Abstract
Myc proteins have long been modeled to operate strictly as classical gene specific transcription factors, however we find that N-Myc has a robust role in the human genome in regulating global cellular euchromatin including that of intergenic regions. Strikingly, 90–95% of the total genomic euchromatic marks histone H3 acetylated at lysine 9 and methylated at lysine 4 is N-Myc dependent. However, Myc regulation of transcription, even of genes it directly binds and at which it is required for maintenance of active chromatin, is generally weak. Thus, Myc has a much more potent ability to regulate large domains of euchromatin than to influence transcription of individual genes. Overall, Myc regulation of chromatin in the human genome includes both specific genes, but also expansive genomic domains that invoke functions independent of a classical transcription factor. These findings support a new dual model for Myc chromatin function with important implications for the role of Myc in cancer and stem cell biology, including that of induced pluripotent stem (iPS) cells.
Keywords: Myc, global chromatin, intergenic, neuroblastoma, stem cell, iPS
Introduction
Myc proteins belong to the basic helix-loop-helix zipper (bHLHz) superfamily of DNA binding proteins that dimerize on CANNTG E-box sequences to function as transcription factors (TF). Myc forms dimers with Max on the CACGTG sequence that activate transcription (1–3), while Myc also has a repressive function as well through association with Miz-1 (4). Myc proteins regulate many aspects of embryonic development as well as normal cell biology including cell cycle, metabolism, differentiation, senescence, apoptosis, and DNA replication (reviewed in (5)). Deregulation of Myc genes, particularly N-Myc, has been strongly linked with many human neural cancers including neuroblastoma and medulloblastoma (6–8). In medulloblastoma, Myc can cooperate with REST to drive tumorigenesis by blocking differentiation (9), which may be a key function in normal stem cells as well. Given the roles of both Myc and REST in ESC biology (10), they may also cooperate in that context. N-Myc amplification is known to correlate with poor prognosis in neuroblastoma (11), but the molecular mechanism by which N-Myc contributes to tumorigenesis is still largely an open question. Myc proteins are atypical bHLHz factors in that they are relatively weak transcriptional activators of potentially thousands of genes (12). This weak, but apparently widespread transcriptional function is a long-standing puzzle.
Intense interest in Myc function has been stimulated more recently by studies linking both c- and N-Myc to the generation of induced pluripotent stem (iPS) cells (reviewed in (13, 14)) as well as to murine embryonic stem cell (mESC) biology (15), together suggesting that Myc has important roles in regulating stem cell self-renewal and pluripotency. The molecular mechanisms by which Myc influences iPS cell formation and ESC biology are open questions. One possibility is that Myc contributes to the process through global chromatin reprogramming, suggested by studies (16, 17) implicating Myc in regulation of widespread histone modifications. Consistent with this idea, a very large Myc-regulated transcriptional program is operative in ESC that may have significance for Myc’s normal and neoplastic functions (18, 19) as well as iPS formation, but how Myc regulates this program at the chromatin level and its impact on cell biology remain unknown.
In neural stem cells, loss of N-Myc is sufficient to cause nuclear condensation most likely due to a global spread of heterochromatin (20). Myc’s recruitment of histone acetyltransferases such as GCN5 (21) and TIP60 (22) as well as its regulation of histone acetylation at a number of genic loci (17, 23, 24), suggests regulation of euchromatin through histone acetylation is involved. Unlike most TF, Myc binds to tens of thousands of genomic sites, both near and far from core promoters (24–29) indicating the euchromatic program may be quite expansive. Also at least at genes Myc may directly impact transcription via binding to PTEF-b (30, 31). Additional evidence suggests the Myc regulated chromatin program involves both histone acetylation but also methylation of lysine 4 of histone H3 (triMeK4) (16) possibly through the demethylase LID (32).
Since Myc’s normal and neoplastic functions depend on its ability to bind DNA and likely in turn to influence chromatin, the specific nature of its activity on cellular chromatin is a critically important open question. To address this key gap we conducted a functional genomics study in which we mapped Myc binding and chromatin function in a global, unbiased manner in the human genome using a conditional N-Myc transgene in neuroblastoma cells, finding that Myc has a global role in euchromatin maintenance. These findings support a new model in which Myc proteins act not only as classical TF but also more broadly to maintain widespread euchromatin.
