Abstract
Neuronal activity regulates the development and maturation of excitatory and inhibitory synapses in the mammalian brain. Several recent studies have identified signalling networks within neurons that control excitatory synapse development. However, less is known about the molecular mechanisms that regulate the activity-dependent development of GABA (γ-aminobutyric acid)-releasing inhibitory synapses. Here we report the identification of a transcription factor, Npas4, that plays a role in the development of inhibitory synapses by regulating the expression of activity-dependent genes, which in turn control the number of GABA-releasing synapses that form on excitatory neurons. These findings demonstrate that the activity-dependent gene program regulates inhibitory synapse development, and suggest a new role for this program in controlling the homeostatic balance between synaptic excitation and inhibition.
Sensory experience controls multiple steps in the development and maturation of synapses in the mammalian brain1–4. Many of the effects of neuronal activity are mediated by the release of glutamate at excitatory synapses and the subsequent influx of calcium (Ca2+) into the postsynaptic neuron. This results in changes in the number and strength of synapses, a process that underlies learning and memory as well as animal behaviour.
Neurons in the central nervous system receive excitatory synaptic input from glutamatergic neurons and inhibitory input from GABA-releasing (GABAergic) interneurons, except during early development when the first GABAergic synapses are depolarizing and provide the excitatory drive critical for the subsequent development of glutamatergic synapses5. The proper balance between excitatory and inhibitory synapses is crucial for representation of sensory information6,7, execution of motor commands8,9 and higher-order cognitive functions10. Neurological disorders such as autism, schizophrenia and epilepsy are associated with an imbalance between excitatory and inhibitory synapses11–13. The number or strength of excitatory synapses can be modified in response to changes in activity, and the molecular mechanisms of these processes have been extensively investigated14–16. Less is known about the activity-dependent regulation of inhibitory synapses.
The density of inhibitory synapses in brain regions such as primary sensory cortex, hippocampus and cerebellum is regulated by the level of excitatory synaptic activity and sensory input17–22. In addition, initiation of the critical period for synaptic plasticity in the visual cortex is dependent on visual activity and strongly influenced by the maturation of inhibitory synapses23, suggesting that the activity-dependent regulation of GABAergic synapses is important for the plasticity of the nervous system. Finally, recent studies indicate that regulation of GABAergic synapses in response to neuronal activity may be a critical component of the homeostatic mechanism that maintains a balance between excitation and inhibition in the face of fluctuations in the level of sensory input into neural circuits24. Despite the accumulating evidence that neuronal activity regulates the development and maintenance of inhibitory synapses, the molecular mechanisms that control these processes remain to be characterized.
Here we identify a transcription factor, Npas4, that is critical for activity-dependent regulation of GABAergic synapse development. Npas4 expression is rapidly activated by excitatory synaptic activity and turns on a program of gene expression that triggers the formation and/or maintenance of inhibitory synapses on excitatory neurons. These findings provide a molecular link between neuronal excitation and GABAergic synapse development, and suggest a new role for the activity-dependent gene program in controlling inhibitory synapse formation/maintenance on excitatory neurons.
Npas4 is regulated by neuronal activity
The formation of inhibitory synapses onto excitatory neurons is regulated by neuronal activity, takes place over several days, and is a cell-wide process that results in the formation of synapses onto both the cell body and dendrites18,25. These features led us to hypothesize that activity-dependent development of inhibitory synapses might be controlled postsynaptically by one or more activity-regulated genes. To test this hypothesis, we used DNA microarrays to identify genes that are induced by membrane depolarization in mouse cortical neurons at the time when inhibitory synapses are developing.
We identified more than 300 genes whose expression levels were altered upon membrane depolarization (Gene Expression Omnibus accession number GSE11256), a third of which were novel activity-regulated genes not seen in previous screens26,27. We looked for genes predicted to encode transcription factors, reasoning that, through genome-wide characterization of the targets of an activity-regulated transcription factor that controls inhibitory synapse number, we could gain insight into the biological program that is important for inhibitory synapse development. Among the approximately 20 known or putative transcription factors identified, we focused on genes that are selectively induced by Ca2+ influx in neurons but not other cell types, that are transcribed in response to excitatory synaptic activity, and that are expressed coincidently with the development of inhibitory synapses. One transcription factor, the bHLH-PAS family member Npas4 (refs 28–31), fulfilled all these criteria (Fig. 1) and was investigated further.
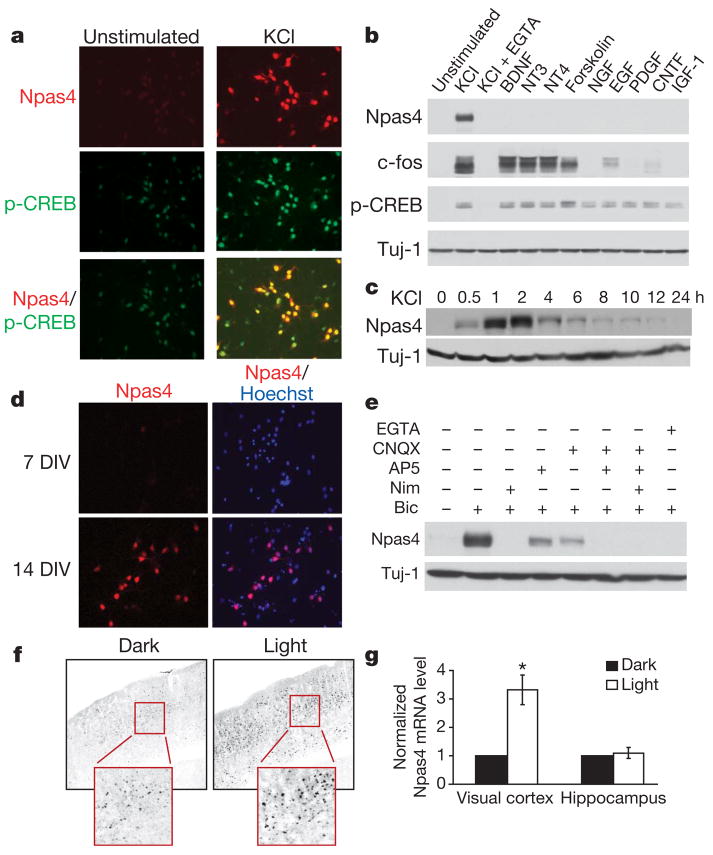
Figure 1. Npas4 expression is regulated by neuronal activity in vitro and in vivo.
