cancer: considering efficacy, safety, and quality of life. Clin J Oncology. 2004;8:629–637. doi: 10.1188/04.CJON.629-637. [DOI] [PubMed] [Google Scholar]
- 2.Shibutani S, Ravindernath A, Suzuki M, Terashima I, Sugarman SM, Grollman AP, Paearl ML. Identification of tamoxifen-DNA adducts in the endometrium of women treated with tamoxifen. Carcinogenesis. 2000;21:1461–1467. [PubMed] [Google Scholar]
- 3.Shibutani S, Reardon JT, Suzuki M, Sancar A. Excision of tamoxifen-DNA adducts by the human nucleotide excision repair system. Cancer Res. 2000;60:2607–2610. [PubMed] [Google Scholar]
- 4.Glatt H, Davis W, Meinl W, Hermersdörfer H, Venitt S, Phillips DH. Rat, but not human, sulfotransferase activates a tamoxifen metabolite to prodice DNA adducts and gene mutations in bacteria and mammalian cells in culture. Carcinogenesis. 1998;19:1709–1713. doi: 10.1093/carcin/19.10.1709. [DOI] [PubMed] [Google Scholar]
- 5.Divi RL, Osborne MR, Hewer A, Phillips DH, Poirier MC. Tamoxifen-DNA adduct formation in rat liver determined by immunoassay and 32P-Postlabeling. Cancer Res. 1999;59:4829–4833. [PubMed] [Google Scholar]
- 6.Hellmann-Blumberg U, Cartner MG, Wurz GT, DeGregorio MW. Intrinsic reactivity of tamoxifen and toremifene metabolites with DNA. Breast Cancer Res Treat. 1998;50:135–141. doi: 10.1023/a:1006002324995. [DOI] [PubMed] [Google Scholar]
- 7.Rajaniemi H, Koskinen M, Mantuyla E, Hemminski K. DNA binding of tamoxifen and its analogues: Identification of the tamoxifen-DNA adducts in rat liver. Toxicol Lett. 1998;102–103:453–457. doi: 10.1016/s0378-4274(98)00338-5. [DOI] [PubMed] [Google Scholar]
- 8.McLuckie KIE, Routledge MN, Brown K, Gaskell M, Farmer PB, Roberts GCK, Martin EA. DNA adducts formed from 4-hydroxytamoxifen are more mutagenic than those formed by α-acetoxytamoxifen in a shuttle vector target gene replicated in human AD293 cells. Biochemistry. 2002;41:8899–8906. doi: 10.1021/bi025575i. [DOI] [PubMed] [Google Scholar]
- 9.Willems MI, van Benthem J. National Institute of Public Health and the Environment (RIVM) Bilthoven. Netherlands Organization for Applied for Scientific Research (TNO) Zeist; AI Zeist, The Netherlands: 2000. pp. 1–47. [Google Scholar]
- 10.Davies R, Gant TW, Smoith LL, Styles JA. Tamoxifen induces G:C→T:A mutations in the cII gene in the liver of lambda/lacI transgenic rats but not at 5′-CpG-3′ dinucleotide sequences as found in the lacI transgene. Carcinogenesis. 1999;20:1351–1356. doi: 10.1093/carcin/20.7.1351. [DOI] [PubMed] [Google Scholar]
- 11.Styles JA, Davies R, Fenwick S, Walker J, White INH, Smith LL. Tamoxifen mutagenesis and carcinogenesis in livers of lambda/lacI transgenic rats: selective influence of phenobarbital promotion. Cancer Lett. 2001;162:117–122. doi: 10.1016/s0304-3835(00)00627-3. [DOI] [PubMed] [Google Scholar]
- 12.Gupta S, Mukhopadhyay A, Ray S, Giri AK. Comparative antimutagenic effects of D- and L-centchroman and their comparison with tamoxifen in Salmonella assay. Mutat Res. 1999;445:1–8. doi: 10.1016/s1383-5718(99)00148-5. [DOI] [PubMed] [Google Scholar]
- 13.Yu H. Environmental carcinogenic polycyclic aromatic hydrocarbons: photochemistry and phototoxicity. J Environ Sci & Health, Part C-Environ Carcinog & Ecotoxic Revs. 2002;C20:149–183. doi: 10.1081/GNC-120016203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onoue S, Tsuda Y. Analytical studies on the prediction of photosensitive/phototoxic potential of pharmaceutical substances. Pharm Res. 2006;23:156–164. doi: 10.1007/s11095-005-8497-9. [DOI] [PubMed] [Google Scholar]