Materials and Methods
Expression microarray studies
RNA samples were prepared from TET21N cells in biological duplicate for expression microarray studies. WG-6 beadchip arrays from Illumina were used. Data was normalized and analyzed using Illumina Beadstudio 3.0 and GeneSpringGX 7.3.1 (Agilent Technologies).
ChIP-chip
Chromatin samples were prepared and PCR conducted as previously described (33), except for some PCR when it initially failed, a PCR enhancer mix (34) was used but in all cases the same PCRs were used for different IPs to be compared at the same gene. For TET21N cells, 4 × 15cm plates (~108 cells) were crosslinked per experiment. Antibodies used for ChIPs include the following: N-Myc (1ug/ml Abcam 16898), AcK9 (2.5 ug/ml Upstate 06-942), and triMeK4 (2.5 ug/ml Upstate 07-473). 2ug Rabbit IgG was used as a background control. Immunoprecipitated chromatin fragments were amplified using the Whole Genome Amplification kit (Sigma) for 2 × 14 cycles, purified, then checked for enrichment over control IgG and total samples before sending for probing of the ENCODE array (Nimblegen Systems).
Chromatin samples were used to probe ENCODE arrays and bioinformatics performed as previously described (28, 33). Briefly, for identification of the N-Myc binding sites on the ENCODE arrays which represent 1% of the human genome and include ~400 genes and intergenic regions, we used the Tamalpais program (28) and chose the L1 set of high confidence peaks for further analysis. Briefly, binding sites are identified as peaks that have a minimum of 6 consecutive probes in the top 2% of all probes on the array with a p-value less than 0.0001. The identified binding sites were then determined to locate to a nearest gene based on GENCODE database (35). Each binding site was then classified as downstream or upstream relative to the distance of an annotated gene in GENCODE. Straight ChIP assays for verification were done using 10ng of input amplicon DNA.
Manual peak counting was conducted by using Signalmap zoomed in to a high enough magnification to count individual peaks per cluster. Thresholds for scoring peaks were determined by the average peak intensity within a cluster at Day 0 for a given ChIP. Log2 value thresholds used were the following for N-Myc, AcK9 and triMeK4 respectively: Leng 9 (> 1.5, 1.5, and 1.4), Luc7L (> 1.0, 2.5, and 2.5), and ZNF259 (>1.3, 2.3, and 1.7).
Motif analysis
The peaks identified from the N-Myc ENCODE array were used to de novo identify putative N-Myc binding motifs and possible binding partner motifs using our previous developed method ChIPMotifs (36). Briefly, the ChIPMotifs approach incorporates a statistical bootstrap re-sampling method to identify the top motifs detected from a set of ChIP-chip training data using ab initio motif-finding programs such as Weeder (37) and MEME (38).
Immunoblotting and staining
Immunoblotting and staining were performed as previously described (16).