a, Immunostaining showing Npas4 protein is induced in rat hippocampal neurons (7 DIV) by depolarization (50 mM KCl, 2 h, right). p-CREB, CREB phosphorylated at Ser 133. b, Western blot showing Npas4 is selectively induced by membrane depolarization (50 mM KCl, 7 DIV rat hippocampal neurons), but not by BDNF (50 ng ml−1), NT3 (50 ng ml−1), NT4 (50 ng ml−1), forskolin (10 μM), NGF (100 ng ml−1), EGF (100 ng ml−1), PDGF (100 ng ml−1), CNTF (100 ng ml−1) or IGF-1 (100 ng ml−1). Induction is prevented by pretreatment with the Ca2+ chelator EGTA (5 mM, 10 min). c, Western blot showing Npas4 (7 DIV rat hippocampal neurons) is transiently induced by membrane depolarization (50 mM KCl, 30 min). d, Basal Npas4 expression increases as neurons mature, presumably because of increased endogenous spontaneous activity: compare immunostaining of 7 and 14 DIV rat hippocampal neurons. e, Western blot showing stimulation of primary hippocampal neurons (14 DIV) with bicuculline (50 μM, 2 h) increases Npas4 expression levels. This is prevented by pretreatment with nimodipine (5 μM, 1 h) or EGTA (5 μM, 5 min) and reduced by pretreatment (1 h) with antagonists to NMDA receptors (100 μM AP5) or AMPA receptors (50 μM CNQX). f, g, Mice dark-reared for one week (P21–P28) and then stimulated with strobe lights have greater Npas4 expression levels in the visual cortex than their dark-reared littermates, but there is no difference in the hippocampus. Light stimulation was applied for 2 h for immunocytochemistry analysis (f) or 1 h for Npas4 mRNA quantification (g). Significance was determined using a one-tailed paired t-test, *P < 0.05. Data are shown as mean ± s.e.m.
Unlike other activity-dependent transcription factors such as CREB and c-fos, Npas4 expression in neurons is selectively induced by Ca2+ influx but not by several neurotrophic factors, growth factors or forskolin, an activator of protein kinase A (Fig. 1b). Furthermore, Npas4 induction is transient (Fig. 1c), occurs selectively in neurons, and predominately in excitatory neurons (data not shown). Npas4 expression is barely detectable in immature primary hippocampal neurons, when there are few synapses32, and increases with the formation and maturation of synapses25 (Fig. 1d).
Npas4 is induced in cultured neurons by the GABAA-receptor antagonist bicuculline, which increases action-potential firing and excitatory synaptic transmission (Fig. 1e). This induction requires an influx of extracellular Ca2+ through L-type voltage-sensitive calcium channels (L-VSCCs) and is partly dependent on the activation of N-methyl-D-aspartate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors (Fig. 1e). Npas4 expression is also induced in pertinent brain regions in vivo in response to specific stimuli: visual stimulation of mice after a period of dark-rearing results in an increase in Npas4 messenger RNA (mRNA) and protein levels specifically in the visual cortex (Fig. 1f, g).
Npas4 regulates the development of inhibitory synapses
The effect of Npas4 on inhibitory synapse development was investigated by RNA interference (RNAi)-mediated knockdown in excitatory neurons of rat dissociated hippocampal cultures, before synapse formation was underway. A small hairpin RNA targeting Npas4 (Npas4-RNAi) effectively reduced Npas4 expression (Fig. 2a) without affecting the overall health of the neurons (Supplementary Fig. 1). To measure inhibitory synapse number, neurons were immunostained for presynaptic GABA-producing enzyme GAD65 and postsynaptic GABAA-receptor γ2 subunit (GABAA-γ2). Co-localization of GAD65 and GABAA-γ2 puncta on a green fluorescent protein (GFP)-transfected glutamatergic neuron was considered indicative of a synapse (Fig. 2b). Expression of Npas4-RNAi, but not a scrambled RNAi (control-RNAi), significantly reduced the number of inhibitory synapses (Fig. 2c). Analysis of synapse number using antibodies that recognize a second pair of inhibitory synaptic proteins (GAD67 and GABAA-β2/3) gave a similar result (data not shown). These data suggest that Npas4 positively regulates the number of inhibitory synapses that form on excitatory neurons.
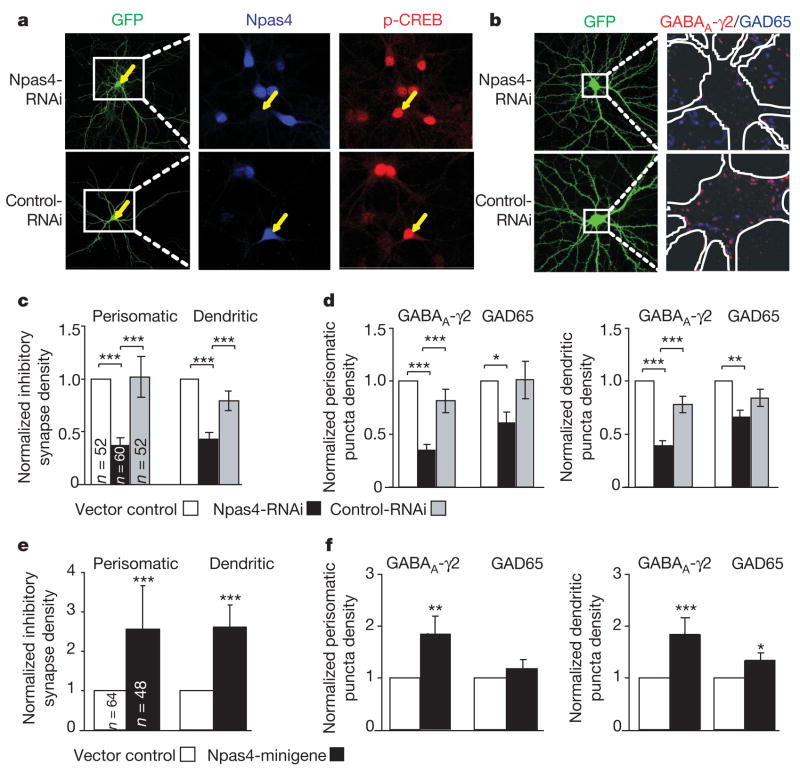
Figure 2. Npas4 regulates the number of GABAergic synapses in cultured hippocampal neurons.
