Abstract
The human voltage-gated sodium channel gene cluster on chromosome 2q24 contains three paralogs, SCN1A, SCN2A, and SCN3A, which are expressed in the central nervous system. Mutations in SCN1A and SCN2A cause several subtypes of idiopathic epilepsy. Furthermore, many SCN1A mutations are predicted to reduce protein levels, emphasizing the importance of precise sodium channel gene regulation. To investigate the genetic factors that regulate the expression of SCN1A, SCN2A, and SCN3A, we characterized the 5′ untranslated region of each gene. We identified multiple noncoding exons and observed brain region differences in the expression level of noncoding exons. Comparative sequence analysis revealed 33 conserved noncoding sequences (CNSs) between the orthologous mammalian genes, and six CNSs between the three human paralogs. Seven CNSs corresponded to noncoding exons. Twelve CNSs were evaluated for their ability to alter the transcription of a luciferase reporter gene, and three resulted in a modest, but statistically significant change.
Keywords: epilepsy, sodium channel, SCN1A, SCN2A, SCN3A, 5′ untranslated region, sequence alignment, conserved sequence, gene expression regulation
Introduction
Voltage-gated sodium channels are transmembrane proteins that activate in response to membrane depolarization, allowing the influx of sodium ions into the cell and the propagation of an action potential. Voltage-gated sodium channels are composed of one α subunit and one or more auxiliary β subunits. The α subunits are large, 260-kDa transmembrane proteins with four extracellular pore-forming loops and an intracellular inactivation gate. The human genome contains ten α subunit isoforms with different temporal and spatial expression patterns and distinct biophysical properties (for review [1]). Eight of these isoforms are localized in two gene clusters on human chromosomes 2q24 and 3p21. These clusters arose from gene duplication events in ancestral genomes [2]. The cluster on chromosome 3p21 contains the cardiac sodium channel SCN5A and the peripheral nervous system sodium channels SCN10A and SCN11A. The cluster on chromosome 2q24 contains three of the four sodium channels that are primarily expressed in the central nervous system, SCN1A, SCN2A, and SCN3A, and the peripheral nervous system sodium channels SCN7A and SCN9A.
Mutations in SCN1A and SCN2A have been identified in autosomaldominant subtypes of human epilepsy. Mutations in both genes lead to Generalized Epilepsy with Febrile Seizures Plus (GEFS+; MIM 604233) [3, 4]. SCN2A mutations also lead to Benign Familial Neonatal-Infantile Seizures (BFNIS, MIM 607745) [5], while SCN1A dysfunction is the major cause of Severe Myoclonic Epilepsy of Infancy (SMEI or Dravet syndrome; MIM 607208) [6]. Mutations in the coding exons or the exon-intron junctions of SCN1A occur in approximately 10% of GEFS+ families and 50% of SMEI individuals [7]. The majority of SCN1A mutations identified in SMEI patients are predicted to abolish channel function, resulting in reduced protein levels. This haploinsufficiency phenotype demonstrates the importance of maintaining normal sodium channel expression levels and suggests that reduced channel expression would lead to altered neuronal excitability.
There is growing recognition that differences in gene expression in humans are, at least in part, caused by sequence variation in functional cis-DNA elements, and an increasing number of disorders are associated with mutations in noncoding elements [8, 9]. However, identifying this important class of genomic elements has been challenging, and the majority of functional noncoding DNA elements are currently unknown. The present interest in the identification and characterization of this class of genomic elements is reflected by the initiation of large-scale studies, such as the ENCODE project (ENCyclopedia of DNA Elements), whose goal is to identify all functional human DNA elements [10]. However, the genomic region that contains SCN1A, SCN2A, and SCN3A is not currently under investigation by the ENCODE consortium. As a first step towards determining if mutations in the noncoding regulatory elements of these genes contribute to disease, we used a combination of bioinformatics and functional analyses to identify potential cis-regulatory elements within the SCN1A, SCN2A, and SCN3A loci.
Results
The genomic organizations of the SCN1A, SCN2A, and SCN3A loci are evolutionarily conserved
To examine the evolutionary conservation of the SCN1A, SCN2A, and SCN3A loci, a contiguous 1.1-Mb genomic region of human chromosome 2q24 containing the three sodium channels and the intervening TAIP-2 and GALNT3 genes was aligned to the orthologous region in mouse (chromosome 2qC1.3), rat (chromosome 3q21), dog (chromosome 36), and chicken (chromosome 7). Both the gene order and orientation of the five genes were conserved in all species examined (Fig. 1), indicating that the architecture of this genomic region has been maintained for at least 310 million years, since the divergence of mammals and birds [11]. The 26 coding exons of human SCN1A, SCN2A, and SCN3A were distributed over 83 kb, 96 kb, and 87 kb of genomic DNA, respectively. The intron-exon structures of the orthologous genes were highly conserved. The 3′ untranslated region (UTR) of each gene was also highly conserved, with approximately 80% sequence identity between the orthologous human and mouse genes.
Figure 1.
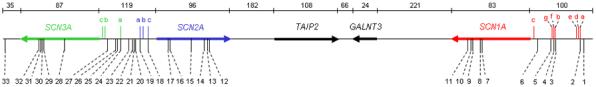
Physical map of the sodium channel gene cluster on human chromosome 2q24, showing the position and orientation of each gene and the location of noncoding exons (lowercase letters) and CNSs (numbered 1-33). Noncoding exons are color-coded as follows: red, SCN1A; blue, SCN2A; green, SCN3A. Genomic distances are in kilobases.
Organization of the 5′ untranslated regions of SCN1A, SCN2A, and SCN3A
Since cis-regulatory elements often are located upstream of coding exons, we first determined the organization of the 5′ UTR of SCN1A, SCN2A, and SCN3A by performing 5′ rapid amplification of cDNA ends (5′ RACE) on total RNA from human and mouse brain. We confirmed the expression of all identified noncoding exons by reverse transcription-polymerase chain reaction (RT-PCR) analysis.
SCN1A
Sequencing of more than 150 5′ RACE clones from human frontal cortex, cerebellum, and hippocampus identified three frequently used noncoding exons, designated exon 1a to exon 1c (GenBank accession nos. DQ993522 to DQ993524), contained in five splice variants with frequencies greater than 5% (Figs. 1 and 2A, Table 1). Transcripts in which exon 1a spliced directly into exon 1 were observed most frequently, representing 54% of clones. The most distal exon, exon 1a, was located 75 kb upstream of the first coding exon, exon 1. Four rare noncoding exons, exon 1d to exon 1g (GenBank accession nos. DQ993525 to DQ993527), present in less than 2% of clones were also identified. We found one clone that contained each rare noncoding exon spliced to exon 1c, and then to exon 1. We also observed two clones containing exon 1b spliced to exon 1g, and then to exon 1.
Figure 2.