- 15.DellaGreca M, Iesce MR, Isidori M, Nardelli A, Previtera L, Rubino M. Phototransformation products of tamoxifen by sunlight in water. Toxicity of the drug and its derivatives on aquatic organisms. Chemosphere. 2007;67:1933–1939. doi: 10.1016/j.chemosphere.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Salamoun J, Macka M, Nechvatal M, Matousek M, Knesel L. Identification of products formed during UV irradiation of tamoxifen and their use for fluorescence detection in high-performance liquid chromatography. J Chromatogr. 1990;514:179–187. doi: 10.1016/s0021-9673(01)89389-4. [DOI] [PubMed] [Google Scholar]
- 17.Campeta AM, Lombardo F, Sharp TS, Horan GJ, Rescek DM. Identification of photodegradants of droloxifene by combined HPLC-MS, NMR spectroscopy and computational chemistry. J Phys Org Chem. 1999;12:881–889. [Google Scholar]
- 18.Wilson S, Ruenitz PC. Structural characterization and biological effects of photocyclized products of tamoxifen irradiation. J Pharm Sci. 1993;82:571–574. doi: 10.1002/jps.2600820605. [DOI] [PubMed] [Google Scholar]
- 19.Tajima M, Saito M, Kato Y, Oh-I T, Tsuboi R. A case of male breast cancer involving a photosensitive reaction induced by doxifluridine. Nishinihon J Dertmat. 2005;67:475–477. [Google Scholar]
- 20.Bostrom A, Sjolin-Forsberg G, Wilking N, Bergh J. Radiation recall: Another call with tamoxifen. Acta Oncologica. 1999;38:955–959. doi: 10.1080/028418699432653. [DOI] [PubMed] [Google Scholar]
- 21.Jhaveri K, Halperin P, Shin SJ. Erythema nodosum secondary to aromatase inhibitor use in breast cancer patients: case reports and review of the literature. Breast Cancer Res Treat. 2007;106:315–318. doi: 10.1007/s10549-007-9518-7. [DOI] [PubMed] [Google Scholar]
- 22.Janzen EG, Hare DL. Two decades of spin trapping. Adv Radic Chem. 1990;1:253. [Google Scholar]
- 23.Rinalducci S, Pedersen JZ, Zolla L. Formation of radical from signet oxygen produced during photoinhibition of isolated light-harvesting protein of photosystem II. Biochim Biophy Acta. 2004;1608:63–73. doi: 10.1016/j.bbabio.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Joseph J, Zhang H, Karoui H, Kalyanaraman B. Synthesis and biochemical applications of a solid cyclic nitrone spin trap: a relatively superior trap for detecting superoxide anions and glutathiyl radicals. Free Rad Biol Med. 2001;31:599–606. doi: 10.1016/s0891-5849(01)00619-0. [DOI] [PubMed] [Google Scholar]
- 25.Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Yan J, Fu PP, Parekh KA, Yu H. Photomutagenicity of cosmetic ingredient chemicals azulene and guaiazulene. Mutat Res. 2003;530:19–26. doi: 10.1016/s0027-5107(03)00131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Wang L, Fu PP, Yu H. Photomutagenicity of 16 polycyclic aromatic hydrocarbons from the US EPA priority pollutant list. Mutat Res. 2004;557:99–108. doi: 10.1016/j.mrgentox.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Hur E, Prager A, Green AP, Rosenthal I. pH dependence of the phototoxic and photomutagenic effects of chlorpromazine. Chem-Biol Interact. 1980;29:223–33. doi: 10.1016/0009-2797(80)90035-6. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Yan J, Hardy W, Mosley C, Wang S, Yu H. Light-induced mutagenicity in Salmonella TA102 and genotoxicity/cytotoxicity in human T-cells by 3,3′-dichlorobenzidiine: a chemical used in the manufacture of dyes and pigments and in tattoo inks. Toxicology. 2005;207:411–418. doi: 10.1016/j.tox.