Results
N-Myc genomic binding is widespread, including domains both near and distant from promoters
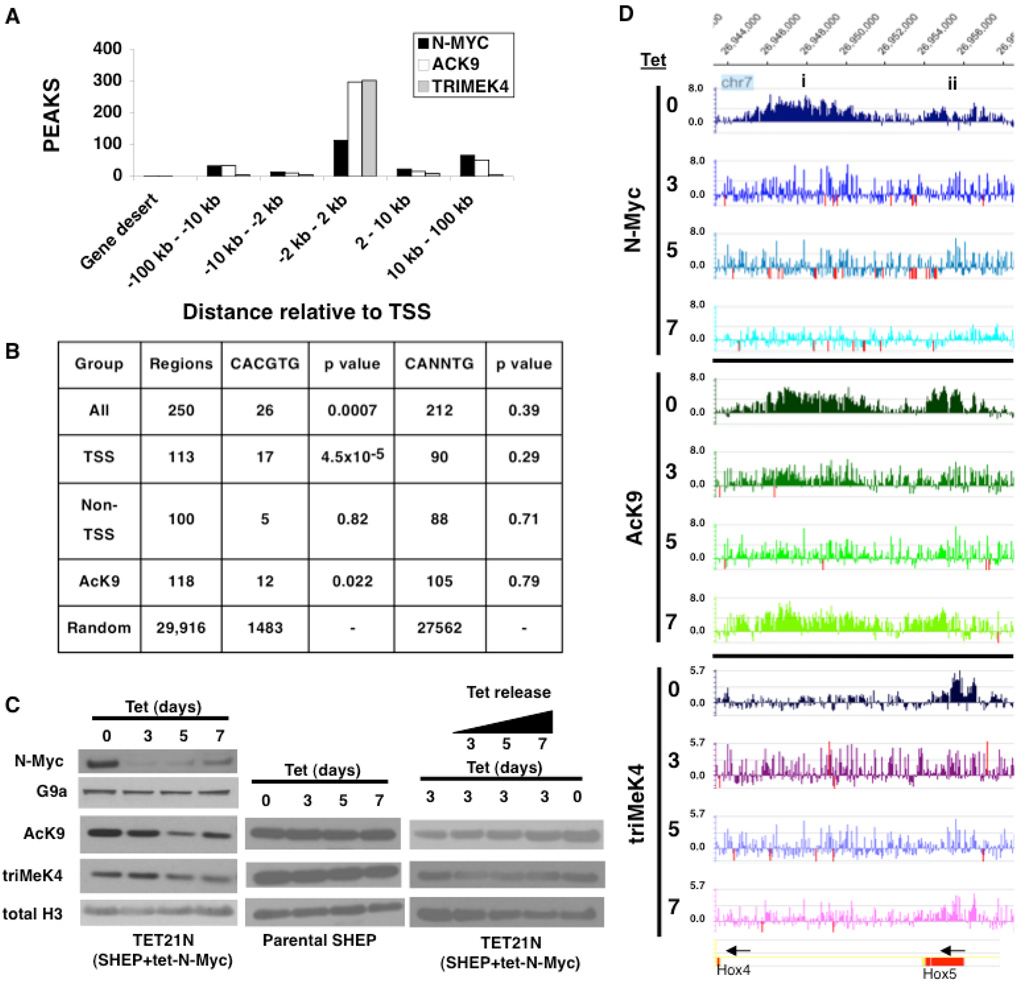
N-Myc regulation of chromatin in the human genome was analyzed using a well-characterized human neuroblastoma cell line containing a Tetracycline (Tet) repressible N-Myc transgene (TET21N) (39). A functional genomics analysis was conducted on N-Myc binding in TET21N cells untreated (day 0) as well as treated with Tet for 3, 5, and 7 days using ChIP-chip with ENCODE arrays (Nimblegen Systems) covering 1% of the human genome (35). N-Myc binding in the day 0 control cells (Fig. 1A) was confirmed as specific by comparison to ChIP-chip conducted on cells treated for 3 days with Tet, in which N-Myc protein was absent. N-Myc bound a total of 250 sites in control (day 0) cells while notably only 3 N-Myc binding sites were evident in cells treated with 3 days of Tet, with both scored at the same, high statistical stringency (L1; (28)). These data demonstrate that in this system strong reduction of N-Myc levels translates to essentially a complete absence of detectable N-Myc binding to chromatin. Even with peak scoring at the much lower L3 stringency 397 binding sites were observed in the control cells and only 28 sites in day 3 cells. Extrapolated to the whole human genome, these findings predict approximately 25,000–40,000 total N-Myc binding sites with almost 40% of the binding to remote regions at least 10kb from Transcriptional Start Sites (TSS; Fig. 1A), very similar to previous predictions for c-Myc (27, 28).
Figure 1. Myc chromatin binding and regulation in human neuroblastoma cells.
