Abstract
The rnpA mutation, A49, in Escherichia coli reduces the level of RNase P at 43°C because of a temperature-sensitive mutation in C5 protein, the protein subunit of the enzyme. Microarray analysis reveals the expression of several noncoding intergenic regions that are increased at 43°C compared with 30°C. These regions are substrates for RNase P, and they are cleaved less efficiently than, for example, tRNA precursors. An analysis of the tna, secG, rbs, and his operons, all of which contain RNase P cleavage sites, indicates that RNase P affects gene expression for regions downstream of its cleavage sites.
Several years ago, it was reported that two of the enzymes in the lac operon were produced in different molar quantities, but no direct explanation was provided for this result (1). General speculative hypotheses for differing molar concentrations of proteins coded for in operons have also been suggested (2). Intergenic noncoding RNAs in operon mRNAs in Escherichia coli may provide endoribonuclease sites as one method of allowing production of operon mRNAs and proteins with differing molar concentrations. Products of endoribonuclease cleavage could be degraded rather quickly by an mRNA nuclease degradation mechanism involving RNase E. The small intergenic RNAs could be substrates for RNase P, and they would indicate that RNase P might process many more RNAs than was originally thought.
RNase P itself is an essential enzyme responsible for the processing of the 5′ ends of precursors to tRNA and other naturally occurring stable RNA molecules in E. coli (3). In Salmonella, there is one region in an operon mRNA that appears to be cleaved by RNase P (4). In this report, the noncoding intergenic regions and genes coding for proteins (ORFs) in several operons were cloned separately and examined as substrates for RNase P. Several of the intergenic transcripts were substrates for RNase P. These transcripts accumulate at the restrictive temperature in an E. coli mutant strain thermosensitive for RNase P function. ORFs in the relevant operons were not substrates. A mechanism of determining gene expression is described in which the action of a specific endoribonuclease controls regions downstream of its cleavage site in polycistronic operon mRNAs.
Materials and Methods
Strains of E. coli. E. coli A49 and several derivatives of this strain are described in ref. 5 and references therein.
Cloning and Nomenclature of Intergenic RNAs. Several intergenic regions, the expression of which was high at 43°C in A49 (5), were chosen for cloning. Clones were amplified by PCR starting generally with the first nucleotide after the stop codon of an upstream gene and the last nucleotide before the start codon of a downstream gene. Clones were numbered according to the length of the sequences cloned and w or c, depending on whether transcription was in the clockwise (w) or counterclockwise (c) direction on the chromosome. The start and stop sites for each fragment on the chromosome are listed below, with the numerology based on that of Blattner et al. (6): 205w (length 205 nt) from 3,886,139 to 3,886,343; 90w from 3,888,775 to 3,887,864; 125w from 3,934,795 to 3,934,919; 145w from 2,088,069 to 2,088,213; 154w from1,091,361 to 1,091,514; and 82c from 3,320,147 to 3,320,228.
Transcription of RNAs. Transcription was carried out with T7 RNA polymerase after clones were digested with BstBI. Transcripts were purified on 7 M urea- 6% gels and exposed to RNase P holoenzyme from E. coli. The substrate concentrations were 100 nM, the enzyme concentration was M1 RNA 1 nM, and C5 protein was 10 nM. Incubation was for 37°C for 15 or 30 min.
Translation of Intergenic RNAs. b0669 and b0671, when assayed as one contiguous clone, translated a peptide in vitro by using the TNT T7 Quick Coupled Transcription/Translation System (Promega). The clones were identified originally by Blattner et al. (6).
