Abstract
Complex secretion machineries mediate protein translocation across cellular membranes. These machines typically recognize their substrates via signal sequences, which are required for proper targeting to the translocon. We report that during posttranslational secretion the widely conserved targeting factor SecA performs a quality-control function that is based on a general chaperone activity. This quality-control mechanism involves assisted folding of signal sequenceless proteins, thereby excluding them from the secretion process. These results suggest that SecA channels proteins into one of two key pathways, posttranslational secretion or folding in the cytoplasm. Implications of this finding for intracellular protein localization are discussed.
Roughly one-third of proteins that are synthesized in the cytoplasm are translocated into other cellular compartments or the extracellular medium. In humans these compartments include, for example, the endoplasmic reticulum, mitochondria, peroxisomes, nucleus, or various membranes. In Gram-negative bacteria, such as Escherichia coli, the periplasmic space resembles the endoplasmic reticulum of eukaryotes and the secretion (sec) machineries serving these compartments are conserved (1, 2). In principle, protein secretion requires secretory proteins and a translocon. The translocon mediates the transfer of proteins across the membrane. For optimal performance, several potential problems need to be solved. First, cells have to distinguish between secretory proteins and proteins that need to remain in the cytoplasm. One major strategy nature has adopted is to supply secretory proteins with a signal sequence (3). Signal sequences are recognized by several components of the secretion apparatus, and they are removed by a leader peptidase during translocation. The second challenge is to efficiently target secretory proteins to the translocon (4). Here, two methods are used. One is cotranslational targeting, where translation of the secretory protein is coupled to translocation (5). As substrates need to remain unfolded during sec-dependent translocation this mechanism elegantly solves the problem of folding before translocation. Because during posttranslational secretion (6) translation and translocation are not coupled, premature folding must be prevented. The widely conserved SecA protein catalyses this important step. SecA recognizes secretory proteins and carries them to the translocon. During its interaction with the translocon, SecA energizes secretion via ATP hydrolysis (7, 8).
Even though protein secretion is well studied, the important question of mechanism remains unanswered. To obtain additional information about the molecular mechanism of protein targeting during posttranslational secretion we studied the interaction of unfolded polypeptides with the targeting factor SecA in vitro. Native substrates of the E. coli sec system with and without their signal sequences were used to investigate how SecA distinguishes between secretory and nonsecretory proteins. We show that via a general molecular chaperone activity SecA stimulates folding of signal sequenceless (ss) proteins into the active conformation. This chaperone activity was, however, not observed for native precursor proteins, which have to be kept unfolded for secretion across the membrane. Therefore, SecA is involved in a control mechanism of intracellular protein localization, which is based on the active and productive exclusion of ss proteins from translocation.
Methods
Protein Production. SecA, premature (pre)-MalS and ΔssMalS, and pre-PhoA and ΔssPhoA were cloned as C- or N-terminal His tag derivatives. Purification was achieved by Ni-affinity chromatography following standard protocols. SecA was purified under native conditions, whereas MalS and PhoA were purified under denaturing conditions in the presence of 8 M urea. The proteins were >95% pure as judged by SDS/PAGE followed by Coomassie blue staining.
As in WT strains significant amounts of pre-MalS and pre-PhoA were exported to the periplasm the temperature-sensitive secA mutant strain MM52 (7) was used to prevent export. By shifting the temperature to nonpermissive 42°C before induction, export was significantly reduced. Optimized overproduction conditions used were to shift cultures at OD600 = 0.8 to 42°C before inducing protein production with 1 mM isopropyl β-D-thiogalactoside for 3 h before harvest.
MalS and PhoA Refolding Assays. Refolding of MalS was done at 30°C as described by using 0.13 μg of MalS per assay (9). The refolding assay for PhoA was similar to the MalS assay except for using 0.2 μg of PhoA and 20 mM Mops buffer, pH 7, containing 0.04% p-nitrophenylphosphate at 25°C.
Enzymatic activity of MalS was determined as described (10). Samples were incubated for 1 h at 30°C in a volume of 100 μl. Amounts of p-nitrophenol released were determined by measuring changes of absorption at 420 nm in a microwell plate spectrophotometer (Molecular Devices). Enzymatic activity of PhoA was determined in the presence of 0.04% p-nitrophenylphosphate. After 10-min incubation at 25°C the released p-nitrophenol was measured by spectrophotometry at 420 nm.