(A) Summary of ChIP-chip results on global distribution of N-Myc, AcK9 and triMeK4 in the human neuroblastoma genome. (B) E-box enrichment in ChIP-chip datasets with p values calculated using the Fisher test to compare specific N-Myc dataset groups to the random ENCODE dataset. (C) Left, Repression of N-Myc levels and total acid extractable active histone modifications with Tet treatment of Tet21N neuroblastoma cells. Immunoblots of nuclear (top two rows) and acid extracts (bottom three rows) of Tet21N cells treated with Tet for 0–7 days. Middle, Tet treatment does not influence AcK9 and triMeK4 in parental SHEP cells. Right, Tet treatment followed by “release” in Tet free media for 3, 5, and 7 days leads to a partial recovery of AcK9. (D) Examples of N-Myc bound domains. In this figure and throughout the study, blue peaks are N-Myc binding, green peaks at AcK9, and purplish peaks are triMeK4. Red peaks are off-scale. Arrows indicate direction of gene transcription of the indicated genes at the bottom. Domains (i) and (ii) are regions of widespread and focal binding/function respectively.
Evidence that N-Myc binding in non-promoter regions is E-box independent
We used the ChIPMotifs software (36) to directly scan for the Myc canonical E-Box motif (CACGTG) and a more degenerate E-Box (CANNTG) within the entire set of 250 N-Myc binding regions (Fig. 1B). CACGTG E-boxes were enriched in the dataset of 250 N-Myc bound sequences (P = 0.0007) while in contrast the degenerate E-box was not significantly enriched (P = 0.39) here nor in any dataset indicating that binding to non-canonical E-boxes is not a common event. The Myc E-box CACGTG was most highly enriched specifically in TSS regions, with 17 of 113 binding regions have at least one E-Box, with a P-value of 0.000045 (Fig. 1B). Intriguingly, while N-Myc binding to non-TSS by ChIP-chip appears just as robust as binding to TSS regions, the CACGTG E-box was not enriched in non-TSS regions as only 5 of 100 binding regions had the CACGTG E-Box (P = 0.82). This finding suggests regulation of non-TSS regions by N-Myc may utilize a distinct method of binding DNA, possibly via a cofactor. We also found enrichment for CACGTG E-boxes in AcK9 regions with 12 of 118 binding regions have an E-Box (P-value of 0.022).
N-Myc genomic binding is strongly linked to AcK9 and triMeK4
In parallel to N-Myc, we also used ChIP-chip to determine the global distribution of AcK9 and triMeK4 in TET21N (Fig. 1A). The majority (92%) of triMeK4 was within TSS regions (−2kb to +2kb) as expected from previous studies (40). While AcK9 was most abundant in TSS regions, it also had substantial binding (28%) elsewhere and the relative distribution of AcK9 peaks in regions progressively further from TSS was quite similar to that of N-Myc (Fig. 1A). Furthermore, in specific domains bound by N-Myc, peaks of binding for N-Myc, AcK9, and triMeK4 were most often remarkably similar in patterns and precise locations (Fig. 1D).
Loss of N-Myc decreases total cellular pools of two key active euchromatin marks in human neuroblastoma
By immunoblotting, Tet treatment of TET21N led to a pronounced global decrease of total acid extractable AcK9 that correlated with decreased levels of N-Myc protein in the nucleus, most prominently at 5 days (Fig. 1C left) with similar, but more modest changes in triMeK4 also evident. Similar decreases in AcK9 and triMeK4 are also evident by immunostaining (Fig. S6). It is notable that at 7 days N-Myc levels modestly rose again due to exhaustion of Tet in the media. The global decreases in AcK9 with N-Myc depletion were partially reversed at day 7 with re-increased N-Myc levels. Tet alone appears to have no effect on global chromatin as the parental SHEP neuroblastoma cells, from which the TET21N were derived, show no changes in global chromatin with the same course of Tet treatment (Fig. 1C middle). This finding also suggests the effects on chromatin observed in TET21N cells are specific to loss of N-Myc. Treating cells with Tet for 3 days, which reduced AcK9 levels, followed by a switch to Tet free media for 3, 5, and 7 days (release) led to a progressive but only partial restoration of AcK9 even at day 7 of release (Fig. 1C right). These data suggest N-Myc may have the ability to establish domains of AcK9 as well as maintain AcK9 levels, but indicate maintenance of AcK9 may be the predominant function.