Results
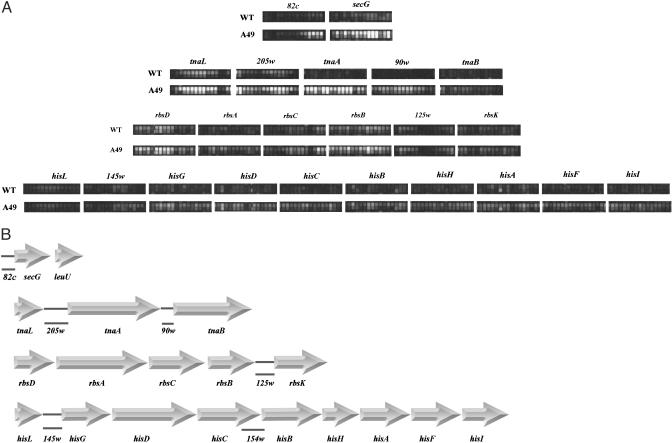
Regions of Interest. To investigate global roles of RNase P, highdensity DNA oligonucleotide microarray analysis was used to examine the appearance of RNAs at restrictive temperatures in an E. coli strain mutant in the protein subunit (C5 protein) of RNase P (5). Forty-nine genes were discovered that were expressed at a low level at 30°C but at a much higher level at 43°C in the mutant strain, A49. Twenty-nine of these genes were precursors to tRNAs. Several genes, besides the ORFs b0669 and b0671 (5), were located in polycistronic operons, tna (typtophan degradation), his (histidine biosynthesis), rbs (D-ribose transport), and secG (protein excretion). A picture of hybridizing transcripts on the chip from regions that are of interest here are portrayed in Fig. 1A, and their organization is shown in Fig. 1B. Functional relationships among these operons, the polycistronic mRNAs of which are unstable and turn over in several seconds or minutes, are discussed below. Finally, we note that the transcripts we analyze here are a direct result of the shutdown in RNase P function in the mutant strain, A49.
Fig. 1.
Polycistronic operons. (A) Microarray images of the expression of polycistronic operons (data were taken from ref. 5). Each probe set for coding sequences or intergenic regions consisted of fifteen 25-mer oligonucleotides. The secG gene is followed immediately by leuU, a gene for a leucine tRNA (5), which is processed by RNase P at the beginning of its 5′ mature sequence at a high rate (B). The operons and cloned intergenic regions used in this study. Large arrows indicate ORFs, which are joined by horizontal bars. The intergenic regions are solid lines directly below the parts of the sequence they represent and are designated by 205w, etc. The regions cloned, 205w and other numbered sequences, were chosen to start at the first downstream nucleotide after the stop codon of the upstream ORF and continuedtothe last nucleotide before the start codon of the downstream ORF. For 82c, the sequence starts in the upstream transcribed region and stops at the last nucleotide before the beginning of the secG ORF.
Certain intergenic regions were cloned under the control of the T7 RNA polymerase promoter (see Materials and Methods). These regions generally were between two ORFs of a polycistronic mRNA or, in one case mentioned here, at the 5′ side of a particular ORF at the start of transcription of an operon. The regions are: 205w, between tnaL(C) and tryptophanase (tnaA); 90w, between tnaA and tnaB; 125w, between rbsB and rbsK; 145w, between hisL and hisG; 154w, overlapping hisC and hisB; 82c, start of transcription of secG to the start codon of secG (Fig. 1B). The resulting plasmids contain the T7 polymerase promoter, the fragment cloned, and a restriction site, BstBI. Three other intergenic regions of operon mRNAs were not substrates for RNase P.
Cleavage by RNase P. After DNA restriction enzyme digest, the cloned DNAs were transcribed by T7 RNA polymerase, and the transcripts were exposed to the RNase P holoenzyme from E. coli (see Materials and Methods). Neither M1 RNA nor C5 protein, the RNA and the protein subunits of RNase P from E. coli, had any activity on these substrates by themselves. All of the transcripts were cleaved by the holoenzyme except for 154w, which had to be shortened to be cleaved (see below).