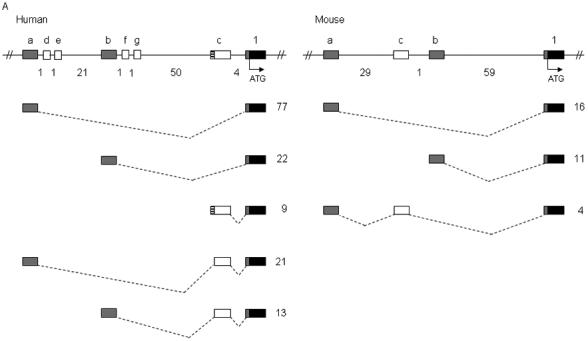
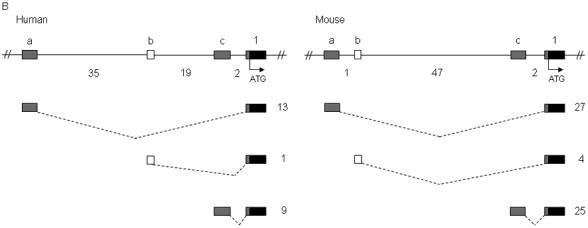
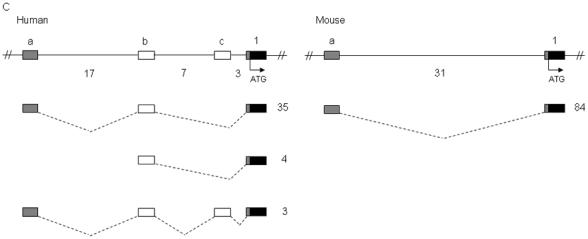
Identification of noncoding exons encoding the 5′ UTR transcripts of SCN1A, SCN2A, and SCN3A. Sequence analysis of 5′ RACE clones identified multiple noncoding exons in (A) human and mouse SCN1A, (B) human and mouse SCN2A, and (C) human and mouse SCN3A. Boxes represent exons: black, coding exons; grey, noncoding exons conserved between human and mouse; white, noncoding exons identified in either human or mouse transcripts. Noncoding exons are named alphabetically and the first coding exon (exon 1) of each gene is indicated. Dashed lines indicate splicing. The number of 5′ RACE clones identified for each transcript is shown to the right of each transcript. Genomic distances between exons are indicated in kilobases. Human SCN1A transcripts with a frequency <2% are not shown.
Table 1.
Characteristics of human and mouse noncoding exons
| Species | Gene | Exon | Length (bp) | % Identity Human/Mouse | 5′ Splice Donor | 3′ Splice Acceptor |
|---|---|---|---|---|---|---|
| Human | SCN1A | 1aa | 231 | 87 | TTGGGGgtaaata | - |
| Human | SCN1A | 1ba | 264 | 84 | AATCAGgtaagcc | - |
| Human | SCN1A | 1c | 139 | - | CCTAAGgtatgca | ccacagGTTATTT |
| Human | SCN1A | 1da | 68 | - | TCCCTTgtaagtg | - |
| Human | SCN1A | 1ea | 86 | - | TTCCAGgtttgat | - |
| Human | SCN1A | 1fa | 39 | - | AATCAGgtttgtt | - |
| Human | SCN1A | 1g | 255 | - | TTTGAGgtatgtg | |
| Human | SCN2A | 1aa | 234 | 76 | TTTCAGgtaagcc | - |
| Human | SCN2A | 1ba | 59 | - | TTTCTGgtatgat | - |
| Human | SCN2A | 1ca | 95 | 86 | ACAGGGgtaatgt | - |
| Human | SCN3A | 1aa | 269 | 80 | TATCAGgtaagct | - |
| Human | SCN3A | 1b | 200 | - | TCTAAGgtaacta | ctacagAGATTAT |
| Human | SCN3A | 1c | 143 | - | AAGAAGgtaaagg | ttgcagGGGAAAA |
| Mouse | Scn1a | 1aa | 153 | 87 | TCGGGGgtaaata | - |
| Mouse | Scn1a | 1ba | 266 | 84 | TATCAGgtaagcc | - |
| Mouse | Scn1a | 1c | 115 | - | AAAGAGgtaaaat | tttcagAAACTAA |
| Mouse | Scn2a | 1aa | 258 | 76 | TTTCAGgtaagca | - |
| Mouse | Scn2a | 1ba | 35 | - | GAAATGgtaacaa | - |
| Mouse | Scn2a | 1ca | 135 | 86 | TCAGGGgtaatgt | - |
| Mouse | Scn3a | 1aa | 268 | 80 | CATCAGgtaagct | - |
Exonic sequences appear in uppercase letters, and intronic sequences appear in lowercase letters.
The exon length was determined by 5′ RACE, and may therefore not be full length.
To determine whether the 5′ noncoding exons were evolutionarily conserved, we performed 5′ RACE on RNA from whole mouse brain. From 31 clones we identified three mouse Scn1a noncoding exons, exon 1a to exon 1c (GenBank accession nos. DQ993528 to DQ993530), contained in three splice variants (Fig. 2A and Table 1). Mouse exons 1a and 1b were orthologous to human exons 1a and 1b with 87% and 84% sequence identity; however mouse exon 1c was not conserved in the human genome. As in humans, mouse transcripts containing exon 1a spliced directly to exon 1 were observed most frequently, accounting for 52% of clones. Genomic sequence orthologous to human exon 1f was identified in the mouse with 95% identity but was not observed in mouse 5′ RACE clones.
SCN2A
Sequence analysis of 23 5′ RACE clones from human frontal cortex RNA revealed three human SCN2A noncoding exons, exon 1a to exon 1c (GenBank accession nos. DQ993531 to DQ993533) (Figs. 1 and 2B, Table 1). Each exon was directly spliced to exon 1, resulting in three transcripts. Transcripts that initiated from exon 1a, located 56 kb upstream of exon 1, were observed most frequently. Analysis of 56 5′ RACE mouse clones identified three noncoding exons, exon 1a to exon 1c (GenBank accession nos. DQ993534 and DQ993535), distributed over 50 kb of genomic sequence. Mouse clones containing exons 1a and 1c were equally represented, whereas only 7% of clones contained exon 1b. Mouse exons 1a and 1c were orthologous to human exon 1a and exon 1c with 76% and 86% sequence identity. Genomic sequence orthologous to mouse exon 1b was conserved in the human genome with 83% identity, but was not identified in any human SCN2A clones.
SCN3A
We examined 42 5′ RACE clones from human frontal cortex and identified three SCN3A noncoding exons, exon 1a to exon 1c (GenBank accession nos. DQ993536 to DQ993538), dispersed over 27 kb of genomic sequence and contained in three splice variants (Figs. 1 and 2C, Table 1). Transcripts in which exon 1a spliced to exon 1b and then to exon 1 were observed most frequently, representing 90% of clones. However, we also identified four clones beginning with exon 1b. Sequence analysis of 84 5′ RACE clones from mouse-brain RNA identified a single Scn3a noncoding exon, exon 1a (GenBank accession no. DQ993539), located 31 kb upstream of exon 1. The orthologous human exon, exon 1a, was 80% identical to the mouse sequence. Although sequence orthologous to human SCN3A exon 1b was conserved with 75% identity in the mouse genome, exon 1b was not identified in mouse 5′ RACE clones.