Citrate Synthase Aggregation Assay. To trigger aggregation, heat shock temperature (45°C) was used. Native citrate synthase from procine heart mitochondria (Roche Molecular Biochemicals) was diluted 1:200 to a final concentration of 0.1125 μM in a temperated buffer (40 mM Hepes, pH 7.5) with SecA in various concentrations or an equal volume of SecA buffer (50 mM Tris, pH 8.0/50 mM KCl/5 mM Mg Cl2/10% glycerol) while stirring. As a negative control BSA replaced SecA in 5-fold molar excess. Light scattering was measured by using a fluorescence spectrometer (Varian Eclipse) with the following settings: λEX/EM = 500 nm, slitEX/EM = 2.5 nm.
Results
After considering the existing models for the targeting process we expected that SecA would bind a precursor protein irreversibly during targeting to the membrane. Tight binding could improve efficient delivery of substrate to the translocon. An additional advantage of substrate fixation would be to keep preproteins in an unfolded state. Based on these considerations we expected SecA to function as a molecular chaperone. Two classes of chaperones are distinguished, foldases and holdases. Foldases actively stimulate protein folding, and classical examples are the Hsp60 and Hsp70 chaperone machines (11). In contrast, holdases, such as Hsp33, tend to bind substrates tightly, thereby interfering with refolding (12). As substrates have to be delivered in an unfolded state to the translocon we expected SecA to have holdase function. We also expected that chaperone assays in the presence of SecA would provide information on whether and perhaps how SecA distinguishes between secretory and nonsecretory proteins.
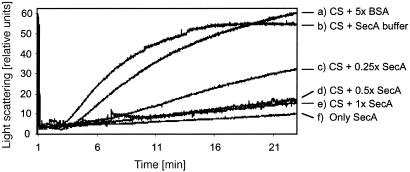
SecA Has a General Chaperone Activity. To address these questions and test whether SecA has a general chaperone activity a non-native substrate was used. A classical assay for general chaperone function involves following thermal aggregation of citrate synthase (13). In this assay aggregation is triggered by incubation at 45°C and the extent of aggregation is measured by light scattering. Over time aggregated particles grow in size and number, causing increased light scattering. The presence of a general molecular chaperone is expected to prevent thermal aggregation and thus to decrease light scattering. During incubation at 45°C thermal aggregation of citrate synthase was observed (Fig. 1b). Aggregation was strongly inhibited with increasing concentrations of SecA (Fig. 1 c-e), whereas a 5-fold molar excess of BSA had only minor effects (Fig. 1a). A heat-stable chaperone is a prerequisite for this experiment. In line with this expectation we detected that SecA itself was not susceptible to aggregation under these conditions (Fig. 1f). Based on these data we conclude that SecA exhibits a general chaperone activity. A molar ratio of SecA dimer/citrate synthase of 0.5:1 saturated the inhibition of aggregation, indicating a 1:1 stoichiometry that is one SecA monomer binds one monomer of citrate synthase. The finding that SecA has chaperone function does not, however, discriminate between foldase and holdase activity. Also, as citrate synthase is a cytoplasmic protein lacking a signal sequence it seemed unlikely that it is a natural substrate of SecA.
Fig. 1.
Thermal aggregation of citrate synthase (CS). Aggregation of CS was induced at 45°C. CS was present in 40 mM Hepes, pH 7.5, containing various concentrations of SecA or BSA. (a) CS + BSA (5-fold molar excess). (b) CS + SecA buffer. (c) CS + SecA (0.25-fold molar excess). (d) CS + SecA (0.5-fold molar excess). (e)CS + SecA (equimolar). (f) Only SecA. The added SecA volume was held constant to exclude effects of the SecA buffer. Light scattering was measured at λEX/EM 500 nm in a fluorescence spectrometer.