N-Myc is essential for global maintenance of triMeK4 and AcK9 in both genic and intergenic regions
We theorized that there were two possible explanations for the tight association of N-Myc genomic binding with AcK9/triMeK4 peaks: 1) N-Myc binding depended on the presence of pre-existing AcK9 and triMeK4 as previously reported (17), or 2) alternatively or in addition that N-Myc played a key role in the establishment or maintenance of these histone marks. To test these possibilities ChIP-chip was also performed for N-Myc, AcK9 and triMeK4 in TET21N cells treated with Tet for 3, 5, and 7 days (Fig. 1D). At days 3 and 5 of N-Myc depletion, nearly all AcK9 and triMeK4 peaks irrespective of relative location to TSS were lost. Interestingly at day 7, with the partial restoration of N-Myc levels, the losses of AcK9 and triMeK4 domains were partially reversed apparently at the same locations and patterns where they were originally mapped at day 0 (Fig. 1D). Although detectable N-Myc binding was not restored at most genomic sites at day 7, the rescue of AcK9 and triMeK4 at this time strongly suggests N-Myc is acting on chromatin but with an affinity below that detectable in our assays. Some domains of N-Myc binding and regulation of AcK9/triMeK4 were expansive (10kb or larger in size), while others were quite focal (Fig. 1D, regions i-ii, Fig. S1).
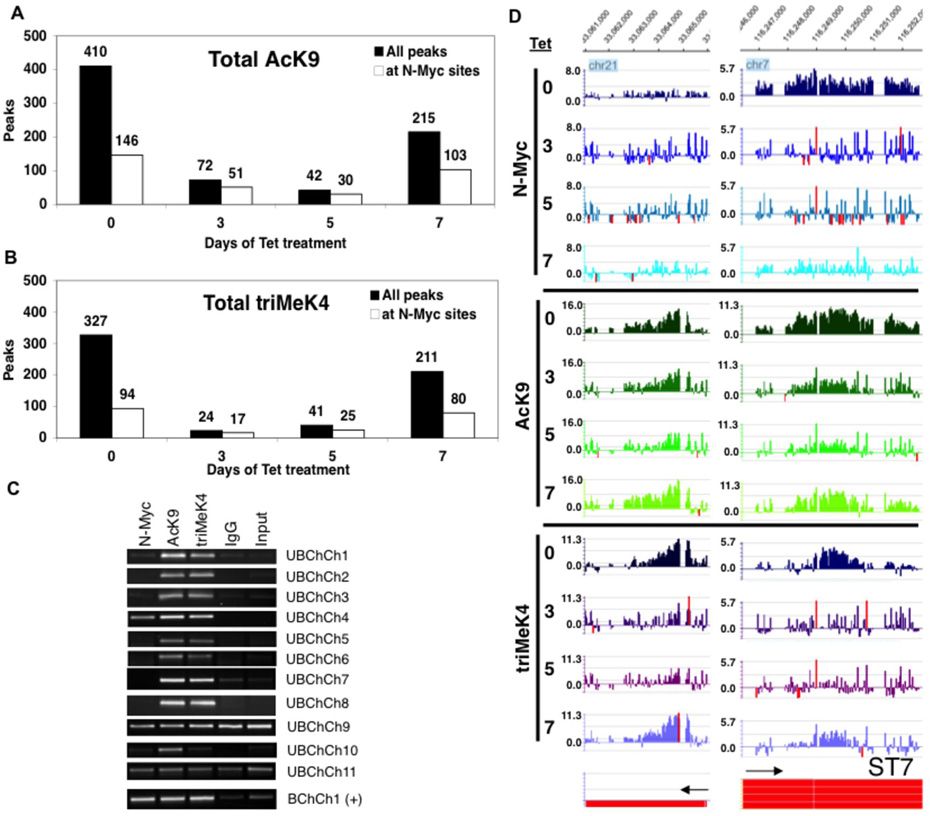
Global bioinformatics analysis of the ChIP-chip data using the Tamalpais program (28) indicates that after 3 and 5 days of N-Myc depletion, 87% and 90% of total genomic AcK9 sites were lost respectively (Fig. 2A). In parallel, greater than 93% and 86% of total genomic triMeK4 were lost (Fig. 2B). Thus just a 3-day deficit of N-Myc was sufficient to induce a nearly complete and global loss of two key euchromatic histone modifications with the loss persisting up to 5 days. Some AcK9 and triMeK4 peaks, even those directly bound by N-Myc, were not detectably affected by N-Myc loss at days 3 and 5 (Fig. S2) and were still scored as peaks at L1 stringency, demonstrating that the 3 and 5 day arrays were successfully probed and that loss of AcK9 and triMeK4 is specific to certain domains despite the overall profound losses of AcK9 and triMeK4. When AcK9 and triMeK4 binding were analyzed at lower stringencies L1–L4 (from highest to lowest; Table S1), we did not find a shift toward lower stringency peaks with loss of N-Myc. However, it appears some bona fide regulated domains remain at day 5 that are likely of even lower affinity than L4.