Brain region differences in Scn1a and Scn2a noncoding exon usage
To determine whether the alternative 5′ UTRs are spatially regulated, we compared the expression profile of total Scn1a and Scn2a transcripts to the expression profile of transcripts initiating from different noncoding exons in nine regions of the adult mouse brain (i.e., cerebellum, brainstem, hippocampus, cortex, thalamus, hypothalamus, striatum, olfactory bulb, and septum) using SYBR-green real-time PCR.
In agreement with Raymond et al. [12], we observed the highest level of total Scn1a expression in the cortex (Fig. 3A). In the cerebellum, brainstem, thalamus, and striatum, total Scn1a expression was 20-35% lower than in the cortex, whereas in the hippocampus, hypothalamus, olfactory bulb, and septum, expression was 50-60% lower than in the cortex.
Figure 3.
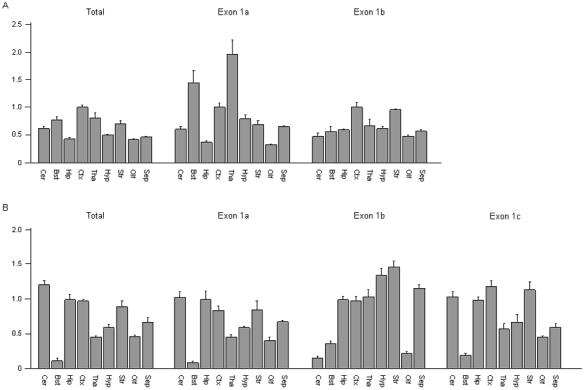
Quantitative differences in the expression levels of distinct Scn1a and Scn2a transcripts. Real-time RT-PCR analysis was used to compare the expression pattern of (A) total Scn1a to transcripts in which exon 1a and exon 1b spliced directly to exon 1, and (B) total Scn2a to transcripts in which exon 1a, exon 1b, and exon 1c spliced directly to exon 1. Brain regions examined: Cer, cerebellum; Bst, brainstem; Hip, hippocampus; Ctx, cortex; Tha, thalamus; Hyp, hypothalamus; Str, striatum; Olf, olfactory; Sep, septum. Scn1a transcripts were normalized to the level of expression in cortex. Scn2a transcripts were normalized to the level of expression in hippocampus. Each bar represents the average value from the analysis of brain tissue from three mice. Error bars represent the SEM.
We quantified transcripts in which Scn1a exon 1a directly spliced to exon 1 using a forward primer positioned over the junction of exon 1a and exon 1 and a reverse primer in exon 1. Unlike the total Scn1a transcripts, which had maximum expression in the cortex, the expression of this transcript in the thalamus and brainstem was approximately 95% and 45% greater than that observed in the cortex (Fig. 3A). We quantified transcripts in which exon 1b was directly spliced to exon 1 using a forward primer located over the junction of exon 1b and exon 1 and a reverse primer in exon 1. The expression profile of the exon 1b transcript was similar to total Scn1a, with the greatest expression observed in the cortex. However, the expression levels of transcripts containing exon 1b were relatively higher in the hippocampus and striatum when compared to the expression profile of total Scn1a (Fig. 3A).
We observed the highest total Scn2a expression in the cerebellum, which is in accord with previous studies (Fig. 3B) [12, 13]. In the hippocampus, cortex, and striatum, expression levels were 20-30% lower than observed in the cerebellum. In the thalamus, hypothalamus, olfactory bulb, and septum, expression levels were 50-70% lower than observed in the cerebellum. We observed the lowest Scn2a expression level in the brainstem.
Since the three mouse Scn2a noncoding exons each spliced directly to exon 1, we were able to quantify each transcript by placing a forward primer in each noncoding exon and a reverse primer in exon 1. The expression profiles observed for exon 1a- and exon 1c-containing transcripts were very similar to total Scn2a expression (Fig. 3B). As observed for total Scn2a expression, the lowest expression of both transcripts was in brainstem. However, transcripts containing exon 1a were not as highly expressed in the cerebellum, and transcripts containing exon 1c were more highly expressed in cortex and striatum when compared to the expression profile of total Scn2a. By contrast, transcripts containing exon 1b showed a distinct pattern, with high expression in the hypothalamus and thalamus and low expression in the cerebellum.
Identification and description of conserved noncoding sequences (CNSs)
To identify additional candidate cis-DNA regulatory elements, we used comparative genomics to detect CNSs. Two segments of the human genome containing 100 kb upstream of SCN1A exon 1 to 35 kb downstream of SCN1A exon 26 and 35 kb downstream of SCN2A exon 26 to 35 kb downstream of SCN3A exon 26 were used to identify human noncoding regions conserved in the mouse, rat, and dog genomes. We identified CNSs with a minimum length of 100 bp and a weighted conservation score greater than 95% using the WebMCS program (http://zoo.nhgri.nih.gov/mcs/) [14]. In addition, we identified CNSs with a minimum length of 100 bp and a score greater than 500 using the phastCon prediction program (http://www.genome.ucsc.edu) (Table 2 and Fig. 1) [15]. In total, 33 CNSs were defined, 20 of which were also conserved in the chicken genome.
Table 2.