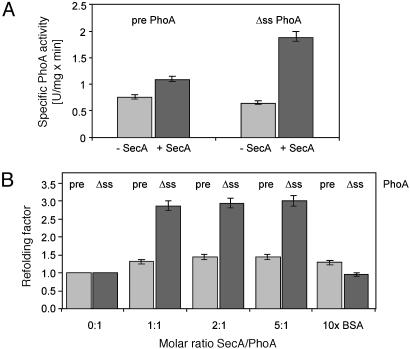
Refolding Assays with Pre-PhoA and ss-Alkaline Phosphatase. To distinguish between foldase and holdase activity by using native substrates of the E. coli sec system, and to determine the effect of signal sequences on SecA function, we expressed and purified periplasmic alkaline phosphatase PhoA (14, 15) and α-amylase MalS (10, 16) with and without their native signal sequence. After purification of substrates under denaturing conditions refolding efficiency was determined after dilution by assaying the recovery of enzymatic activity. The first set of experiments monitored refolding of PhoA in the presence or absence of stoichiometric amounts of SecA. Surprisingly, SecA stimulated refolding of ss-PhoA, whereas the effects on refolding of the precursor were small (Fig. 2A). These data indicated that SecA does not function as a holdase as we had expected. Rather, SecA exhibited a foldase activity, which primarily affected the ss substrate.
Fig. 2.
Refolding of PhoA. (A) Refolding of pre- and ss-PhoA in the presence or absence of SecA. Chemically denatured PhoA was diluted 1:50 in 20 mM Mops, pH 7, containing 0.04% PhoA substrate p-nitrophenyl phosphate and a 2-fold molar excess of SecA or an equal volume of SecA buffer. After refolding for 10 min specific PhoA activity was determined. (B) Refolding of PhoA in the presence of various concentrations of SecA. Refolding was done as described in A except that SecA/PhoA ratios varied as indicated and 10-fold molar excess of BSA was used as a negative control. The added SecA volume was held constant to exclude possible salt effects of the SecA buffer. The refolding factors given represent the enzymatic PhoA activity after refolding in the presence of SecA divided by the PhoA activity after refolding without SecA. Results of samples containing ss-PhoA (Δss) are shown in black and samples containing pre-PhoA (pre), in gray. Bars indicate standard deviation.
To exclude unspecific effects of SecA we carried out a titration experiment. In these assays the concentration of substrate is kept constant while various concentrations of refolding factor (SecA) are added. Unspecific assistance is detected when increased concentrations of protein correlate with increased yields of refolded substrate. In contrast, a saturation of the reaction in the presence of stoichiometric amounts of refolding factor indicates specific effects. Saturation of the SecA-dependent refolding activity was observed at a molar ratio of ss-PhoA/SecA of 1:1. Also, a 10-fold excess of BSA had no or only minor effects on the refolding of pre-PhoA and ss-PhoA (Fig. 2B). These initial results implicate that one mechanism by which cells distinguish between secretory and nonsecretory proteins involves SecA-dependent folding of signal sequenceless proteins, thereby excluding them from translocation.
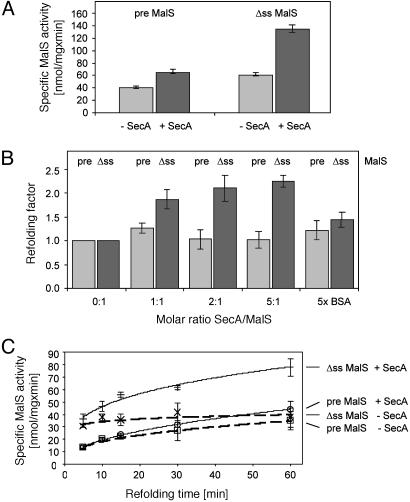
Refolding Assays with Pre-MalS and ss-Amylase. To confirm the results obtained with PhoA we used another periplasmic protein, the α-amylase MalS. As seen with PhoA, SecA exhibited refolding activity at stoichiometric amounts providing additional evidence for a foldase activity of SecA toward ss substrates (Fig. 3A). To further control the validity of these data we carried out a titration experiment that was similar to that described for PhoA. The data of this experiment revealed that a saturation of the SecA-dependent refolding activity occurred at a ratio of MalS/SecA of 1:2 (Fig. 3B). This ratio was expected because the active conformation of SecA is believed to be a dimer (8, 17). We conclude that SecA is directly supporting refolding of ss-MalS. Also, a 5-fold excess of BSA had only a small and unspecific effect on the refolding of pre-MalS and ss-MalS (Fig. 3B). Therefore, the effects of SecA on refolding of the MalS and PhoA derivatives were very similar.