a, Npas4-RNAi, but not control-RNAi, reduces the expression of Npas4 in primary hippocampal neurons. Cultures were transfected at 6 DIV and stimulated with bicuculline (50 μM, 2 h) at 14 DIV. b, The number of GABAergic synapses is significantly reduced by Npas4-RNAi, as illustrated by two representative rat hippocampal neurons. Cultures were co-transfected (6 DIV) with GFP and either Npas4-RNAi (top) or control-RNAi (bottom). Cultures were subsequently immunostained (25 DIV) with antibodies against GAD65 (blue) and GABAA-γ2 (red). c, Quantification of the normalized density of co-localized GABAA-γ2 and GAD65 puncta in 14 DIV rat hippocampal neurons transfected with vector control, Npas4-RNAi or control-RNAi. d, Separate quantification of perisomatic and dendritic GABAA-γ2 and GAD65 puncta measured in c. e, Npas4-minigene increases the density of co-localized GABAA-γ2 and GAD65 puncta. f, Separate quantification of perisomatic and dendritic GABAA-γ2 and GAD65 puncta measured in e. See Methods for details of data normalization and error propagation. Significance was determined using multifactorial analysis of variance. *P < 0.05; **P < 0.005; ***P < 0.0005. Data are presented as mean ± s.e.m. from three(c,d)or four (e,f)independent experiments; total numbers of neurons analysed (n) are indicated.
Different classes of inhibitory neurons synapse onto distinct perisomatic or dendritic regions of pyramidal neurons33. We found that Npas4-RNAi leads to a reduction in the number of inhibitory synapses formed on both the perisomatic and dendritic regions, suggesting that Npas4 regulates the number of inhibitory synapses formed by multiple classes of inhibitory neurons (Fig. 2c). In both regions, Npas4-RNAi significantly reduced the density of post-synaptic GABAA-γ2 puncta but had less effect on presynaptic GAD65 puncta, compared with the control-RNAi (Fig. 2d). These findings suggest that Npas4 regulates inhibitory synapse number by controlling the number of postsynaptic specializations, resulting in subsequent remodelling or retraction of the presynaptic terminals.
To test whether Npas4 is important for the development of functional inhibitory synapses in a more intact neural circuit, rat organotypic hippocampal cultures were biolistically co-transfected with GFP and either a control vector or Npas4-RNAi, and whole-cell recordings were performed on GFP-positive CA1 pyramidal neurons to measure spontaneous miniature inhibitory postsynaptic currents (mIPSCs, Fig. 3a). Knocking down Npas4 expression in organotypic cultures significantly increased the inter-event interval and decreased the amplitude of mIPSCs (Fig. 3b, c). This effect of Npas4-RNAi in organotypic slices may be indicative of a decrease in inhibitory synapse number, consistent with our observation that Npas4-RNAi reduces inhibitory synapse number in dissociated cultures. Together, these findings indicate that Npas4 plays an important role in regulating the number of functional inhibitory synapses received by an excitatory neuron.
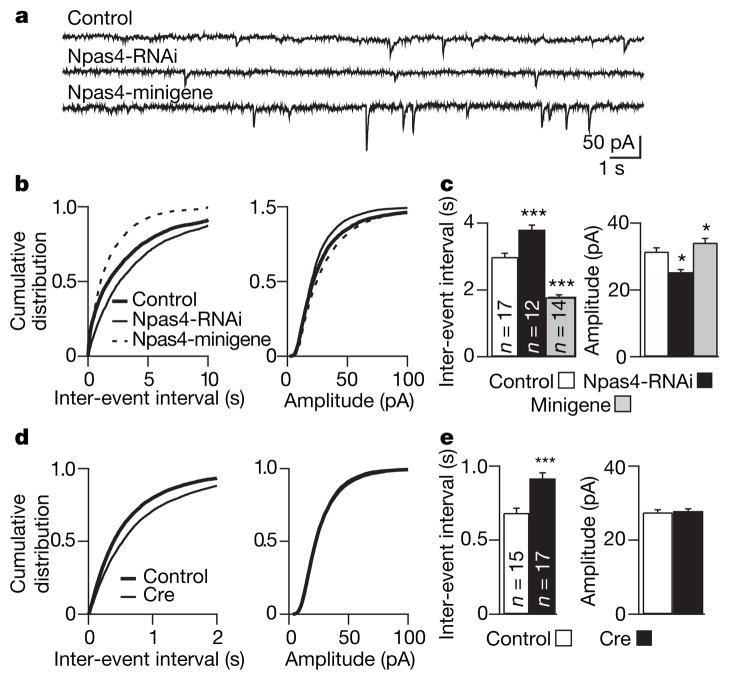
Figure 3. Npas4 regulates GABAergic synapse development in organotypic hippocampal slices.
a, Representative mIPSCs recorded from CA1 pyramidal neurons in organotypic hippocampal slices biolistically co-transfected with GFP and either vector control, Npas4-RNAi or Npas4-minigene. b, Cumulative distributions of mIPSC inter-event intervals and amplitudes recorded from neurons transfected with vector control, Npas4-RNAi or Npas4-minigene. c, Mean ± s.e.m. of data from b. mIPSC inter-event intervals: 2986.3 ± 105.7, 3803.0 ± 136.9 and 1776.9 ± 75.1 ms; amplitudes: 31.5 ± 1.1, 25.7 ± 0.8 and 34.1 ± 1.3 pA; for vector control, Npas4-RNAi and Npas4-minigene, respectively. d, Cumulative distributions of mIPSC inter-event intervals and amplitudes recorded from Npas4flx/flx neurons co-transfected with GFP and either vector control or Cre recombinase. e, Mean ± s.e.m. of data from d. mIPSC inter-event intervals: 684.2 ± 31.2 and 917.4 ± 37.7 ms; amplitudes: 27.5 ± 0.7 and 27.9 ± 0.6 pA; for vector control and Cre, respectively. Total numbers of neurons analysed in each condition (n) are indicated in c and e. *P < 0.05; ***P < 0.001.
Consistent with a role for Npas4 in inhibitory synapse development, Npas4 knockout mice (Npas4−/−, Supplementary Fig. 2) appear anxious and hyperactive, are prone to seizures and have a shortened lifespan compared with their wild-type littermates, phenotypically resembling other knockout mice lacking genes that control the formation or function of inhibitory circuits34–36. However, we found that the frequencies of mIPSCs in acute hippocampal slices prepared from wild-type and Npas4−/− mice were similar (Supplementary Fig. 3), in contrast to the clear change in mIPSC frequency caused by disrupting Npas4 expression acutely in organotypic slices. This difference may be due to the activation of compensatory pathways during development in the absence of Npas4. Alternatively, it may reflect the fact that Npas4 is absent in both pre- and postsynaptic cells in acute slices from Npas4−/− mice, whereas in the experiments with organotypic slices the disruption of Npas4 expression occurs in postsynaptic neurons only.