Noncoding sequences conserved between the orthologous mammalian SCN1A, SCN2A, and SCN3A genes
| CNS | Locus | Location | Length (bp) | % Identity Human/Mouse | % Identity Human/Chicken | Coordinates |
|---|---|---|---|---|---|---|
| CNS1c | SCN1A | 5′ | 213 | 75 | - | chr2:166,842,083-166,842,296 |
| CNS2a | SCN1A | 5′ | 526 | 81 | 67 | chr2:166,830,939-166,831,465 |
| CNS3ac | SCN1A | 5′ | 404 | 83 | 54b | chr2:166,809,777-166,810,181 |
| CNS4a | SCN1A | 5′ | 142 | 91 | 42 | chr2:166,809,487-166,809,629 |
| CNS5c | SCN1A | 5′ | 200 | 88 | - | chr2:166,808,083-166,808,283 |
| CNS6d | SCN1A | 5′ | 161 | 85 | - | chr2:166,800,604-166,800,765 |
| CNS7d | SCN1A | Intron 11 | 113 | 90 | 83 | chr2:166,725,531-166,725,644 |
| CNS8c | SCN1A | Intron 11 | 265 | 82 | - | chr2:166,724,939-166,725,204 |
| CNS9c | SCN1A | Intron 20 | 195 | 85 | 63b | chr2:166,691,462-166,691,657 |
| CNS10 | SCN1A | Intron 20 | 609 | 86 | 64b | chr2:166,689,133-166,689,742 |
| CNS11 | SCN1A | Intron 20 | 365 | 85 | 59b | chr2:166,685,142-166,685,507 |
| CNS12 | SCN2A | Intron 16 | 310 | 85 | 64 | chr2:166,046,497-166,046,807 |
| CNS13 | SCN2A | Intron 16 | 204 | 82 | 57 | chr2:166,046,115-166,046,319 |
| CNS14 | SCN2A | Intron 16 | 299 | 86 | 76 | chr2:166,039,754-166,040,053 |
| CNS15 | SCN2A | Intron 12 | 341 | 90 | - | chr2:166,010,303-166,010,604 |
| CNS16 | SCN2A | Intron 5 | 346 | 79 | 58 | chr2:165,991,994-165,992,340 |
| CNS17 | SCN2A | Intron 5 | 175 | 84 | 52 | chr2:165,991,605-165,991,780 |
| CNS18ac | SCN2A | 5′ | 592 | 81 | 58b | chr2:165,975,883-165,976,475 |
| CNS19ac | SCN2A | 5′ | 335 | 79 | 55b | chr2:165,921,347-165,921,682 |
| CNS20c | SCN2A or 3A | 5′ | 189 | 80 | - | chr2:165,911,346-165,911,535 |
| CNS21d | SCN2A or 3A | 5′ | 153 | 80 | - | chr2:165,909,714-165,909,867 |
| CNS22 | SCN2A or 3A | 5′ | 448 | 88 | 71b | chr2:165,907,878-165,908,326 |
| CNS23d | SCN2A or 3A | 5′ | 271 | 78 | - | chr2:165,898,175-165,898,446 |
| CNS24ac | SCN3A | 5′ | 597 | 81 | 61b | chr2:165,885,613-165,886,210 |
| CNS25c | SCN3A | 5′ | 190 | 89 | - | chr2:165,882,665-165,882,855 |
| CNS26 | SCN3A | 5′ | 301 | 79 | - | chr2:165,881,079-165,881,380 |
| CNS27 | SCN3A | 5′ | 250 | 84 | - | chr2:165,875,531-165,875,781 |
| CNS28d | SCN3A | Intron 10 | 202 | 93 | - | chr2:165,825,443-165,825,645 |
| CNS29 | SCN3A | Intron 16 | 564 | 80 | 66b | chr2:165,804,405-165,804,969 |
| CNS30d | SCN3A | Intron 16 | 212 | 90 | 56 | chr2:165,802,407-165,802,619 |
| CNS31d | SCN3A | Intron 16 | 210 | 86 | 57b | chr2:165,802,029-165,802,239 |
| CNS32c | SCN3A | Intron 16 | 1031 | 78 | 52b | chr2:165,801,700-165,802,731 |
| CNS33d | SCN3A | 3′ | 248 | 80 | - | chr2:165,737,238-165,737,486 |
CNS corresponds to a noncoding exon: CNS2, SCN1A exon 1a; CNS3, SCN1A exon 1b; CNS4, SCN1A exon 1f; CNS18, SCN2A exon 1c; CNS19, SCN2A exon 1a; CNS24, SCN3A exon 1a
Chicken homology does not extend the entire length of the CNS
CNS identified by WebMCS only
CNS identified by phastCon only
We identified 11 CNSs (CNS1-CNS11) in the genomic segment containing SCN1A (Table 2). Over half of these CNSs (CNS1-CNS6) were located upstream of exon 1, with the most distal CNS (CNS1) positioned 86 kb upstream of exon 1 (Fig. 1). CNS2, CNS3, and CNS4 corresponded to human SCN1A noncoding exons 1a, 1b, and 1f, respectively, demonstrating the utility of bioinformatic approaches for identifying functional elements. The conservation of these three CNSs in the chicken genome suggests that they may function as noncoding exons in multiple species. Three CNSs (CNS9-CNS11), also conserved in the chicken genome, were identified in intron 20.
SCN2A and SCN3A are arranged in a head-to-head orientation, with 119 kb of genomic DNA separating the first coding exon of each gene in the human genome (Fig. 1). Ten CNSs (CNS18-CNS27) were identified in this intragenic region, raising the possibility that these genes may share common regulatory elements (Table 2). CNS18 and CNS19 corresponded to human and mouse SCN2A exons 1a and 1c, and CNS24 corresponded to human and mouse SCN3A exon 1a. We identified six intronic CNSs in the coding region of SCN2A (CNS12-CNS17). Multiple CNSs conserved in the chicken genome were identified in SCN2A intron 5 (CNS16-CNS17) and intron 16 (CNS12-CNS14). Five CNSs were found within the introns of the SCN3A coding region (CNS28-CNS32), four of which were clustered in intron 16. In addition, CNS 33 was found 32 kb downstream of SCN3A exon 26.
We also performed comparative sequence analysis between the human paralogs SCN1A, SCN2A, and SCN3A to identify conserved elements that may potentially regulate the expression of multiple brain sodium channels. We identified six CNSs (CNSA-CNSF) with a minimum length of 100 bp and at least 50% sequence identity across two or more paralogs (Table 3). CNSB and CNSC were conserved among all three sodium channel genes and may contain common regulatory elements. CNSA and CNSB were located upstream of the first coding exon of each gene. CNSB corresponded to noncoding exons conserved between SCN1A (exon 1b), SCN2A (exon 1a), and SCN3A (exon 1a), illustrating that an ancestral form of this exon was present before the gene duplication events gave rise to the cluster of sodium channels (Supplementary Fig. 1). CNSD was conserved between SCN1A and SCN2A, while CNSE and CNSF were conserved between SCN2A and SCN3A. CNSE was the only region not identified from the analysis of the orthologous sequences.
Table 3.
Noncoding sequences conserved between the paralogous human SCN1A, SCN2A, and SCN3A genes
| CNS | Locus | Location | Length (bp) | % Identity SCN1A/SCN2A | % Identity SCN1A/SCN3A | % Identity SCN2A/SCN3A | Corresponding Orthologous CNS(s) | SCN2A Coordinates |
|---|---|---|---|---|---|---|---|---|
| CNSA | SCN1A, 2A, or 3A | 5′ | 393 | 56 | - | - | CNS22 | chr2:165,790,680-165,791,073 |
| CNSBa | SCN1A, 2A, 3A | 5′ | 360 | 50 | 51 | 58 | CNS2, 19, 24 | chr2:165,804,097-165,804,457 |
| CNSC | SCN1A, 2A, 3A | Intron 5 | 239 | 40 | 40 | 56 | CNS17 | chr2:165,874,200-165,874,439 |
| CNSD | SCN1A, 2A | Intron 5 | 622 | 44 | - | - | CNS16 | chr2:165,874,440-165,875,062 |
| CNSE | SCN2A, 3A | Intron 13 | 247 | - | - | 51 | - | chr2:166,024,067-166,024,314 |
| CNSF | SCN2A, 3A | Intron 16 | 464 | - | - | 70 | CNS14, 29 | chr2:166,039,634-166,040,098 |
CNS corresponds to noncoding exons: SCN1A exon 1b, SCN2A exon 1a, and SCN3A exon 1a
Partial duplication of human SCN2A exon 24
While analyzing the MultiPipMaker data, we observed a 78-bp segment of human SCN2A intron 24, located 1.3 kb downstream of the splice donor site of exon 24 that was 96% identical to the last 48 bp of SCN2A exon 24 and the first 30 bp of intron 24 (Fig. 4). The splice donor site of exon 24 was conserved in the duplicated segment, raising the possibility that it may function as an alternative exon. Several consensus splice acceptor sites were identified upstream of the duplicated segment, making it difficult to predict the 5′ junction of the duplication. However, maintenance of the SCN2A open reading frame across the duplicated segment would result in the introduction of a stop codon.