Fig. 3.
Refolding of MalS. (A) Refolding of pre- and ss-MalS in the presence or absence of SecA. Chemically denatured MalS was diluted 1:50 in 100 mM Tris, pH 7.5. SecA was added in a 2-fold molar excess (black) or an equivalent of SecA buffer (gray) for 1 h. After refolding, 2 mM MalS substrate p-nitrophenylhexaoside was added to determine MalS activity. (B) Refolding of MalS in the presence of various concentrations of SecA. Refolding was done as described in A except that SecA/MalS ratios varied as indicated and 5-fold molar excess of BSA was used as a negative control. The added SecA volume was held constant to exclude possible salt effects of the SecA buffer. The refolding factors given represent the enzymatic activity of MalS after refolding in the presence of SecA divided by the enzymatic activity after refolding without SecA. Results of samples containing ss-MalS (Δss) are shown in black and samples containing pre-MalS (pre), in gray. (C) Time-dependent refolding of MalS in the presence or absence of SecA. Refolding was done as described in A except that MalS substrate was added after various times before determining MalS activity. Bars indicate standard deviation.
To investigate the observed effects in greater detail we followed refolding of MalS in the absence and presence of SecA over time (Fig. 3C). As MalS folds more slowly compared with PhoA, time dependence of refolding can be conveniently monitored. It should be noted that the amylase substrate p-nitrophenylhexaoside completely inhibits refolding of chemically denatured MalS (9). This feature was instrumental for time-dependent chaperone assays because refolding of MalS can be readily terminated by the addition of p-nitrophenylhexaoside and provides a starting point of the reactions where the specific MalS activity was <0.1 nmol/mg per min. The potential endpoint of the refolding reaction corresponds to the specific activity of purified WT and nondenatured MalS, which was 227 nmol/mg per min under these conditions.
Two observations can be made from these data. First, in the absence of SecA refolding of the precursor is slower than refolding of ss-MalS. This result is in agreement with earlier models proposing that signal sequences reduce premature folding of secretory proteins (18). Second, SecA refolds ss-MalS more efficiently compared with pre-MalS over most time points investigated. These observations indicate that a signal sequence is not required for the interaction of unfolded proteins and SecA. However, SecA does distinguish between proteins containing or not containing a signal sequence. It refolds ss proteins but not precursor proteins.
As SecA contains two nucleotide binding domains conferring ATPase activity we asked whether the observed chaperone activity is energy dependent. As the addition of either 10 mM ADP, ATP, or Na-azide had no effects on the refolding efficiency we concluded that the chaperone activity of SecA does not require ATP hydrolysis (data not shown).
Discussion
We describe a chaperone function of the SecA protein from E. coli. SecA interacts with unfolded polypeptides even if they do not contain a signal sequence. Via its molecular chaperone activity SecA stimulates folding of ss proteins and is preventing the aggregation of heat-denatured proteins. However, SecA does not promote the folding of proteins containing a signal sequence. Therefore, a mechanism must exist allowing SecA to distinguish between precursor and ss proteins. This mechanism is unknown but it is plausible that a specific binding site for signal sequences is critically involved. Cross-linking studies revealed that SecA binds signal sequences and preproteins (19, 20). The binding site for preproteins was mapped between amino acid residues 267 and 340 of SecA (20), whereas the capacity to bind a signal sequence has recently been assigned to the N-terminal 263 residues. Also, a deletion of residues 219-244 eliminated binding of signal sequences (21). Therefore future inspection could focus on a region of SecA including residues 219-340.
The chaperone function of SecA suggests that posttranslational targeting to the translocon at the cytoplasmic membrane is a dynamic process involving binding and release of substrates. The targeting mechanism includes a quality-control element, which is based on active exclusion of unfolded ss proteins from the translocation process. As there is a constant equilibrium of folded and unfolded polypeptides in the cytoplasm, cells could face significant problems if unfolded cytoplasmic proteins were not efficiently excluded from export. Therefore there are clear benefits of this quality-control step for individual cells and whole organisms.