Figure 2. ChIP-chip indicates N-Myc is essential for global maintenance of AcK9 and triMeK4 throughout the human neuroblastoma genome including genic and non-genic regions.
(A) Total AcK9 and (B) triMeK4 peaks at 0–7 days of Tet treatment. (C) 11 regions that had values of <2 fold enrichment for N-Myc peaks (Unbound by ChIP-chip; UBChCh) but had AcK9 and triMeK4 sensitive to N-Myc reduction were tested for potential false negative N-Myc binding status by straight ChIP assays on control day 0 TET21N amplicons. A strongly N-Myc bound region, >4 fold enrichment, was included in parallel as a positive control, BChCh1(+). (D) Example of N-Myc dependent AcK9 and triMeK4 domains unbound and bound by N-Myc (left and right). Gene on bottom left is not annotated yet.
Evidence for both direct and indirect chromatin functions for N-Myc
While N-Myc maintains 90% or more of total genomic AcK9 and triMeK4 modifications in human neuroblastoma, many of these N-Myc dependent marks are nonetheless localized at sites without detectable N-Myc direct binding (Figs. 2A–B). For example, out of 410 AcK9 peaks on the ENCODE array in TET21N cells, 368 (90%) are lost with 5 days of N-Myc depletion, but of those 368 only 116 are N-Myc bound. Thus, most (252) AcK9 peaks that are N-Myc dependent are nonetheless not detectably bound by N-Myc and the same applies to triMeK4 peaks. A key question is whether these domains reflect indirect N-Myc activity or if they are false negatives for N-Myc binding. To test these possibilities, we conducted direct ChIP verification assays on regions, both genic and >10kb from TSS, that by ChIP-chip were apparently N-Myc unbound, but had N-Myc dependent AcK9 and triMeK4 (Fig. 2C; UBChCh 1–11). Only 1 out of 11 of the tested UbChCh domains (#4) clearly showed N-Myc binding by straight ChIP assay, indicating that most regions scored as unbound by N-Myc are not false negatives and are indeed not bound by N-Myc. The intensity of AcK9 and triMeK4 peaks at unbound regions as well as their degree of dependence on N-Myc are just as robust as at bound regions (Fig. 2D). The loss of AcK9 and triMeK4 at these domains does not appear to be due to effects of Tet alone as Tet has no effect on their levels in parental SHEP cells (Fig. 1C). Thus, N-Myc appears to have the ability to regulate AcK9 and triMeK4 at regions it does not bind, indicating it possesses both direct and indirect mechanisms of regulating chromatin.
The N-Myc regulated chromatin program is hierarchical and reversible
N-Myc exhibited a total of 146 and 94 binding sites coinciding with AcK9 and triMeK4 respectively in untreated TET21N cells (Figs. 2A–B). Only 30 N-Myc bound AcK9 domains remained at day 5, while only 17 N-Myc bound triMeK4 domains remained at day 3, representing approximately 5 fold reductions in both cases. Thus, N-Myc bound domains of AcK9 and triMeK4 are highly dependent on N-Myc levels for maintenance. However, N-Myc dependent AcK9 and triMeK4 domains that are not detectably bound by N-Myc are even more sensitive to loss of N-Myc with approximately 22 and 33 fold reductions in AcK9 (day 5) and triMeK4 (day 3) domains respectively (Fig. 2A–B). Thus, a hierarchy of N-Myc regulated chromatin domains appears to exist, with sites not detectably bound by N-Myc being lowest affinity and hence most sensitive to reduced N-Myc levels.