To determine conclusively the involvement of Npas4 in inhibitory synapse development, we generated an Npas4 conditional knockout mouse (Npas4flx/flx) in which the coding region of Npas4 is flanked by loxP sites and can be acutely removed by Cre-mediated recombination (Supplementary Fig. 4). Organotypic hippocampal slices were prepared from Npas4flx/flx mice and whole-cell recordings made from CA1 pyramidal neurons transfected with GFP and either a control vector or a vector encoding Cre recombinase. Cre expression had no effect on mIPSCs in wild-type neurons (Supplementary Fig. 5). However, compared with transfection with the control construct, transfection of Npas4flx/flx neurons with Cre led to a significant increase in the mIPSC inter-event interval (Fig. 3d, e). This finding provides further evidence that CA1 pyramidal neurons lacking Npas4 receive fewer inhibitory synaptic inputs.
We next investigated whether the number of inhibitory synapses forming onto a cell is controlled by the amount of Npas4 expressed in response to excitatory stimuli. If this is the case, increasing the level of Npas4 should lead to an increase in the number of inhibitory synapses formed on the Npas4-expressing neuron. Additional copies of the Npas4 gene were introduced into neurons using an Npas4-minigene cassette consisting of all Npas4 introns and exons as well as 5 kilobases (kb) of genomic sequence 5′- and 3′- to the coding region. We verified that the Npas4-minigene drives ectopic expression of Npas4 that is activity regulated and functions similarly to the endogenous gene (Supplementary Fig. 6). We found that significantly more inhibitory synapses were formed onto cultured hippocampal neurons expressing the Npas4-minigene than onto neurons transfected with a control vector (Fig. 2e), largely because of an increase in the number of postsynaptic GABAA-γ2 puncta (Fig. 2f). Furthermore, expression of the Npas4-minigene in CA1 pyramidal neurons of hippocampal slices significantly decreases the mIPSC inter-event interval and significantly increases the amplitude (Fig. 3b, c), consistent with the presence of additional and stronger inhibitory synapses on neurons expressing higher levels of Npas4.
Effect of Npas4 on excitatory synapses
The appropriate balance between excitation and inhibition is critical for the function of neural circuits. To maintain this balance, changes in the number or strength of inhibitory synapses are often coupled to changes in excitatory synapses24. We next asked whether excitatory synapse number or function is also affected when Npas4 expression is perturbed.
We first determined whether Npas4 regulates the number of excitatory synapses in dissociated hippocampal cultures. As before, neurons were transfected with either Npas4-RNAi or control constructs, before synaptogenesis was underway and thus before the balance between excitation and inhibition was established. Cultures were immunostained for the presynaptic marker synapsin1 and the excitatory postsynaptic marker PSD95, and the numbers of co-localized synapsin1 and PSD95 puncta on transfected glutamatergic neurons were quantified. Neither the total numbers of excitatory synapses nor the individual numbers of pre- or postsynaptic markers changed significantly upon expression of Npas4-RNAi (Fig. 4a, b), under conditions that significantly decreased the number of inhibitory synapses. Likewise, expression of the Npas4-minigene had no effect on excitatory synapse number (Fig. 4c, d). Because perturbation of the level of Npas4 expression occurred before synaptic connections were established, these experiments suggest that Npas4 is not a major contributor to excitatory synaptogenesis.
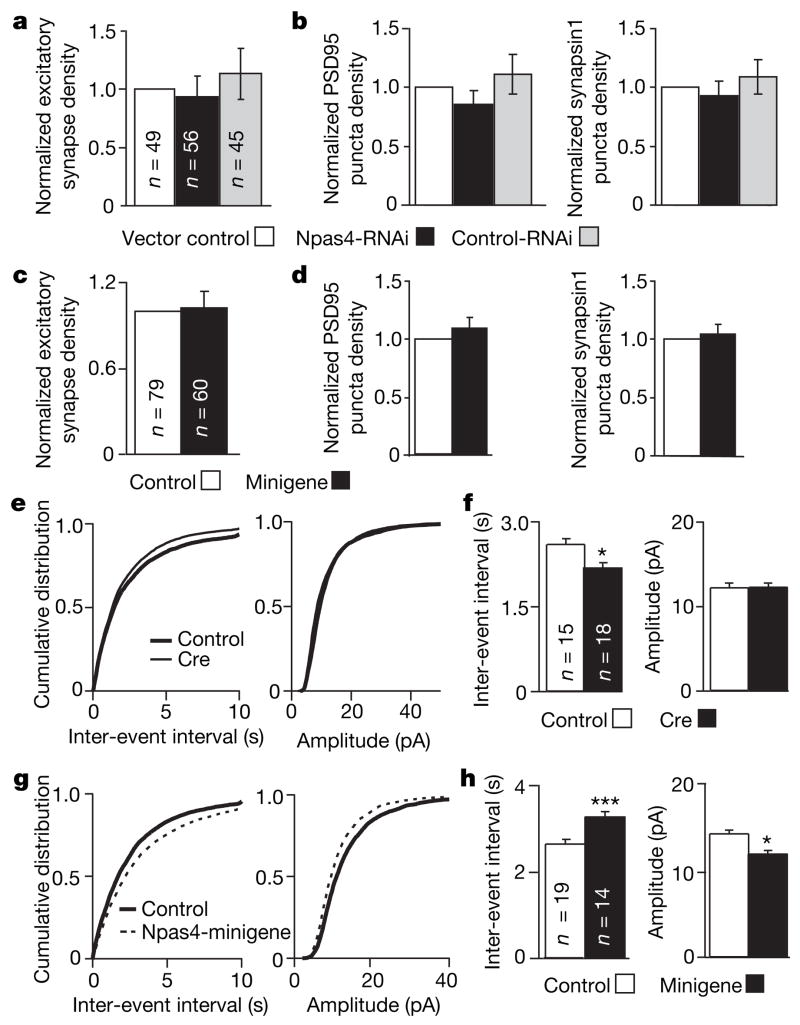
Figure 4. Npas4 has no effect on excitatory synaptogenesis but affects excitatory/inhibitory balance in neural circuits.