Figure 4.
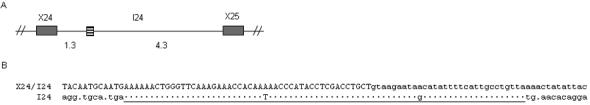
Partial duplication of human SCN2A. (A) A 78-bp duplication (hatched box) consisting of the last 48 bp of exon 24 and the first 30 bp of intron 24 was identified 1.3 kb downstream of the splice donor site of exon 24. (B) The duplicated segment (underlined) is 96% identical to the corresponding sequence at the exon 24/intron 24 junction. Filled boxes, exons; hatched box, duplicated segment. Distances are shown in kilobases. Exonic sequences appear in uppercase and intronic sequences appear in lowercase. Dots indicate sequence identity between the exon 24-intron 24 (X24/I24) junction and intron 24 (I24).
To examine the conservation of the duplicated segment in the human population, the region containing the duplication was PCR amplified from the genomic DNA of 40 individuals from the HGDP-CEPH diversity panel (specifically, seven Africans, seven Middle Easterners, nine Asians, seven Europeans, four Russians, and six South Americans), and then sequenced. The duplicated segment was present in all the individuals examined, and the sequence was invariant. We did not observe the duplicated segment in Scn2a intron 24 in the chimpanzee, mouse, rat, dog, or chicken genomes. However, the current build of the chimpanzee genome (March 2006) contains two gaps in intron 24, which we could not fill by PCR amplification of chimpanzee genomic DNA.
To determine if the duplicated segment functions as an alternative exon, we performed RT-PCR analysis on first-strand cDNA from both human fetal brain and adult frontal cortex using a forward primer in exon 22 and a reverse primer in exon 26. Sequence analysis of 100 clones from each sample failed to reveal the presence of the duplicated segment in the SCN2A transcripts.
Alteration of transcriptional activity by CNSs
A growing body of evidence suggests that CNSs may function as enhancers or repressors of transcription [9, 16]. To determine whether the identified CNSs could alter transcription, we analyzed seven orthologous CNSs within the SCN1A locus (CNS1, CNS4, CNS5, CNS8-CNS11) and the paralogous CNSs (CNSA, CNSC-CNSF) by luciferase reporter gene assays (Fig. 5). The orthologous CNSs were cloned into the pGL3-promoter vector (Promega) containing the SV40 promoter, which served as a proxy for the SCN1A promoter since it has not been characterized. The paralogous CNSs were cloned into a pGL3 vector containing the minimal human SCN2A promoter region [17]. Recombinant vectors were transfected into the human neuroblastoma cell line, SH-SY5Y, and the human cervical carcinoma cell line, HeLa.
Figure 5.
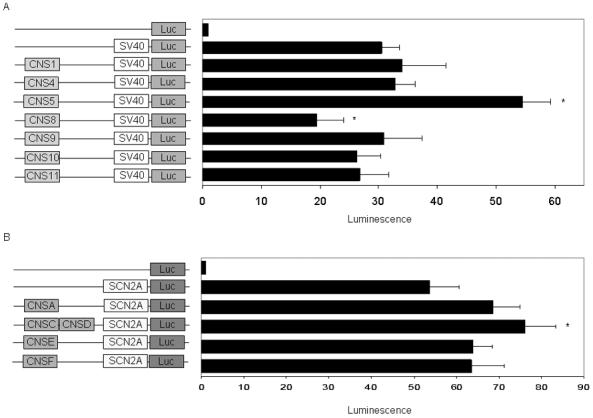
In-vitro functional characterization of CNSs in SH-SY5Y cells. (A) Orthologous CNSs within the SCN1A locus were cloned into a vector containing the firefly luciferase gene under the control of the SV40 promoter (pGL3 promoter). (B) Human paralogous CNSs were cloned into a vector containing the firefly luciferase gene under the control of a human SCN2A promoter. All values are normalized to the empty vector (pGL3 basic). Error bars represent the SEM, and asterisks indicate statistically significant changes in luminescence when compared to the vector containing only the promoter.
CNS5 reproducibly increased transcriptional activity by 80% (P=1.0×10-4) in SH-SY5Y cells, whereas CNS8 decreased transcriptional activity by 36% (P=1.8×10-3) (Fig. 5A). Similar results for CNS5 and CNS8 were observed in HeLa cells (data not shown). CNSC and CNSD together increased transcriptional activity in SH-SY5Y cells by 50% (P=2.6×10-3) (Fig. 5B). The remaining CNSs did not have a statistically significant effect on transcriptional activity.
Materials and Methods
Genomic sequences and sequence alignments
We obtained orthologous human (May 2004), mouse (March 2005), dog (July 2004), and chicken (February 2004) genomic sequences encompassing SCN1A, SCN2A, and SCN3A from the UCSC genome Web site (http://www.genome.ucsc.edu). We aligned the 1.1-Mb human genomic interval spanning 100 kb upstream of the first coding exon of SCN1A to 35 kb downstream of the last coding exon of SCN3A to the orthologous regions from mouse, rat, and dog. The alignment was then repeated with the inclusion of the orthologous region from the chicken genome. To determine the conservation among the human paralogs, the same genomic region was divided into three segments containing SCN1A, SCN2A, or SCN3A, and then aligned. The SCN2A and SCN3A intragenic region was divided at the point halfway between the most distal noncoding exons. We conducted the paralogous and orthologous sequence comparisons with MultiPipMaker using both the “single coverage” and “show all matches” alignment options (http://pipmaker.bx.psu.edu/pipmaker) [18]. We masked the repetitive sequences for the reference human sequence of each alignment with RepeatMasker (http://www.repeatmasker.org).
Identification of conserved noncoding sequences
We submitted the output from the mammalian sequence comparison to WebMCS to identify sequence blocks of at least 25 bp with a conservation score of at least 95% (http://research.nhgri.nih.gov/MCS) [14]. The conservation score is weighted to account for the baseline neutral substitution rate for each species, and this score is different from percent sequence identity. From the WebMCS data, conserved noncoding sequence blocks with a minimum length of 100 bp were identified in the segments of the human genome containing 100 kb upstream of SCN1A exon 1 to 35 kb downstream of SCN1A exon 26 and 35 kb downstream of SCN2A exon 26 to 35 kb downstream of SCN3A exon 26.
From the same segments of the genome, we also identified CNSs using the phastCon prediction program, displayable on the UCSC “most conserved” comparative genomics track (http://www.genome.ucsc.edu) [15]. Conserved phastCon elements of at least 100 bp in length and with a score greater than 500 which did not intersect RefSeq gene sequences, were identified. Sequence blocks identified from both programs were then extended to include adjacent conserved sequence blocks within 50 bp. The conservation of each CNS in the chicken genome was then determined.