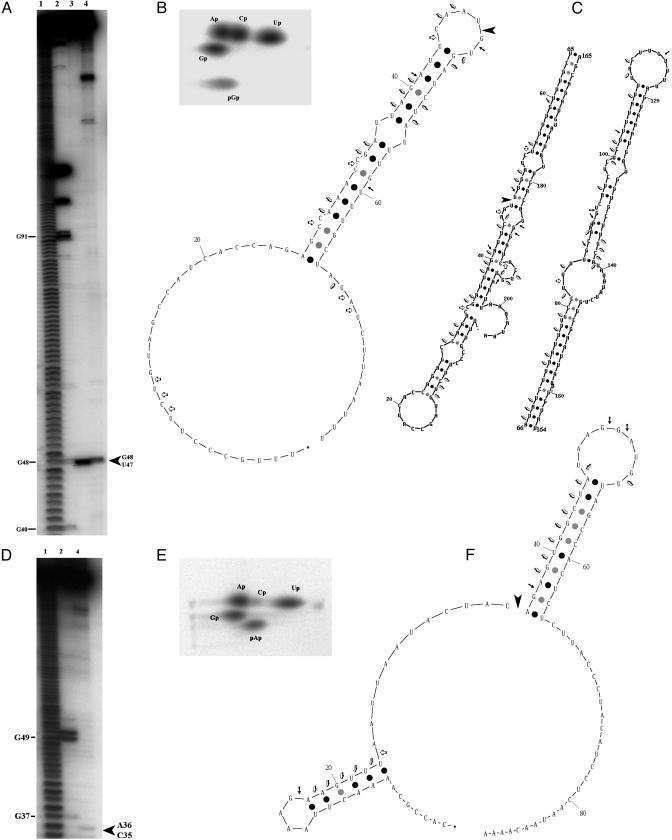
The tna operon transcripts were cleaved in 205w (between U47 and G48; Fig. 2A), producing a 5′ phosphate on the 3′ cleavage product and a hydroxyl on the 5′ cleavage product (Fig. 2B). The secondary structure is unknown, although one can draw structures using a Zuker program (ref. 7; www.bioinfo.rpi.edu/applications/mfold) that might be correct. The theoretical structures depend on the number of nucleotides included, which are limited to the intergenic regions in our cases and are reliable in calculations with the Zuker program. Yanofsky and colleagues (8, 9) have also drawn simpler structures of this region using a more limited number of downstream nucleotides than we have (Fig. 2C). Examination of the cleavages by RNase T1 (cleavage after accessible G residues) or RNase V1 (cleaves double-stranded RNA) in 205w indicates that the Yanofsky structure (Fig. 2C) is more clearly correct, but it includes fewer nucleotides of the total sequence we cloned. The presence of RNase V1 hits in the loop adjacent to the RNase P cleavage site has yet to be shown to portray an experimentally proven double-stranded RNA structure. An examination of 90w (Fig. 2F) indicates that this secondary structure may be similar to the experimental result with RNases T1 and V1.
Fig. 2.
RNase P cleavage of the tna operon intergenic regions. (A) 205w RNA cleaved by RNase P and RNase T1. RNA transcribed by T7 RNA polymerase was labeled with [γ-32P]ATP (lanes 1–3) or internally labeled with [α-32P]GTP (lane 4). Lane 1, alkaline ladder; lane 2, RNase T1; lanes 3 and 4, RNase P. The large arrow (Right) indicates the cleavage site. (B) Determination of the 5′ end group of the 3′ product of RNase P cleavage. The RNase P substrates, labeled with [α-32P]GTP (205w) or [α-32P]ATP (90w), were treated with RNase P and then with RNase T2. The 3′ product of cleavage was run in a 2D thin-layer chromatography system for analysis (10). (C) Theoretical schemes of the secondary structure of 205w RNA. (Left) A scheme based on diagrams by Gong and Yanofsky (9). (Right) A scheme based on the mfold program of Zuker (7). The right side represents the same RNA that is divided to allow a representation. Cleavage by RNase P ( ), RNase T1 (→) and RNase V1(
), RNase T1 (→) and RNase V1( ; Ambion, Austin, TX) is shown. (D–F) Same as in A–C for 90w RNA, except that D has only three lanes.
; Ambion, Austin, TX) is shown. (D–F) Same as in A–C for 90w RNA, except that D has only three lanes.
The cleavage site determined by RNase P in 205w does not correspond to the classic model RNase P site at the 5′ base of a helix in the structure we have drawn (1), although the cleavage site in 90w does (between C35 and A36; Fig. 2). Kinetic constants of cleavage by RNase P indicate why these sites are less favored than the well known cleavage sites.
The kcat and Km of the cleavages in 205w and 90w are shown in Table 1 and are compared with published values for cleavage of a tRNA precursor and the precursor to 4.5S RNA (p4.5). The values of the enzymatic constants for the other substrates are also shown. The Km values are higher than the published values, and the kcat/Km values are ≈10-fold or more lower than the values for tRNA precursors and for p4.5S RNA (3). These data indicate that, as expected, RNase P processes precursors to stable RNA at a high rate and allows low-efficiency processing of polycistronic mRNAs. In the last case, the enzyme functions as a means of determining the amount of downstream sequences in an operon by promoting degradation after cleavage of the polycistronic mRNA by itself. The large Km values of the new transcripts indicate that recognition of these structures is a significant obstacle in the enzyme cleavage reaction. One also notes that the amount of the RNA transcripts of tnaL (5–300 depending on induction), tnaA (0–10), and tnaB (0–5) per cell per generation is significantly smaller in vivo as calculated from our microarray data than that of, for example, total tRNA (2 × 105).