a, The number of excitatory synapses is not affected by Npas4-RNAi. Quantification of the normalized density of co-localized PSD95 and synapsin1 puncta in 14 DIV rat hippocampal neurons transfected with vector control, Npas4-RNAi or control-RNAi. b, Separate quantification of PSD95 and synapsin1 puncta measured in a. c, Npas4-minigene has no effect on the density of excitatory synapses. Quantification of co-localized synapsin1 and PSD95 puncta is shown. d, Separate quantification of PSD95 and synapsin1 puncta measured in c. In a–d, data are presented as mean ± s.e.m. e, Cumulative distribution of mEPSC inter-event intervals and amplitudes recorded from Npas4flx/flx neurons co-transfected with GFP and either vector control or Cre. f, Mean ± s.e.m. of data from e. mEPSC inter-event intervals: 2581.4 ± 104.1 ms and 2140.5 ± 79.7 ms; amplitudes: 12.1 ± 0.4 and 12.4 ± 0.3 pA; for vector control and Cre, respectively. g, Cumulative distribution of mEPSC inter-event intervals and amplitudes recorded from neurons transfected with GFP and either vector control or Npas4-minigene. h, Mean ± s.e.m. of data from g. mEPSC inter-event intervals: 2686.4 ± 89.3 and 3320.7 ± 117.2 ms; amplitudes: 14.3 ± 0.3 and 12.0 ± 0.3 pA; for vector control and Npas4-minigene, respectively. Total numbers of neurons analysed in each condition (n) are indicated. a, b, Three independent experiments; c, d, four independent experiments. *P < 0.05; ***P < 0.001.
We next examined whether Npas4 affects excitatory synapse function in organotypic hippocampal slices, where homeostatic mechanisms are known to control excitatory/inhibitory balance within neural circuits. Deletion of the Npas4 gene from neurons in Npas4flx/flx organotypic hippocampal slices by Cre recombination significantly decreased the inter-event interval of spontaneous miniature excitatory postsynaptic currents (mEPSCs, Fig. 4e, f), whereas elevating the level of Npas4 with the Npas4-minigene significantly increased the inter-event interval and decreased the amplitude of mEPSCs (Fig. 4g, h), compared with control-transfected neurons. Thus, abolishing Npas4 expression in a pyramidal neuron increases, whereas enhancing the level of Npas4 decreases, the number and/or presynaptic release probability of excitatory synapses that form on the neuron. Therefore, the net result of Npas4 activation in an intact neural circuit is an increase in synaptic inhibition and a decrease in excitation of a neuron. We conclude that Npas4 induction in response to increased excitatory input acts to reduce the level of activity, and therefore may function as a negative feedback mechanism to maintain the homeostatic balance between excitation and inhibition.
Npas4 regulates genes that control inhibitory synapse development
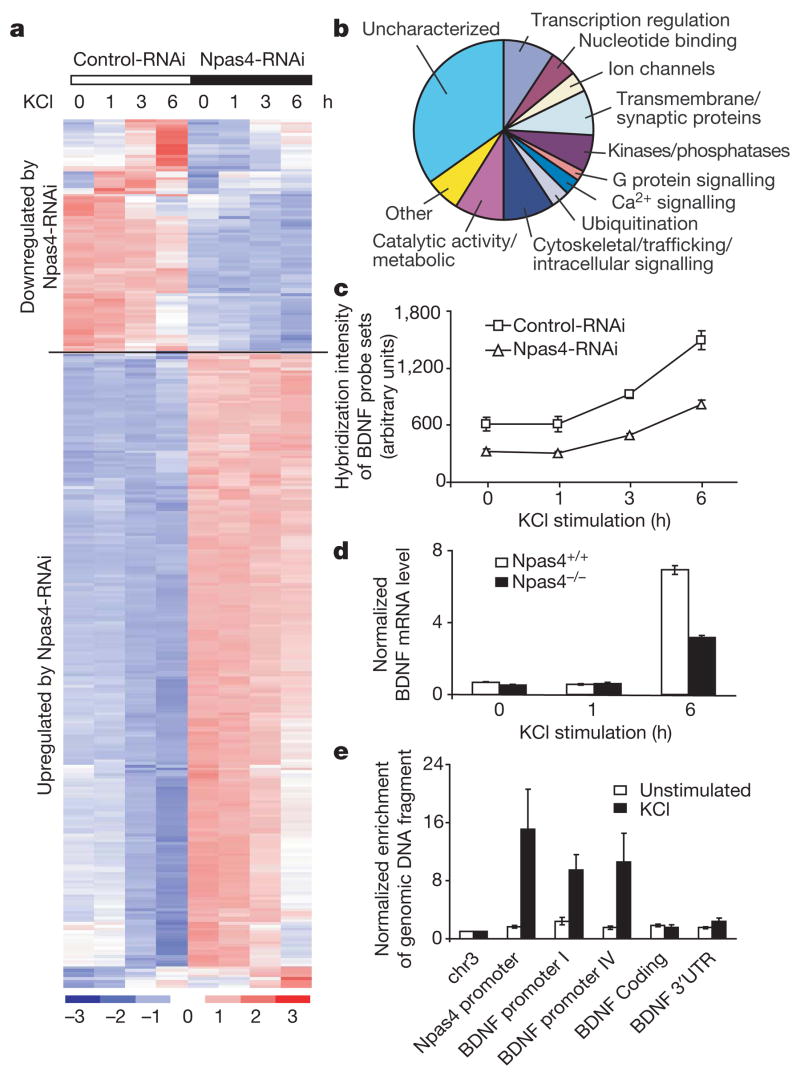
To uncover the program of gene expression controlled by Npas4, we acutely knocked down Npas4 expression in a high percentage of wild-type neurons using a lentivirus expressing Npas4-RNAi (Supplementary Fig. 7) and performed a second DNA microarray experiment to identify activity-regulated genes that are misregulated in the absence of Npas4. The expression levels of 327 microarray probe sets representing 270 unique genes were significantly different in cultures expressing Npas4-RNAi compared with those expressing the control virus (Gene Expression Omnibus accession number GSE11258, Supplementary List). The expression of 182 of these genes was also acutely regulated by membrane depolarization in the absence of Npas4-RNAi, indicating that many putative Npas4-regulated genes are activity regulated (Fig. 5a). Although Npas4 has been shown to function as a transcriptional activator (Supplementary Fig. 6b)28, we found that many genes are negatively regulated by Npas4 (Fig. 5a). This may reflect the fact that Npas4 functions as a transcriptional repressor as well as an activator, and/or that Npas4 indirectly affects gene expression by altering neuronal excitation.