We anticipate that a similar mechanism exists in human cells. As they are lacking SecA other proteins are expected to have a similar function. The chaperone systems of eukaryotic cells might be good candidates as luminal BiP and Kar2p that are homologs of the Hsp70 chaperone of E. coli are thought to provide one SecA function that is energy for translocation via ATP hydrolysis. In any case, because deletion of the cotranslational targeting system SRP in yeast is not lethal the existence of soluble factors performing other SecA functions is very likely (22).
Acknowledgments
We thank Dana Boyd and Rob John for reading the manuscript. Financial support by the Biotechnology and Biological Sciences Research Council and Altana Pharma AG is acknowledged.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: pre, premature; ss, signal sequenceless.
References
- 1.Rapoport, T. A., Jungnickel, B. & Kutay, U. (1996) Annu. Rev. Biochem. 65, 271-303. [DOI] [PubMed] [Google Scholar]
- 2.Schnell, D. J. & Hebert, D. N. (2003) Cell 112, 491-505. [DOI] [PubMed] [Google Scholar]
- 3.Blobel, G. & Dobberstein, B. (1975) J. Cell. Biol. 67, 835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fekkes, P. & Driessen, A. J. (1999) Microbiol. Mol. Biol. Rev. 63, 161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herskovits, A. A., Bochkareva, E. S. & Bibi, E. (2000) Mol. Microbiol. 38, 927-939. [DOI] [PubMed] [Google Scholar]
- 6.Mori, H. & Ito, K. (2001) Trends Microbiol. 9, 494-500. [DOI] [PubMed] [Google Scholar]
- 7.Oliver, D. B. & Beckwith, J. (1981) Cell 25, 765-772. [DOI] [PubMed] [Google Scholar]
- 8.Hunt, J. F., Weinkauf, S., Henry, L., Fak, J. J., McNicholas, P., Oliver, D. B. & Deisenhofer, J. (2002) Science 297, 2018-2026. [DOI] [PubMed] [Google Scholar]
- 9.Spiess, C., Beil, A. & Ehrmann, M. (1999) Cell 97, 339-347. [DOI] [PubMed] [Google Scholar]
- 10.Spiess, C., Happersberger, H. P., Glocker, M. O., Spiess, E., Rippe, K. & Ehrmann, M. (1997) J. Biol. Chem. 272, 22125-22133. [DOI] [PubMed] [Google Scholar]
- 11.Bukau, B. & Horwich, A. (1998) Cell 92, 351-366. [DOI] [PubMed] [Google Scholar]
- 12.Jakob, U., Muse, W., Eser, M. & Bardwell, J. C. (1999) Cell 96, 341-352. [DOI] [PubMed] [Google Scholar]
- 13.Buchner, J., Grallert, H. & Jakob, U. (1998) Methods Enzymol. 290, 323-338. [DOI] [PubMed] [Google Scholar]
- 14.Malamy, M. H. & Horecker, B. L. (1964) Biochemistry 3, 1889-1893. [DOI] [PubMed] [Google Scholar]
- 15.Strauch, K. L., Kumamoto, C. A. & Beckwith, J. (1986) J. Bacteriol. 166, 505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prinz, W. A., Spiess, C., Ehrmann, M., Schierle, C. & Beckwith, J. (1996) EMBO J. 15, 5209-5217. [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma, V., Arockiasamy, A., Ronning, D. R., Savva, C. G., Holzenburg, A., Braunstein, M., Jacobs, W. R. J. & Sacchettini, J. C. (2003) Proc. Natl. Acad. Sci. USA 100, 2243-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, S., Liu, G., Topping, T. B., Cover, W. H. & Randall, L. L. (1988) Science 239, 1033-1035. [DOI] [PubMed] [Google Scholar]
- 19.Miller, A., Wang, L. & Kendall, D. (1998) J. Biol. Chem. 273, 11409-11412. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, E., Akita, M., Matsuyama, S. & Mizushima, S. (1991) J. Biol. Chem. 266, 6600-6606. [PubMed] [Google Scholar]
- 21.Baud, C., Karamanou, S., Sianidis, G., Vrontou, E., Politou, A. S. & Economou, A. (2002) J. Biol. Chem. 277, 13724-13731. [DOI] [PubMed] [Google Scholar]
- 22.Pohlschroder, M., Prinz, W. A., Hartmann, E. & Beckwith, J. (1997) Cell 91, 563-566. [DOI] [PubMed] [Google Scholar]