Table 1. Enzymatic kinetics of RNase P with various substrates.
| Substrates | Km, μM | kcat, min-1 | kcat/Km, min-1·μM-1 |
|---|---|---|---|
| 205w | 0.42 | 14 | 33 |
| 90w | 0.25 | 29 | 116 |
| 125w | 6.8 | 31 | 4.6 |
| 145w | 11 | 2.1 | 0.19 |
| 154w | 0.54 | 8.6 | 16 |
| 82c | 13 | 2.2 | 0.17 |
| ptRNATyr | 0.033 | 29 | 879 |
| p.4.5S RNA | 0.055 | 56 | 1,006 |
These results represent an experiment in which the enzyme parameters for new substrates were measured together. Data for pTyr and p4.5S RNA are published in ref. 3.
When a normal precursor tRNA, pSupS1, was exposed with different concentrations to RNase P in the presence of an intergenic region, 205w, the cleavage of 205w was inhibited as the concentration of pSupS1 was raised (unpublished data). The 205w transcript was cleaved at a rate at least 1,000-fold less than pSupS1 at the same concentration of substrate.
All of the other cleavages are shown in Figs. 7A,8A,9A, and 10A, which are published as supporting information on the PNAS web site, and corresponding 2D thin-layer chromatographs (10), demonstrating the appearance of 5′ phosphates in the 3′ terminal product, are shown in Figs. 7B and 8B. Secondary structures are shown in Figs. 7C,8C,9B, and 10B. The cleavage sites are as follows: 125w, two sites between C82 and G83 and between U112 and G113; 145w, between A122 and C123; 154w, between C69 and U70; 154w shortened by different restriction sites: only the short fragment of 86 nt was cleaved as shown in Fig. 9; 82c between G54 and C55 (Fig. 10).
Other Properties of the tna Operon. The microarray data indicated all of the operons we studied with cells grown in LB broth are elevated in amounts at 43°C in A49 (Fig. 1A). Tryptophan was present in the cell growth medium, and catabolite repressors that affected the tna operon were absent. The transcription of tnaL was, therefore, not terminal, in the sense that transcription stopped after tnaL. Transcription continued after tnaL, as expected.
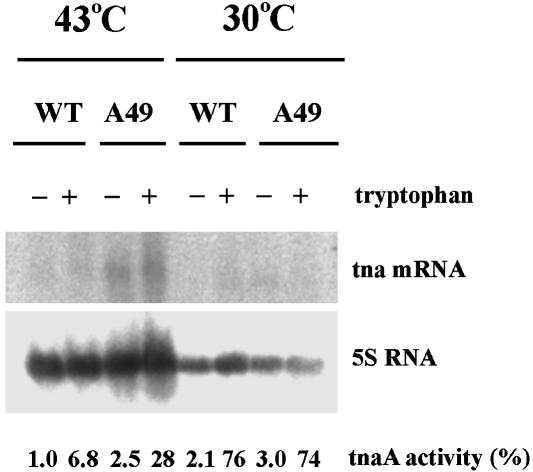
To determine whether the basal level of expression of a protein in an operon was affected by RNase P, cells were grown in minimal medium, and cell extracts were probed by Northern blots and by a tnaA activity assay. The mRNA of tnaA was elevated when A49 was shifted from 30 to 43°C, even without tryptophan induction. Similarly, tnaA activity was assayed by using the substrate S-O-nitrophenyl-L-cysteine (11). The enzyme activity of A49 is lower at 43°C than at 30°C in the same strain but higher than that of the wild type at the restrictive temperature (Fig. 3). These data show that the absence of RNase P activity in A49 at 43°C allows an increase of tnaA activity compared with the wild type. At 43°C in A49, there is a scarcity of aminoacylated tRNAs because of a general shutdown in biosynthesis of tRNA molecules (12). [These assays were done in E. coli that also carries the A49 mutation. The mutation has been moved by phage transduction into BL21(DE3) (13)].
Fig. 3.