We examined the results from the PIP analysis of the human SCN1A, SCN2A, and SCN3A paralogs for CNSs that were at least 100 bp in length with greater than 50% sequence identity between two or more paralogs. These blocks were then extended to include adjacent conserved sequence blocks within 50 bp with greater than 50% sequence identity. CNSs that contained identified noncoding exons were excluded from further analysis.
RNA isolation
Total RNA was isolated from postmortem human brain tissue or from mouse brain tissue using the RNeasy Lipid Tissue Mini Kit (Qiagen) with DNase 1 to eliminate DNA contamination. We obtained the human brain tissue from the Emory University Brain Bank and mouse brain tissue from 5-week-old FVB mice. RNA was quantified by UV spectrometry and qualitatively assessed on 1% agarose gels.
Primer sequences
The sequences of all primers used in this study are listed in Supplementary Table 1.
5′ RACE
We performed 5′ RACE with the GeneRacker Kit (Invitrogen) using 5 μg of either human or mouse brain total RNA. We generated first-strand cDNA using reverse primers complimentary to SCN1A exon 1 of human (h1R) and mouse (m1R), SCN2A exon 1 of human (h2R) and mouse (m2R), and SCN3A exon 1 of human (h3R) and mouse (m3R). PCR and nested PCR of the human genes were performed with the primers h1R2 and h1R3 (SCN1A), h2R2 and h2R3 (SCN2A), and h3R2 and h3R3 (SCN3A). PCR and nested PCR of the mouse genes were performed with the primers m1R2 and m1R3 (Scn1a), m2R2 and m2R3 (Scn2a), and m3R2 and m3R3 (Scn3a). PCR amplification was performed for 32 cycles of 1 min at 94°C, 45 s at 60°C, and 1 min at 72°C. Mixed PCR products were cloned into the pCR4-TOPO vector using the TOPO TA cloning kit (Invitrogen), and individual clones were sequenced.
Quantitative real-time PCR
Adult mouse brain was dissected to generate tissue from nine regions: cerebellum, brainstem, hippocampus, cortex, thalamus, hypothalamus, striatum, olfactory bulb, and septum. First-strand cDNA was synthesized from 5 μg of total RNA using SuperScript III Reverse Transcriptase and oligo (dT) primers (Invitrogen). Quantitative PCR was performed from 1 μl first-strand cDNA using the Platinum SYBR Green qPCR master mix (Invitrogen) with the Roche LightCycler detection system. Primers pairs were: 1F, 1R (total Scn1a); 1aF, 1aR (Scn1a exon 1a); 1bF, 1bR (Scn1a exon 1b); 2F, 2R (total Scn2a); 2aF, 2aR (Scn2a exon 1a); 2bF, 2bR (Scn2a exon b); 2cF, 2cR (Scn2a exon c). PCR amplification was performed for one cycle at 50°C for 2 min, one cycle at 95°C for 2 min, and 45 cycles of 94°C for 5 s, 60°C for 20 s, and 72°C for 40 s. PCR results were normalized against the Hprt housekeeping gene. We employed the ΔΔcTmethod to quantify the relative expression of each gene or transcript. The total Scn1a and Scn2a expression pattern was then compared to the expression pattern of distinct transcripts containing noncoding exons spliced to exon 1.
RT-PCR analysis of duplicated SCN2A sequence
We synthesized first-strand cDNA from 5 μg total RNA from human frontal cortex (Ambion) and human fetal brain (26-40 weeks) (Clontech) using a primer (2A3′R) complementary to the 3′ UTR of SCN2A. Exon 22 to exon 26 of SCN2A was PCR-amplified from 1 μl of cDNA using the primer pair 22F, 26R. PCR was carried out with one cycle at 94°C for 2 min and 32 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min. The PCR product was cloned into the pCR4-TOPO TA cloning vector (Invitrogen) and transformed into DH5α chemically competent cells (Invitrogen). For each RNA source, we purified plasmid DNA from 100 clones by column purification (GenElute Plasmid Purification, Sigma) and sequenced via automated sequencing.
The genomic DNA of 40 individuals from the CEPH panel (7 Africans, 7 Middle Easterners, 9 Asians, 7 Europeans, 4 Russians, and 6 South Americans) was PCR-amplified with the primer pair 2AF, 2AR to produce a product containing the 78-bp duplicated segment. PCR was carried out with one cycle at 94°C for 2 min and 32 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s. PCR products were purified by column purification (Millipore) and sequenced.
RNA secondary structure prediction
All possible RNA secondary structures of the CNSs were predicted using Mfold 3.2 (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) [19, 20]. CNSs greater than 200 bp in length were divided into 200-bp fragments with a minimum of 100-bp overlap between adjacent fragments. Hairpin structures characterized by arms >23 nucleotides in length, arms with minimal bulges (containing no more than 3 nucleotides from one arm in a bulge), symmetric loops, and a ΔG <-23 kcal/mol were considered possible miRNA precursors [21, 22].
Reporter constructs and transfections
The CNSs within the SCN1A locus were PCR-amplified from human genomic DNA using primers containing SalI sites. Each PCR product was digested with SalI and cloned into the SalI site of the pGL3-promoter vector containing the SV40 promoter (Promega). The SCN2A promoter was PCR-amplified from human genomic DNA with primers 2ApromF and 2ApromR and cloned into the HindIII site of the pGL3-basic vector [17]. CNSs identified from sequence comparisons of the human SCN1A, SCN2A, and SCN3A paralogs were cloned into the NheI or BglII site of the pGL3-basic vector containing the human SCN2A promoter. All PCR reactions were carried out with one cycle at 94°C for 2 min and 32 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min. Plasmid DNA for transient transfections was isolated using the Endofree Maxiprep kit (Qiagen) and verified by restriction digestion and sequencing.
Cell culture and transfection
We purchased SH-SY5Y and HeLa cells from the American Type Culture Collection (ATCC). Cells were cultured under standard conditions with DMEM (Cellgro) containing 10% fetal bovine serum (Invitrogen). Cells were grown at 37°C in the presence of 5% CO2. Transfections were performed by first plating 5×105 to 8×105 cells into each well of a 6-well culture dish and incubating overnight. SH-SY5Y and HeLa cells were transfected at 70% confluence with 1 μg of the recombinant plasmids and 100 ng of Renilla (pRL-TK) (Promega) using FuGENE 6 (Roche). Transfection efficiencies of 15% and 30% were achieved in the SH-SY5Y and HeLa cell lines respectively. Firefly luminescence for each cell lysate was normalized to Renilla luminescence, to control for transfection variability. Luminescence was measured using the dual-luciferase reporter assay (Promega). Transfections were conducted in triplicate on three separate days with three independent plasmid DNA preparations. All P values were calculated using a paired t-test.
Discussion
Diversity of sodium channel mRNA transcripts
5′ RACE analysis of SCN1A, SCN2A, and SCN3A identified complex 5′ UTRs consisting of multiple, alternatively spliced noncoding exons distributed over large genomic intervals. The observed complexity of the 5′ UTR of these genes appears to be a general feature of the voltage-gated sodium channels, since the 5′ UTR of the fourth brain sodium channel SCN8A and the heart sodium channel SCN5A also contain multiple noncoding exons [24-25]. Human SCN1A appeared to have the most complex organization, with seven noncoding exons distributed over a 75-kb interval upstream of exon 1. However, this greater complexity may reflect the more thorough analysis of SCN1A.