Figure 5. Npas4 controls a program of gene expression that regulates GABAergic synapses.
a, Hierarchical clustering of 327 probe sets (270 putative Npas4 target genes) based on their expression profiles using dChip49. The expression level of each probe set is normalized to a mean of 0 and a standard deviation of 1. Expression values are displayed within the range [−3, 3] with levels above, equal to or below the mean displayed in red, white and blue, respectively. Dark red represents 3 or higher, and dark blue −3 or lower. b, Biological functions of 270 putative Npas4 target genes based on Gene Ontology information provided by Affymetrix (http://www.affymetrix.com). c, BDNF expression is reduced by Npas4-RNAi (1.9 ± 0.11-fold reduction, P < 0.01, one-tailed paired t-test). Mean ± s.e.m. from three independent experiments is shown, and each data point was generated by averaging the hybridization intensity of two BDNF probe sets (1422168_a_at and 1422169_a_at). d, BDNF levels are consistently reduced in neurons from Npas4−/− mice compared with their wild-type littermates (58.46 ± 6.49% decrease, 95% confidence interval 30.5–86.4%). Cortical cultures prepared from Npas4+/+ and Npas4−/− littermates (7 DIV) were stimulated with KCl (55 mM), and BDNF mRNA levels were measured by quantitative reverse transcriptase PCR using primers in BDNF coding region. Three littermate pairs from three different litters were analysed; data (mean ± s.e.m.) from one representative pair is shown. e, Npas4 interacts directly with BDNF promoters I and IV in an activity-dependent manner, as shown by chromatin immunoprecipitation. The Npas4 promoter is used as a positive control. Data are normalized to a control region on chromosome 3 and are presented as mean ± s.e.m. from five independent experiments.
Npas4 appears to regulate a wide variety of genes, such as activity-regulated immediate early genes, various classes of transcription factors, channel proteins, G-protein signalling molecules, kinases and phosphatases, and genes involved in pathways that modulate synaptic functions, such as ubiquitination, trafficking and receptor endocytosis (Fig. 5b and Supplementary List). Interestingly, the functions of 94 of the 270 putative Npas4-regulated genes are uncharacterized (Fig. 5b), suggesting that Npas4 regulates many genes that could affect inhibitory synapses in novel ways.
As a first step towards understanding the genetic program regulated by Npas4, we focused on targets that might be directly involved in the development of GABAergic synapses. Brain-derived neurotrophic factor (BDNF) stood out because it had previously been shown to regulate GABAergic synapse maturation and function18,20,37–39. BDNF expression is consistently reduced by almost twofold in cultures expressing Npas4-RNAi compared with control cultures (Fig. 5c). Primary cultures from Npas4−/− mice showed a similar decrease in depolarization-induced BDNF expression compared with their wild-type littermates (Fig. 5d).
The BDNF gene has many promoters and the activity-dependent BDNF mRNA transcripts are controlled by promoters I and IV40–42. Using a chromatin immunoprecipitation assay, we asked whether Npas4 binds to these promoters in primary cortical cultures that were left untreated or membrane-depolarized to trigger Npas4 expression. Although no Npas4 binding was detected in untreated neurons, in membrane-depolarized neurons Npas4 was found to be associated with BDNF promoters I and IV, but not with the coding region or 3′ untranslated region (Fig. 5e), suggesting that Npas4 may directly regulate activity-dependent expression of BDNF.
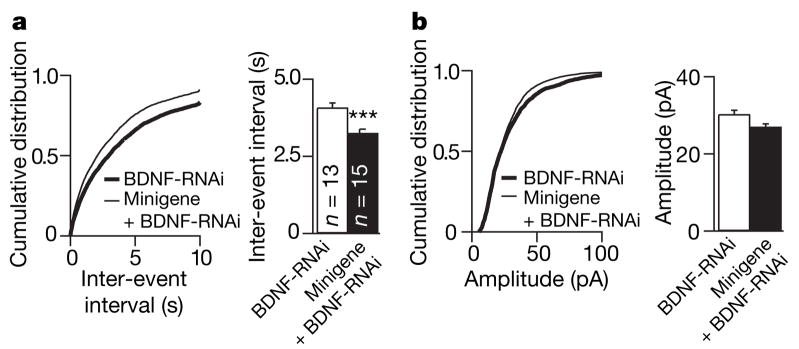
To determine whether BDNF contributes to the effects of Npas4 on GABAergic synapse development, we tested whether the ability of the Npas4-minigene to increase the number of inhibitory synapses is attenuated by knockdown of BDNF expression, using a previously validated small hairpin RNA that targets the BDNF coding region (BDNF-RNAi)43. Confirming that BDNF increases inhibitory synapse number44, BDNF-RNAi increased mIPSC inter-event intervals compared with control values (2986.3 ± 109.7 and 4086.1 ± 140.7 ms, control and BDNF-RNAi, respectively, P < 0.01). Expression of the Npas4-minigene alone led to an approximately 40% decrease in the inter-event interval of mIPSCs recorded from CA1 neurons (Fig. 3b, c), as previously observed, but this was partly attenuated to approximately 20% by the presence of BDNF-RNAi (Fig. 6a). In addition, in the presence of BDNF-RNAi, the effect of the Npas4-minigene on the amplitude of mIPSCs was completely reversed (Fig. 6b). These findings suggest that BDNF mediates a portion of the effect of Npas4 on inhibitory synapse number, but that additional Npas4 targets may also be involved.
Figure 6. Knockdown of BDNF partially attenuates the ability of the Npas4-minigene to elevate GABAergic synapses.
a, Cumulative distributions (left) and mean ± s.e.m. (right) of mIPSC inter-event intervals in neurons transfected with BDNF-RNAi (4086.1 ±140.7 ms) or BDNF-RNAi 1Npas4-minigene (3257.9 ± 117.7 ms). b, Cumulative distributions (left) and mean ± s.e.m. (30.1 ± 0.9 and 27.1 ± 0.7 pA, BDNF-RNAi versus BDNF-RNAi 1Npas4-minigene, right) of mIPSC amplitudes. Total numbers of neurons analysed in each condition (n) are indicated. ***P < 0.001.