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng B, Hwang HM, Yu H, Ekunwe S. DNA damage produced in HaCat cells by combined fluoranthenne exposure and ultraviolet A irradiation. Environ Mol Mutagen. 2004;44:151–155. doi: 10.1002/em.20040. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Sheng YH, Feng M, Leszczynski J, Wang L, Tachikawa H, Yu H. Light-induced cytotoxicity of 16 polycyclic aromatic hydrocarbons on the US EPA Priority Pollutant List in human skin HaCaT keratinocytes: Relationship between phototoxicity and excited state properties. Environ Toxicol. 2007;22:318–327. doi: 10.1002/tox.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong S, Fu PP, Shirsat RN, Hwang HM, Leszczynski J, Yu H. UVA light-induced DNA cleavage by isomeric methybena[a]anthracenes. Chem Res Toxicol. 2002;15:400–407. doi: 10.1021/tx015567n. [DOI] [PubMed] [Google Scholar]
- 33.Xia Q, Yin JJ, Cherng SH, Wamer WG, Boudreau M, Howard PC, Fu PP. UVA photodecomposition of retinyl palmitate - formation of singlet oxygen and superoxide, and their role in induction of lipid peroxidation. Toxicol Lett. 2006;163:30–43. doi: 10.1016/j.toxlet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Takita M, Marita M. Identification of new geometric isomers of methyl linoleate hydroperoxide and their chromatographic behavior. Biosci Biotechnol Biochem. 2000;64:1044–6. doi: 10.1271/bbb.64.1044. [DOI] [PubMed] [Google Scholar]
- 35.Cherng SH, Xia Q, Blankenship LR, Freeman JP, Wamer WG, Howard PC, Fu PP. Photodecomposition of retinyl palmitate in ethanol by UVA light - formation of photodecomposition products, reactive oxygen species, and lipid peroxides. Chem Res Toxicol. 2005;18:129–138. doi: 10.1021/tx049807l. [DOI] [PubMed] [Google Scholar]
- 36.Majumdar SK, Valdellon JA, Brown KA. In vitro investigations on the toxicity and cell death induced by tamoxifen on two non-breast cancer cell types. J Biomed Biotech. 2001;1:99–107. doi: 10.1155/S1110724301000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong S, Hwang HM, Shi X, Holloway L, Yu H. UVA-induced DNA single-strand cleavage by 1-hydroxypyrene and formation of colvalent adducts between DNA and 1-hydroxypyrene. Chem Res Toxicol. 2000;13:585–593. doi: 10.1021/tx990199x. [DOI] [PubMed] [Google Scholar]
- 38.Agon VV, Bubb WA, Wright A, Hawkins CL, Davies MJ. Sensitizer-mediated photooxidation of histidine residues: Evidence for the formation of reactive side-chain peroxides. Free Rad Biol Med. 2006;40:698–710. doi: 10.1016/j.freeradbiomed.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 39.Pazos M, Anderson ML, Skibsted LH. Amino acid and protein scavenging of radicals generated by iron/hydrogenperoxide system: An electron spin resonance spin trapping study. J Agr Food Chem. 2006;54:10215–10221. doi: 10.1021/jf062134n. [DOI] [PubMed] [Google Scholar]
- 40.Saeed S, Fawthrop SA, Howell NK. Electron spin resonance (ESR) study on free radical transfer in fish lipid-protein interaction. J Sci Food Agr. 1999;79:1809–1816. [Google Scholar]
- 41.Park J, Troxel AB, Harvey RG, Penning TM. Polycyclic aromatic hydrocarbon (PAH) o-quinones produced by the Aldo-keto-reductases (AKRs) generate abasic sites, oxidized pyrimidines, and 8-oxo-dGuo via reactive oxygen species. Chem Res Toxicol. 2006;19:719–728. doi: 10.1021/tx0600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka K, Tokumaru S, Kojo S. Possible involvement of radical reactions in desialylation of LDL. FEBS Lett. 1997;413:202–204. doi: 10.1016/s0014-5793(97)00917-4. [DOI] [PubMed] [Google Scholar]