Several recent studies in various organisms have demonstrated that the spatial-specific expression of genes is driven by distinct promoter elements [26-28]. The complexity of the sodium channel 5′ UTRs, together with the variation in expression levels of distinct Scn1a and Scn2a transcripts in different regions of the mouse brain, is consistent with the usage of multiple promoters. Two SCN2A promoters located upstream of exons 1a and 1c have been identified [17, 23]; however, the distinct expression pattern of Scn2a exon 1b raises the possibility of an additional Scn2a promoter. Similarly, differences in the expression patterns of Scn1a exons 1a and 1b suggest that distinct promoter elements upstream of these exons may contribute to the regulation of these transcripts. Three human SCN3A noncoding exons were identified; however, only the ortholog of exon 1a was identified in the mouse, suggesting the presence of a primary promoter upstream of this exon. Interestingly, SCN2A exons 1a and 1c, SCN1A exons 1a and 1b, and SCN3A exon 1a are conserved in the chicken genome, suggesting that these noncoding exons and any associated promoter elements may be functional in a variety of species. Further characterization of these putative promoter regions will be a subject of future analysis.
In addition to the temporal and spatial regulation of channel expression levels provided by the usage of multiple promoters, the noncoding exon composition of different transcripts may also regulate channel levels by altering translation efficiency, mRNA stability, and mRNA splicing [29-31]. Brain region differences in transcript usage may therefore result in spatial differences in sodium channel composition and, consequently, cell type-specific electrophysiological characteristics.
CNSs correspond to potential transcriptional regulatory elements
Recent studies have demonstrated the effectiveness of evolutionary conservation as a tool for identifying biologically functional CNSs [16, 32, 33]. To identify CNSs that may regulate the expression of SCN1A, SCN2A, or SCN3A, we used both the WebMCS and phastCon prediction programs. A total of 33 noncoding sequences conserved across mammals were identified. Of these, 14 CNSs were identified by both methods; 11 CNSs were identified by the WebMCS program alone; and eight CNSs were identified by the phastCon program alone. This discrepancy demonstrates that more than one bioinformatic approach will be necessary to identify all CNSs in the genome. Sixteen of the mammalian CNSs are located upstream of a first coding exon, and 20 are conserved in the chicken genome. Interestingly, there were six CNSs conserved among the human paralogs, possibly representing regulatory motifs common to two or more genes. Six of the identified orthologous CNSs corresponded to noncoding exons, which highlights the ability of comparative genomics to identify functional elements.
Since CNSs have been shown to act as enhancers or repressors of both near or distal genes [16, 33], we tested a subset of CNSs that were identified in the SCN1A locus and five paralogous CNSs via reporter gene assays. Of the 12 CNSs examined, two increased transcription to a level that was statistically significant. An 80% increase in luciferase activity was observed in the presence of CNS5, located 53 kb upstream of SCN1A exon 1. CNSC and CNSD, located in intron 5, together increased the activity of the SCN2A promoter by 50%. While modest, these observed changes in transcription could potentially reflect a biologically relevant in vivo effect. Given that greater-order chromatin structure, surrounding DNA elements, and noncoding RNAs are required for the activity of some regulatory elements, further analysis of these CNSs in mouse models or stably transfected cells will be required to more accurately determine their biological relevance.
CNS10 may correspond to a noncoding RNA
MicroRNAs (miRNAs) are a large class of 21- to 22-nucleotide noncoding RNAs that regulate the translation of messenger RNAs (mRNAs). miRNAs are processed from highly conserved precursor transcripts (pre-miRNAs) that form a distinct hairpin structure [34]. Several recent studies have made use of evolutionary conservation along with RNA secondary structure prediction to identify potential pre-miRNAs [21, 35].
To determine whether any of the CNSs might correspond to a miRNA, we used the Mfold 3.2 program to predict possible RNA secondary structures of the 39 identified CNSs [19]. An 80-bp segment of CNS10, located in intron 20 of SCN1A, was predicted to form a stable hairpin structure characteristic of pre-miRNAs (Fig. 6A). The RNA secondary structure has a ΔG of -27 kcal/mol, arms that are 34 nucleotides in length, and a loop region of 10 nucleotides in length. The 80-bp segment is 96% identical in human and mouse, which agrees with the level of conservation reported for miRNAs in previous studies (Fig. 6B) [21]. At present this 80-bp sequence is not included in the miRBase miRNA registry (http://microrna.sanger.ac.uk) [36]. Further studies will be necessary to determine whether this predicted pre-miRNA is biologically functional.
Figure 6.
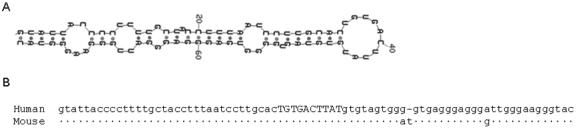
Predicted RNA secondary structure and evolutionary conservation of CNS10. (A) Mfold 3.2 predicted an 80-bp pre-miRNA hairpin structure with a ΔG of -27 kcal/mol. (B) Alignment of the human and mouse sequences across the predicted pre-miRNA sequence. Three mismatches are present in the 3′ arm of the hairpin. Arms are illustrated by lowercase letters and the loop is illustrated by uppercase letters. Dots indicate sequence identity.
Partial duplication in SCN2A intron 24
The developmental regulation of sodium channel function is influenced by the usage of alternatively spliced exons [37-40]. Previous studies have identified two pairs of alternatively spliced exons in the voltage-gated sodium channel gene family, exons 5N and 5A and exons 18N and 18A, expressed in the neonate and the adult. Although exons 5N and 5A differ at only a few amino acid residues, they result in channels with distinct biophysical properties [41]. Exon 18N, identified in SCN8A, introduces an in-frame stop codon, which may serve as a mechanism to developmentally regulate protein levels.
The identification of a 78-bp duplication in human SCN2A intron 24 that was 96% identical to the last 48 bp of exon 24 and the first 30 bp of intron 24, raised the possibility that it may similarly function as an alternative exon. This was particularly intriguing, since exon 24 of SCN2A encodes the inactivation gate, a structure of critical importance to the channel’s function. The duplication event may therefore represent a recent evolutionary attempt, unique to the human lineage, to generate a SCN2A channel with an alternate inactivation gate and different biophysical properties.
Although we did not identify transcripts containing the duplicated segment, we cannot exclude the possibility that it may be expressed in specific cell populations or at specific times during development. The duplication may also serve as a template for meiotic or mitotic gene conversion event with exon 24.
In conclusion, we have examined the SCN1A, SCN2A, and SCN3A loci for functional regulatory elements by characterizing their 5′ UTRs in both human and mouse. We have demonstrated spatial specific usage of 5′ UTR transcripts initiating from different noncoding exons across nine brain regions, suggesting the presence of multiple cell-type specific promoters. Finally, we have demonstrated the usefulness of comparative genomics in the identification of noncoding exons, and defined conserved genomic elements that will be the focus of future functional analysis.