The level of tnaA mRNA and the tnaA enzymatic activities of wild-type and A49 derivatives of BL21(DE3). The cells were grown in minimal medium (Vogel and Bonner medium supplemented with 0.2% glucose and 0.05% casein) with or without tryptophan (100 μg/ml) at 30°C until their OD 600 nm reached 0.25. Culture was then continued for 45 min at either 30 or 43°C. Cells were collected for RNA extraction and Northern blots (3) or prepared for extraction for the tnaA activity assay. The tnaA activity is standardized to 1.0, which means 35 nmol product formed per minute per milligram protein of cell extract.
Tryptophan induces tnaA activities for both the wild-type and A49 strains >30-fold at 30°C but only 7- to 10-fold at 43°C (Fig. 3). In fact, it has been suggested that tnaA activity is not thermostable unless a sufficient quantity of pyridoxal phosphate (PLP) is present in solution with it (14). The activity of tnaA was assayed with and without PLP at 30 and 43°C. No activity was lost in PLP at the higher temperature, but ≈80% was lost without PLP, as was the case in our initial assays.
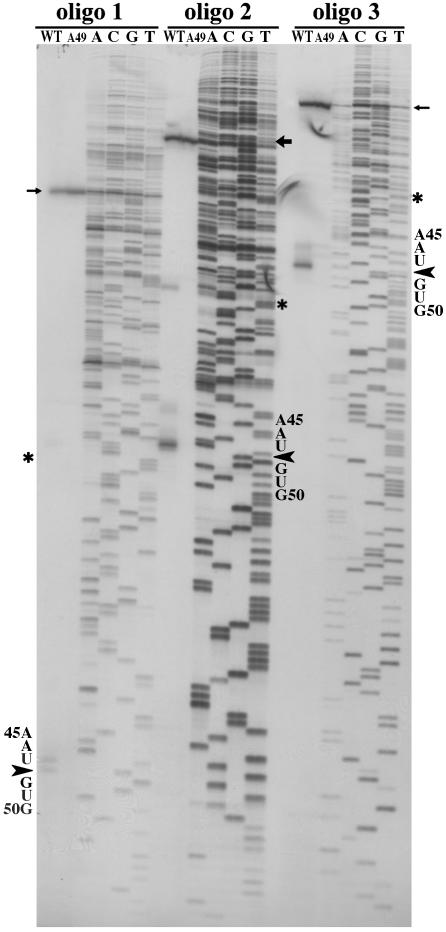
Finally, in the wild-type strain, there should be the remnant of RNase P cleavage of the 205w fragment. This fragment should be absent from the A49 derivative at 43°C. RNA was extracted from these strains and reverse transcribed with three different primers complementary to 205w.
The results as shown in Fig. 4 indicate that only in the wild-type lanes is there any indication of a fragment that resulted from cleavage by RNase P. As listed in Table 2, which is published as supporting information on the PNAS web site, all other relevant RNases are about at the same level in the wild-type and A49 strains: only RNase P transcripts are lowered in the A49 derivative, and no RNase P activity could be detected therein at 43°C after 45 min of growth. The enzyme is fully active in the wild-type parent.
Fig. 4.
The cleavage product of RNase P cleavage of 205w in vivo. Primer extension of total RNA was used to detect the product of RNase P cleavage. Wild-type (200 μg) and A49 (50 μg) RNAs from the cells cultured at 43°C for 45 min were annealed to three oligonucleotides separately and reverse transcribed by SuperScript II enzyme (Invitrogen). The mixtures were then layered onto an 8% denaturing PAGE adjacent to sequencing reactions with the three oligonucleotides and the 205w plasmid DNA as template:  , transcription starting site; *, the 5′ end of the sequence of 205w, itself;
, transcription starting site; *, the 5′ end of the sequence of 205w, itself;  , RNase P cleavage site. Oligo 1 covered 69–92 of 205w; oligo 2 covered 116–142 of 205w; oligo 3 covered 185–205 of 205w. The RNase P cleavage product is in the wild-type lane only. The scales, base names, and cleavage sites are expanded for oligos 2 and 3.
, RNase P cleavage site. Oligo 1 covered 69–92 of 205w; oligo 2 covered 116–142 of 205w; oligo 3 covered 185–205 of 205w. The RNase P cleavage product is in the wild-type lane only. The scales, base names, and cleavage sites are expanded for oligos 2 and 3.