Discussion
We have identified the activity-regulated transcription factor Npas4 as a key regulator of GABAergic synapse development. Excitatory synaptic activity induces Npas4 in a Ca2+-dependent manner, and the level of Npas4 determines the number of functional GABAergic synapses by controlling a program of activity-dependent gene expression. Future characterization of Npas4 target genes will help to determine whether Npas4 acts by initiating inhibitory synapse formation, stabilizing nascent inhibitory synapses or promoting the maturation of weak inhibitory synapses. It is possible that different subsets of Npas4 targets control the development of GABAergic synapses formed by distinct classes of interneurons, providing a mechanism for the independent regulation of inhibition received by subregions of a neuron. Another intriguing possibility is that Npas4 controls experience-dependent developmental processes, such as critical period plasticity in the visual cortex, which depend on the function and maturation of GABAergic synapses.
Although Npas4 is not required for the initial formation of excitatory synapses, activation of Npas4 in excitatory neurons within a neural circuit appears to diminish the excitatory synaptic input they receive. It is not known whether this effect is mediated directly by Npas4 or is an indirect consequence of changes in inhibitory input. In either case, our findings suggest that Npas4 functions as part of the homeostatic mechanism that stabilizes the activity of a neuron in the face of changing glutamatergic input. Further investigation of the function of Npas4 and other activity-dependent regulators of inhibitory synapses45 will provide insight into the mechanism by which neuronal activity controls the balance between excitation and inhibition in the brain, and how the disruption of this balance leads to neurological disorders such as autism, epilepsy and schizophrenia.
METHODS SUMMARY
Dissociated neuron culture and transfection
Dissociated cortical and hippocampal neurons were prepared from E18 rat or E16 mouse embryos as previously described46. Cultures were maintained in Neurobasal Medium supplemented with B27 (Invitrogen), penicillin–streptomycin and glutamine. Neurons were plated at 100,000–150,000 per well in a 24-well plate or 15,000,000 per 10-cm plate. For synapse assays, hippocampal neurons (60,000 per well, 24-well plate) were plated on a monolayer of astrocytes47. Neurons were transfected at 5–6 days in vitro (DIV) using the calcium phosphate method46.
Organotypic slice culture and transfection
Organotypic hippocampal slice cultures were prepared from P6 rats or mice as previously described48. Slices were biolistically transfected with a Helios Gene Gun (Biorad) after 2 days. Bullets for the gene gun were 1.6-μm gold particles coated with 15 μg eGFP and either 5 or 10 μg RNAi construct, 45 μg Npas4-minigene or 30 μg Cre. Empty plasmid was added to bring the total DNA to 60 μg in each case.
Synapse density assay
Hippocampal neurons (14–18 DIV) were immunostained for synaptic markers and imaged on a Zeiss LSM5 Pascal microscope using a ×63 objective lens. Image acquisition and synapse quantification were performed in a blinded manner. Glutamatergic neurons were identified based on morphology and the absence of cytosolic GAD65 staining. Synapse density was measured using Metamorph software as previously described47. Within each experiment, density of puncta was normalized against the control, and the error of the control propagated into each experimental condition. Statistical analysis was performed on the raw data (without normalization) using multifactorial analysis of variance in StatView 4.5 (Abacus Concepts).
Electrophysiology
Whole-cell patch clamp recordings were made at room temperature from CA1 pyramidal neurons 7–10 days after transfection. Data were analysed in IgorPro (Wavemetrics) using custom-written macros. Statistical significance was determined by Kolmogorov–Smirnov test and Monte Carlo simulation.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank members of the Greenberg laboratory for suggestions; S. Paradis, J. M. Gray, S. S. Margolis, J. Zieg and C. M. Fletcher for reading the manuscript; S. Vasquez for preparing primary neuronal cell cultures; M. Thompson, Y. Zhou and H. Ye for assistance in generating Npas4−/− mice; T. Diefenbach and the Neurobiology Program Imaging Center for assistance with confocal microscopy; M. Fagiolini for help with dissection of the visual cortex; and X. J. Liu and C. Chen for help with electrophysiology. M.E.G. acknowledges the generous support of the F. M. Kirby Foundation to the Neurobiology Program of the Children’s Hospital and support from the Nancy Lurie Marks Family Foundation. This work was supported by a Lefler Foundation postdoctoral fellowship (Y.L.), a Ruth L. Kirschstein National Research Service Award and a Helen Hay Whitney postdoctoral fellowship (B.L.B.), a National Science Foundation Graduate Research Fellowship (A.D.L.), the Jane Coffin Childs Memorial Fund (T.-K.K.) and Mental Retardation Research Center grant HD18655 and National Institutes of Health grants NS27572 and NS48276 (M.E.G.).
Footnotes
Author Information Data have been placed in the GEO database under accession numbers GSE11256 and GSE11258. Reprints and permissions information is available at www.nature.com/reprints.
Author Contributions Y.L. and M.E.G. conceived and designed the experiments and wrote the manuscript. Y.L. performed or participated in each of the experiments described in the manuscript. B.L.B. performed the electrophysiological recordings and contributed to the writing of the manuscript. J.L.H. quantified Npas4 mRNA levels for the light stimulation experiment, generated the Npas4-minigene construct and performed the luciferase assay to characterize it, managed the Npas4 animal colony and provided extensive technical support. A.D.L. performed immunocytochemistry for the light stimulation experiment and confocal imaging of neurons in the synapse assay with Npas4-RNAi. A.C.K. provided technical support during the early phase of the study and helped generate many reagents used in this study including the Npas4 antibody, Npas4 knockout construct and Npas4-RNAi lentivirus. T.-K.K. performed the chromatin immunoprecipitation experiments. L.S.H. helped generate the Npas4 antibody. A.N.M. performed the initial chromatin immunoprecipitation experiments.