Supplementary Material
Supplementary Figure 1. Alignment of CNSB across human SCN1A, SCN2A, and SCN3A. The SCN1A/SCN2A sequences are 50% identical across the 360bp region. The SCN1A/SCN3A sequences are 51% identical, and the SCN2A/SCN3A sequences are 58% identical. Exonic sequences appear in uppercase and intronic sequences appear in lowercase. Dots indicate sequence identity and dashes represent gaps in the alignment.
Acknowledgments
This work was supported by NIH Research Grant NS051834 (A.E.). We thank Cheryl Strauss (Emory University), James Thomas (Emory University), Tamara Caspary (Emory University), and Michael Zwick (Emory University) for critically reading the manuscript. We also thank Howard Rees (Emory University) for his help with the dissection of mouse brain regions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Auld VJ, et al. Neuron. Vol. 1. 1988. A rat brain Na+ channel alpha subunit with novel gating properties; pp. 449–461. [DOI] [PubMed] [Google Scholar]
- [2].Strong M, Chandy KG, Gutman GA. Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol. Biol. Evol. 1993;10:221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- [3].Sugawara T, et al. A missense mutation of the Na+ channel alpha II subunit gene Na(v)1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proc Natl Acad Sci U S A. 2001;98:6384–6389. doi: 10.1073/pnas.111065098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Escayg A, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat. Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- [5].Heron SE, et al. Sodium-channel defects in benign familial neonatal-infantile seizures. Lancet. 2002;360:851–852. doi: 10.1016/S0140-6736(02)09968-3. [DOI] [PubMed] [Google Scholar]
- [6].Claes L, et al. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mulley JC, et al. SCN1A mutations and epilepsy. Hum. Mutat. 2005;25:535–542. doi: 10.1002/humu.20178. [DOI] [PubMed] [Google Scholar]
- [8].Pant PV, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Emison ES, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- [10].The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- [11].Hedges SB. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- [12].Raymond CK, et al. Expression of alternatively spliced sodium channel alpha-subunit genes. Unique splicing patterns are observed in dorsal root ganglia. J. Biol. Chem. 2004;279:46234–46241. doi: 10.1074/jbc.M406387200. [DOI] [PubMed] [Google Scholar]
- [13].Whitaker WR, et al. Distribution of voltage-gated sodium channel alpha-subunit and beta-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J. Comp. Neurol. 2000;422:123–139. doi: 10.1002/(sici)1096-9861(20000619)422:1<123::aid-cne8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- [14].Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Loots GG, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- [17].Schade SD, Brown GB. Identifying the promoter region of the human brain sodium channel subtype II gene (SCN2A) Brain Res. Mol. Brain Res. 2000;81:187–190. doi: 10.1016/s0169-328x(00)00145-5. [DOI] [PubMed] [Google Scholar]
- [18].Schwartz S, et al. PipMaker--a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- [21].Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- [23].Maue RA, Kraner SD, Goodman RH, Mandel G. Neuron-specific expression of the rat brain type II sodium channel gene is directed by upstream regulatory elements. Neuron. 1990;4:223–231. doi: 10.1016/0896-6273(90)90097-y. [DOI] [PubMed] [Google Scholar]
- [24].Drews VL, Lieberman AP, Meisler MH. Multiple transcripts of sodium channel SCN8A (Na(V)1.6) with alternative 5′- and 3′-untranslated regions and initial characterization of the SCN8A promoter. Genomics. 2005;85:245–257. doi: 10.1016/j.ygeno.2004.09.002. [DOI] [PubMed] [Google Scholar]
- [25].Shang LL, Dudley SC., Jr. Tandem promoters and developmentally regulated 5′- and 3′-mRNA untranslated regions of the mouse Scn5a cardiac sodium channel. J Biol. Chem. 2005;280:933–940. doi: 10.1074/jbc.M409977200. [DOI] [PubMed] [Google Scholar]
- [26].Itani OA, Campbell JR, Herrero J, Snyder PM, Thomas CP. Alternate promoters and variable splicing lead to hNedd4-2 isoforms with a C2 domain and varying number of WW domains. Am. J. Physiol. Renal. Physiol. 2003;285:F916–F929. doi: 10.1152/ajprenal.00203.2003. [DOI] [PubMed] [Google Scholar]
- [27].Choi J, Newman AP. A two-promoter system of gene expression in C. elegans. Dev. Biol. 2006;296:537–544. doi: 10.1016/j.ydbio.2006.04.470. [DOI] [PubMed] [Google Scholar]
- [28].Barker DF, et al. Functional properties of an alternative, tissue-specific promoter for human arylamine N-acetyltransferase 1. Pharmacogenet. Genomics. 2006;16:515–525. doi: 10.1097/01.fpc.0000215066.29342.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- [30].Mancl ME, et al. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J. Biol. Chem. 2005;280:21078–21090. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- [31].Gauss KA, et al. Variants of the 5′-untranslated region of human NCF2: expression and translational efficiency. Gene. 2006;366:169–179. doi: 10.1016/j.gene.2005.09.012. [DOI] [PubMed] [Google Scholar]
- [32].Farhadi HF, et al. A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. J. Neurosci. 2003;23:10214–23. doi: 10.1523/JNEUROSCI.23-32-10214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- [34].Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- [35].Lim LP, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sarao R, Gupta SK, Auld VJ, Dunn RJ. Developmentally regulated alternative RNA splicing of rat brain sodium channel mRNAs. Nucleic Acids Res. 1991;19:5673–5679. doi: 10.1093/nar/19.20.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gustafson TA, Clevinger EC, O’Neill TJ, Yarowsky PJ, Krueger BK. Mutually exclusive exon splicing of type III brain sodium channel alpha subunit RNA generates developmentally regulated isoforms in rat brain. J. Biol. Chem. 1993;268:18648–18653. [PubMed] [Google Scholar]
- [39].Plummer NW, et al. Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics. 1998;54:287–296. doi: 10.1006/geno.1998.5550. [DOI] [PubMed] [Google Scholar]
- [40].Belcher SM, Zerillo CA, Levenson R, Ritchie JM, Howe JR. Cloning of a sodium channel alpha subunit from rabbit Schwann cells. Proc. Natl. Acad. Sci. U S A. 1995;92:11034–11038. doi: 10.1073/pnas.92.24.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dietrich PS, et al. Functional analysis of a voltage-gated sodium channel and its splice variant from rat dorsal root ganglia. J. Neurochem. 1998;70:2262–2272. doi: 10.1046/j.1471-4159.1998.70062262.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Alignment of CNSB across human SCN1A, SCN2A, and SCN3A. The SCN1A/SCN2A sequences are 50% identical across the 360bp region. The SCN1A/SCN3A sequences are 51% identical, and the SCN2A/SCN3A sequences are 58% identical. Exonic sequences appear in uppercase and intronic sequences appear in lowercase. Dots indicate sequence identity and dashes represent gaps in the alignment.