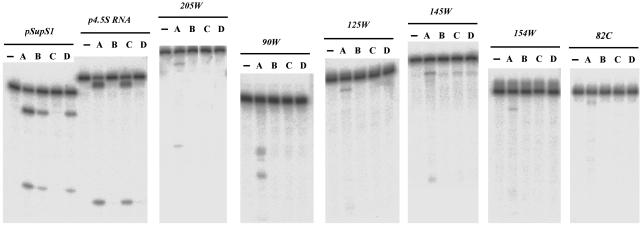
Mutants of M1 RNA and Their Function on New Substrates. Wild-type RNase P and mutants thereof were assayed on precursor to yeast serine tRNA (pSupS1), the precursor to E. coli tyrosine tRNA and 4.5S RNA (p4.5), and some of the new substrates we identified. Δ (94–204) M1 RNA with C5 protein is not active on precursor tRNA. Δ65 M1 RNA and Δ245 M1 RNA with C5 protein are not active on p4.5 (refs. 15, 16; unpublished data). In every case of the new substrates, none of the mutants was active (Fig. 5). This result demonstrates that none of the new substrates was very similar in tertiary structure to either precursor tRNA or p4.5.
Fig. 5.
Intergenic RNAs cleaved by RNase P with different mutants in M1 RNA. Shown is RNase P using different M1 RNA and wild-type C5 protein. Lane 1, control; lane A, wild-type M1 RNA; lane B, M1 RNA Δ65; lane C, M1 RNA Δ (94–204); lane D, M1 RNA Δ245.
Discussion
RNase P Influences Subsequent Degradation of Polycistronic Substrates. Although we have shown that several polycistronic mRNAs are cleaved by RNase P in intergenic regions, the results we have obtained with the tna operon are indicative of a functional role in vivo. In this case, the amount of tna mRNA is increased at restrictive temperature in an RNase P temperature sensitive mutant and the appearance of intermediate products of RNase P cleavage are apparent only in a wild-type strain but not in the mutant strain at 43°C. Assays of tnaA activity are also higher in the mutant strain at high temperature compared with the wild-type strain. These data are all compatible with RNase P affecting the amount of tnaA and tnaB products in the wild-type strain. That influence is regulated by the apparent enzyme kinetics of RNase P on the cleavage sites in the tna mRNA, which allow for a lower rate of processing than, for example, tRNA precursors.
There appears to be no correlation of RNase P function with transcriptional control, initiation, or termination of tnaA. Subsequent to RNase P cleavage of the tna operon, RNase E degrades the rest of the cleaved products, via the degradasome, because this enzyme attacks preferentially molecules with a 5′ monophosphate, the actual product of RNase P cleavage (17). Experiments in vitro indicated that RNase E can degrade the products of RNase P cleavage (unpublished data). Our experiments have no relevance to the finding that the half-life of some mRNAs depends on the 3′ terminal sequence of these mRNAs and their removal by exonucleases.
The tna Operon. The expression of the tna operon depends on concentration of cAMP-receptor protein (CRP) and tryptophan induced transcription termination by the Rho protein (18). CRP and Rho are present in about equal concentrations in wild-type A49 derivatives (Table 2). Experiments in vitro show that only RNase P holoenzyme is involved in these cleavages so the action of Rho is not necessary for RNase P activity. Similarly, the action of Rho does not require the action of RNase P as one can assume by examining recent results on transcription controls from Gong and Yanofsky (personal communication). Other data indicate that in the presence or absence of tryptophan, RNA polymerase pausing and restarting or basal level transcription occur, neither appears to affect the location and efficiency of specific cleavages by RNase P.
Five bacteria aside from E. coli contain the tna operon. (Shigella flexneri, Haemophilus influenzae, Proteus vulgaris, Enterobacter aerogenes, and Pasteurella multiocida; refs. 19–23). All contain similar structural features (e.g., a Rho site) even though some intergenic sequences are changed in size. There is a long perfect sequence stem between tnaA and tnaB among these bacteria. Disrupting latter feature in 90w of E. coli prevents RNase P from cleaving fragment (unpublished data).
The his and rbs Operons. In histidine biosynthesis, phosphoribosyl pyrophosphate and ATP are initial substrates, and they link pathway to the biosynthesis of pyrimidine, purine, pyridine, tryptophan (24–26). The elevated level of his operon RNA in RNase P defective strain, A49, is illustrated in Fig. 1A. The level hisL and the first half of 145w are comparable to that in the wild type, but the level of the transcript downstream neighboring RNase P cleavage was elevated in A49. This high level is caused by transcription initiation or attenuation, because there are no other factors that affect the downstream sequence, but by the inability RNase P to cleave the his operon substrate. The hisC mRNA could be processed by RNase P but only when considerably reduced size at its 3′ end, suggesting that it does get cleaved after the action at a downstream site by RNase E (4).