References
- 1.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 2.Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- 3.Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nature Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- 4.Zito K, Svoboda K. Activity-dependent synaptogenesis in the adult mammalian cortex. Neuron. 2002;35:1015–1017. doi: 10.1016/s0896-6273(02)00903-0. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Ari Y. Excitatory actions of GABA during development: the nature of the nurture. Nature Rev Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- 6.Kenet T, Froemke RC, Schreiner CE, Pessah IN, Merzenich MM. Perinatal exposure to a noncoplanar polychlorinated biphenyl alters tonotopy, receptive fields, and plasticity in rat primary auditory cortex. Proc Natl Acad Sci USA. 2007;104:7646–7651. doi: 10.1073/pnas.0701944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maffei A, Nelson SB, Turrigiano GG. Selective reconfiguration of layer 4 visual cortical circuitry by visual deprivation. Nature Neurosci. 2004;7:1353–1359. doi: 10.1038/nn1351. [DOI] [PubMed] [Google Scholar]
- 8.Buschges A, Manira AE. Sensory pathways and their modulation in the control of locomotion. Curr Opin Neurobiol. 1998;8:733–739. doi: 10.1016/s0959-4388(98)80115-3. [DOI] [PubMed] [Google Scholar]
- 9.Brown P, Ridding MC, Werhahn KJ, Rothwell JC, Marsden CD. Abnormalities of the balance between inhibition and excitation in the motor cortex of patients with cortical myoclonus. Brain. 1996;119:309–317. doi: 10.1093/brain/119.1.309. [DOI] [PubMed] [Google Scholar]
- 10.Cline H. Synaptogenesis: a balancing act between excitation and inhibition. Curr Biol. 2005;15:R203–R205. doi: 10.1016/j.cub.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Mohler H. GABAA receptors in central nervous system disease: anxiety, epilepsy, and insomnia. J Recept Signal Transduct Res. 2006;26:731–740. doi: 10.1080/10799890600920035. [DOI] [PubMed] [Google Scholar]
- 12.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wassef A, Baker J, Kochan LD. GABA and schizophrenia: a review of basic science and clinical studies. J Clin Psychopharmacol. 2003;23:601–640. doi: 10.1097/01.jcp.0000095349.32154.a5. [DOI] [PubMed] [Google Scholar]
- 14.Burrone J, O’Byrne M, Murthy VN. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature. 2002;420:414–418. doi: 10.1038/nature01242. [DOI] [PubMed] [Google Scholar]
- 15.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 16.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 17.Benevento LA, Bakkum BW, Cohen RS. Gamma-aminobutyric acid and somatostatin immunoreactivity in the visual cortex of normal and dark-reared rats. Brain Res. 1995;689:172–182. doi: 10.1016/0006-8993(95)00553-3. [DOI] [PubMed] [Google Scholar]
- 18.Chattopadhyaya B, et al. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foeller E, Feldman DE. Synaptic basis for developmental plasticity in somatosensory cortex. Curr Opin Neurobiol. 2004;14:89–95. doi: 10.1016/j.conb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Marty S, Wehrle R, Sotelo C. Neuronal activity and brain-derived neurotrophic factor regulate the density of inhibitory synapses in organotypic slice cultures of postnatal hippocampus. J Neurosci. 2000;20:8087–8095. doi: 10.1523/JNEUROSCI.20-21-08087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micheva KD, Beaulieu C. Development and plasticity of the inhibitory neocortical circuitry with an emphasis on the rodent barrel field cortex: a review. Can J Physiol Pharmacol. 1997;75:470–478. [PubMed] [Google Scholar]
- 22.Seil FJ, Drake-Baumann R. Activity-dependent changes in “transplanted” cerebellar cultures. Exp Neurol. 1996;138:327–337. doi: 10.1006/exnr.1996.0071. [DOI] [PubMed] [Google Scholar]
- 23.Hensch TK. Critical period plasticity in local cortical circuits. Nature Rev Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- 24.Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nature Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- 25.Benson DL, Cohen PA. Activity-independent segregation of excitatory and inhibitory synaptic terminals in cultured hippocampal neurons. J Neurosci. 1996;16:6424–6432. doi: 10.1523/JNEUROSCI.16-20-06424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- 27.Worley PF, Cole AJ, Saffen DW, Baraban JM. Regulation of immediate early genes in brain: role of NMDA receptor activation. Prog Brain Res. 1990;86:277–285. doi: 10.1016/s0079-6123(08)63184-2. [DOI] [PubMed] [Google Scholar]
- 28.Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol Cell Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flood WD, Moyer RW, Tsykin A, Sutherland GR, Koblar SA. Nxf and Fbxo33: novel seizure-responsive genes in mice. Eur J Neurosci. 2004;20:1819–1826. doi: 10.1111/j.1460-9568.2004.03646.x. [DOI] [PubMed] [Google Scholar]
- 30.Hester I, et al. Transient expression of Nxf, a bHLH-PAS transactivator induced by neuronal preconditioning, confers neuroprotection in cultured cells. Brain Res. 2007;1135:1–11. doi: 10.1016/j.brainres.2006.11.083. [DOI] [PubMed] [Google Scholar]
- 31.Shamloo M, et al. Npas4, a novel helix-loop-helix PAS domain protein, is regulated in response to cerebral ischemia. Eur J Neurosci. 2006;24:2705–2720. doi: 10.1111/j.1460-9568.2006.05172.x. [DOI] [PubMed] [Google Scholar]
- 32.Fletcher TL, Cameron P, De Camilli P, Banker G. The distribution of synapsin I and synaptophysin in hippocampal neurons developing in culture. J Neurosci. 1991;11:1617–1626. doi: 10.1523/JNEUROSCI.11-06-01617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang ZJ, Di Cristo G, Ango F. Development of GABA innervation in the cerebral and cerebellar cortices. Nature Rev Neurosci. 2007;8:673–686. doi: 10.1038/nrn2188. [DOI] [PubMed] [Google Scholar]
- 34.Gomeza J, et al. Deletion of the mouse glycine transporter 2 results in a hyperekplexia phenotype and postnatal lethality. Neuron. 2003;40:797–806. doi: 10.1016/s0896-6273(03)00673-1. [DOI] [PubMed] [Google Scholar]
- 35.Kash SF, et al. Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA. 1997;94:14060–14065. doi: 10.1073/pnas.94.25.14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kash SF, Tecott LH, Hodge C, Baekkeskov S. Increased anxiety and altered responses to anxiolytics in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA. 1999;96:1698–1703. doi: 10.1073/pnas.96.4.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang ZJ, et al. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- 38.Rutherford LC, DeWan A, Lauer HM, Turrigiano GG. Brain-derived neurotrophic factor mediates the activity-dependent regulation of inhibition in neocortical cultures. J Neurosci. 1997;17:4527–4535. doi: 10.1523/JNEUROSCI.17-12-04527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seil FJ, Drake-Baumann R. TrkB receptor ligands promote activity-dependent inhibitory synaptogenesis. J Neurosci. 2000;20:5367–5373. doi: 10.1523/JNEUROSCI.20-14-05367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu QR, et al. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhou P, et al. Polarized signaling endosomes coordinate BDNF-induced chemotaxis of cerebellar precursors. Neuron. 2007;55:53–68. doi: 10.1016/j.neuron.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohara K, et al. A local reduction in cortical GABAergic synapses after a loss of endogenous brain-derived neurotrophic factor, as revealed by single-cell gene knock-out method. J Neurosci. 2007;27:7234–7244. doi: 10.1523/JNEUROSCI.1943-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paradis S, et al. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 49.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.