- 43.Graham MJ, Freier SM, Crooke RM, Ecker DJ, Masslova RN, Lesnik EA. Trtiium labeling of antisense oligonucleotides by exchange with tritiated water. Nucl Acids Res. 1993;21:2737–3743. doi: 10.1093/nar/21.16.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans E, Baskevitch PP, Rochefort H. Estrogen receptor-DNA interaction: Differences between activation by estrogen and antiestrogen. Eur J Biochem. 1982;128:185–191. [PubMed] [Google Scholar]
- 45.Snyder RD, Brown JE. Evidence for and role of the dimethylamino group in tamoxifen DNA intercalation in intact Chinese Hamster V79 cells. Drug Chem Toxicol. 2002;25:473–479. doi: 10.1081/dct-120014797. [DOI] [PubMed] [Google Scholar]
- 46.Wozniak K, Kolacinska A, Blasinska-Morawiec M, Morawiec-Bajda A, Morawiec Z, Zadrozny M, Blasiak J. The DNA-damaging potential of tamoxifen in breast cancer. Arch Toxicol. 2007;81:519–527. doi: 10.1007/s00204-007-0188-3. [DOI] [PubMed] [Google Scholar]
- 47.Terashima I, Suzuki M, Shibutani S. Mutagenic potential of α-(N2-deoxyguanosinyl)tamoxifen lesions, the major DNA adducts detected in endometrial tissues of patients treated with tamoxifen. Cancer Res. 1999;59:2091–2095. [PubMed] [Google Scholar]
- 48.Kim SY, Suzuki N, Laxmi YRS, McGarrigle BP, Olson JR, Sharma M, Sharma M, Shibutani S. Formation of tamoxifen-DNA adducts in human endometrial explants exposed to a-hydroxytamoxifen. Chem Res Toxicol. 2005;18:885–895. doi: 10.1021/tx050019l. [DOI] [PubMed] [Google Scholar]
- 49.Umemoto A, Monden Y, Lin C, Momen MA, Ueyama Y, Komaki K, Laxmi YRS, Shibutani S. Determination of tamoxifen-DNA adducts in leukocytes from breast cancer patients treated with tamoxifen. Chem Res Toxicol. 2004;17:1577–1583. doi: 10.1021/tx049930c. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Yan J, Jiao Y, Fu P. Photochemical reaction of 7,12-dimethylbenz[a]anthracene and formation of DNA covalent adducts. Int J Environ Res Public Health. 2005;2:114–122. doi: 10.3390/ijerph2005010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Nat Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 52.Decensi A, Gradini D, Guerrieri-Gonzaga A, Johansson H, Mananni B, Bonanni B, Sandri MT, Barreca A, Costa A, Robertson C, Lien EA. Effects of blood tamoxifen concentrations on surrogate biomakers in a trial of dose reduction in healthy women. J Clin Oncology. 1999;17:2633–2638. doi: 10.1200/JCO.1999.17.9.2633. [DOI] [PubMed] [Google Scholar]
- 53.Bergan RC, Reed E, Myers CE, Headlee D, Brawley O, Cho HK, Figg WD, Tompkins A, Linehan WM, Kohler D, Steinberg SM, Blagosklonny MV. A phase II study of high-dose tamoxifen in patients with hormone-refractory prostate cancer. Clin Cancer Res. 1999;5:2366–2373. [PubMed] [Google Scholar]
- 54.Kisanga ER, Gjerde J, Guerrieri-Gonzaga A, Pigatto F, Pesci-Feltri A, Robertson C, Serrano D, Pelosi G, Decensi A, Lien EA. Tamoxifen and metabolite concentrations in serum and breast cancer tissue during three dose regimens in a randomized preoperative trial. Clin Cancer Res. 2004;10:2336–2343. doi: 10.1158/1078-0432.ccr-03-0538. [DOI] [PubMed] [Google Scholar]
- 55.Nieder M, Jaeger H. Quantification of tamoxifen and N-desmethyltamoxifen in human plasma by high-performance liquid chromatography, photochemical reaction and fluorescence detection, and its application to biopharmaceutical investigation. J Chromatogr. 1987;413:207–217. doi: 10.1016/0378-4347(87)80228-1. [DOI] [PubMed] [Google Scholar]