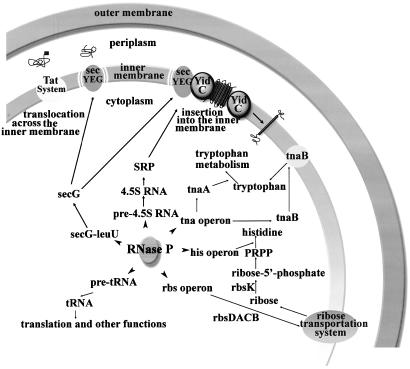
The connection between the rbs and the his operon is also interest. Ribose is first phosphorylated by rbsK to ribose 5′ phosphate, which is then converted to phosphoribosyl pyrophosphate (PRPP) by ribose 5′ phosphate kinase (24–26). The genes of rbsD, rbsA, rbsC, rbsB (all involved in transporting D-ribose), and rbsK are all affected by RNase P (Fig. 6). These data imply that the level of PRPP is determined by the nuclease, RNase P. Thus, ribose transportation and utilization and histidine biosynthesis are related at the posttranscriptional level by the action of RNase P.
Fig. 6.
Schematic of some global functions of RNase P in E. coli.
RNase P in Transport. In Gram-negative bacteria, secY, secE, and secG form the membrane-embedded core complex of the protein transport apparatus (27). Largely hydrophobic proteins are selectively recognized by the RNA component of which the signal recognition particle (SRP), 4.5S RNA, depends on RNase P for its biosynthesis. The excretion of secretory proteins involves the posttranslational binding of protein by the chaperone secB and its subsequent transfer to secA, which then translocates the protein across the secYEG channel (Fig. 6). The function of RNase P in these transport processes can be summarized in the following way: (i) maturation of SRP RNA depends on RNase P; (ii) the5′ UTR of secG, an important stimulator of protein translocation, can be cleaved by RNase P leading to its lowered expression; (iii) all of the genes involved in ribose transport (rbsA–D and rbsK) are reduced in expression by the action of RNase P; and (iv) the low expression of tnaB, a low-affinity tryptophan permease, is controlled by RNase P.
Finally, b0669 and b0671, two putative ORFs identified in the E. coli genome (5), which are not part of an operon, are coded on the complementary strand to a tRNA polycistronic RNA, and they are part of the high-level expression group in the microarrays in the A49 mutant strain used as a prelude to this study. RNase P does not cleave these transcripts. b0669 shares homology with an intracellular alkaline protease of Bacillus clausii, whereas b0671 does so with a cross-ER membrane fragment of rat InsP3 receptor (6). We propose that b0669 and b0671 might form one ORF that encodes a protein across or embedded in the inner membrane of E. coli (Materials and Methods and unpublished data).
RNase P in E. coli is under the control of the rel locus and ppGpp (3). Little direct data have been shown that these factors actually do affect the amount of subunits in culture. However, if this were the case, then RNase P would regulate gene control in operons. Additional experiments must be performed in this case. In the examples studied here, there is no doubt that RNase P behaves in intergenic regions adjacent to ORFs exactly as it does on precursor tRNAs at the permissive temperature and at the restrictive temperature.
Other Roles. Some operons are not induced under the conditions used here to prepare for microarray analysis. For example, the β-galactosidase operon is induced only in the presence of lactose and some of its derivatives. This latter operon transcript does show RNase P cleavage between lacY and lacA (1) but no cleavage between lacZ and lacY in preliminary experiments in vitro. In eukaryotes, there are no operons of the kind we studied in a prokaryote, E. coli. Although we have not tested flanking regions of eukaryotic genes involved in metabolism as substrates for RNase P, a distinct mechanism in which these regions are not substrates for the enzyme may prove important in further delineating eukaryotes from prokaryotes.
Supplementary Material
Acknowledgments
We are extremely grateful to Dr. C. Yanofsky for supportive advice and encouragement. We thank Donna Wesolowski for technical assistance and discussions with the members of our laboratory. Dr. K. Cole (Affymetrix, Santa Clara, CA) kindly assisted with preparation of Fig. 1 A. We thank Drs. C. Yanofsky (Stanford University, Stanford, CA) and R. Phillips (University of Georgia, Athens) for generous gifts of the substrate for the tnaA assay and Dr. G. Mackie (Department of Biochemistry, University of British Columbia, Vancouver) for RNase E. This research was supported by National Institutes of Health Grant GM-19422 (to S.A.).
Abbreviations: w, clockwise; c, counterclockwise.
References
- 1.Zabin, I. & Fowler, A. V. (1978) in The Operon, eds. Miller, J. H. & Reznikoff, W. S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 111-112.
- 2.Beckwith, J. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol. Press, Washington, DC), p. 1230.
- 3.Altman, S. & Kirsebom, L. (1999) in The RNA World, eds. Gesteland, R., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 351-380.
- 4.Alifano, P., Rivellini, F., Piscitelli, C., Arriano, C. M., Bruni, C. B. & Carlomagno, M. S. (1994) Genes Dev. 8, 3021-3031. [DOI] [PubMed] [Google Scholar]
- 5.Li, Y., Cole, K. & Altman, S. (2003) RNA 9, 518-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., Plunkett, G, III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Zuker, M. (2003) Nucleic Acids Res. 31, 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart, V., Landick, R. & Yanofsky, C. (1986) J. Bacteriol. 166, 217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, F. & Yanofsky, C. (2002) J. Biol. Chem. 277, 17095-17100. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura, S. (1972) Prog. Nucleic Acid Res. Mol. Biol. 12, 49-85. [PubMed] [Google Scholar]
- 11.Stewart, V. & Yanofsky, C. (1985) J. Bacteriol. 164, 731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schedl, P., Primakoff, P. & Roberts, J. (1974) in Brookhaven Symposia in Biology, ed. Dunn, J. J. (Brookhaven Natl. Lab., Brookhaven, NY), Vol. 26, pp. 31-76. [Google Scholar]
- 13.Guerrier-Takada, C., Li, Y. & Altman, S. (1995) Proc. Natl. Acad. Sci. USA 92, 11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton, W. A., Morino, Y. & Snell, E. E. (1965) J. Biol. Chem. 240, 1211-1218. [PubMed] [Google Scholar]
- 15.Guerrier-Takada, C., Lumelsky, N. & Altman, S. (1989) Science 246, 1578-1584. [DOI] [PubMed] [Google Scholar]
- 16.Guerrier-Takada, C. & Altman, S. (1992) Proc. Natl. Acad. Sci. USA 89, 1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coburn, G. A. & Mackie, G. A. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 55-108. [DOI] [PubMed] [Google Scholar]
- 18.Gong, F., Ito, K., Nakamura, Y. & Yanofsky, C. (2001) Proc. Natl. Acad. Sci. USA. 98, 8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, Q., Yuan, Z., Xu, J., Wang, Y., Shen, Y., Lu, W., Wang, J., Liu, H., Yang, J., Yang, F., et al. (2002) Nucleic Acids Res. 30, 4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May, B. J., Zhang, Q., Li, L. L., Paustian, M. L., Whittam, T. S. & Kapur, V. (2001) Proc. Natl. Acad. Sci. USA 98, 3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, K., Morlin, G., Smith, A., Nordyke, A., Eisenstark, A. & Golomb, M. (1998) J. Bacteriol. 180, 107-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki, K., Yokota, A., Oita, S., Kobayashi, C., Yoshikawa, S., Kawamoto, S., Takao, S. & Tomita, F. (1993) J. Gen. Microbiol. 139, 3275-3281. [DOI] [PubMed] [Google Scholar]
- 23.Kamath, A. V. & Yanofsky, C. (1992) J. Biol. Chem. 267, 19978-19985. [PubMed] [Google Scholar]
- 24.Winkler, M. (1996) in Esherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol. Press, Washington, DC), pp. 485-505.
- 25.Iida, A., Harayama, S., Iino, T. & Hazelbauer, G. L. (1984) J. Bacteriol. 158, 674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopilato, J. E., Garwin, J. L., Emr, S. D., Silhavy, T. J. & Beckwith J. R. (1984) J. Bacteriol. 158, 665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valent, Q. A., Scotti, P. A., High, S., deGier, J.-W. L., von Heijne, G. & Luirink, J. (1998) EMBO J. 17, 2504-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.