AcK9 and triMeK4 are dynamically regulated by N-Myc and importantly their loss is reversed by re-increasing N-Myc levels as evidenced by IB of acid extracts of histones (Fig. 1C). At a genomic level, the partial restoration of N-Myc protein levels at day 7 of Tet treatment also has clearly discernable effects on histone modifications detectable by ChIP-chip leading to a strong restoration of AcK9 and triMeK4 sites (Figs. 2A–B). Remarkably, possibly reflecting an epigenetic memory, approximately 97% of total day 7 peaks of both AcK9 (209/215) and triMeK4 (204/211) at day 7 were at sites where these marks existed previously in day 0 control cells. These data also indicate that the loss of AcK9 and triMeK4 is specifically caused by depletion of N-Myc and is not due to an intermediate irreversible change in cell biology.
N-Myc regulates AcK9 and triMeK4 at remote domains large distances from TSS
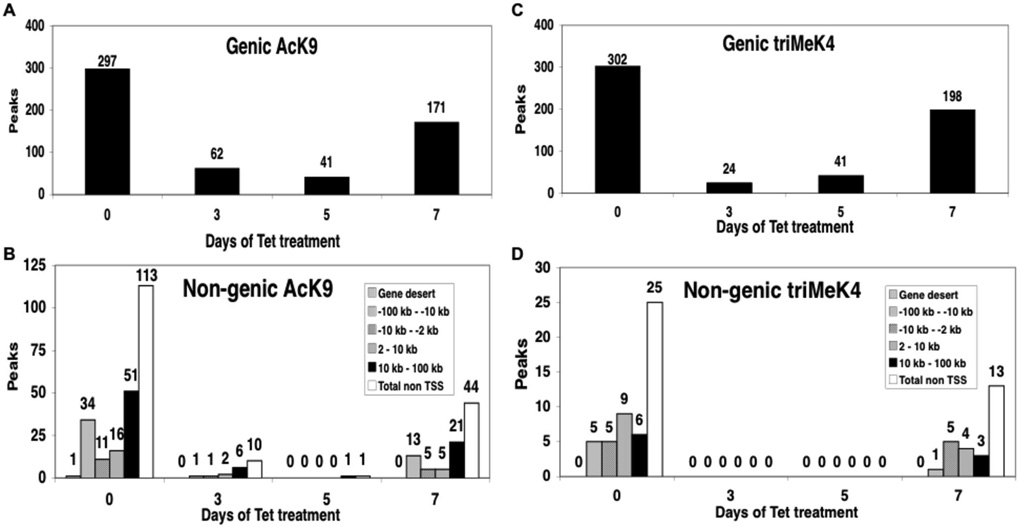
A substantial subset, about 40%, of N-Myc binding is at non-genic sites (Fig. 1B) from 2kb to >100kb from TSS. While such binding is not typically associated with TF function, AcK9 and triMek4 at these sites is almost completely dependent on N-Myc (Fig. 3), suggesting N-Myc is mediating these euchromatic modifications despite their distance from promoters. Interestingly, the largest subset of non-genic domains is at least 10–100kb from TSS. These remote from start site domains (RSS), defined here as domains >10kb away from the nearest TSS, are very rarely bound by most TF. For example, only 8% of E2F1 and only 3% of PolII binding sites are in such regions (28). Nonetheless AcK9 and triMeK4 at RSS domains are essentially 100% dependent on N-Myc for their maintenance (Figs. 3C–D), much more sensitive than TSS domain euchromatin (Figs. 3A–B), suggesting RSS are bona fide sites of Myc activity.
Figure 3. N-Myc regulation of AcK9 and triMeK4 includes both genic and non-genic regions.
Bioinformatics analysis of ChIP-chip data subdivided into (A) Genic and (B) Non-genic AcK9 domains. (C) Genic and (D) Non-genic are triMeK4 domains. Non-genic peaks are presented as subgroups relative to the distance to the TSS. Peaks scored at L1 stringency.
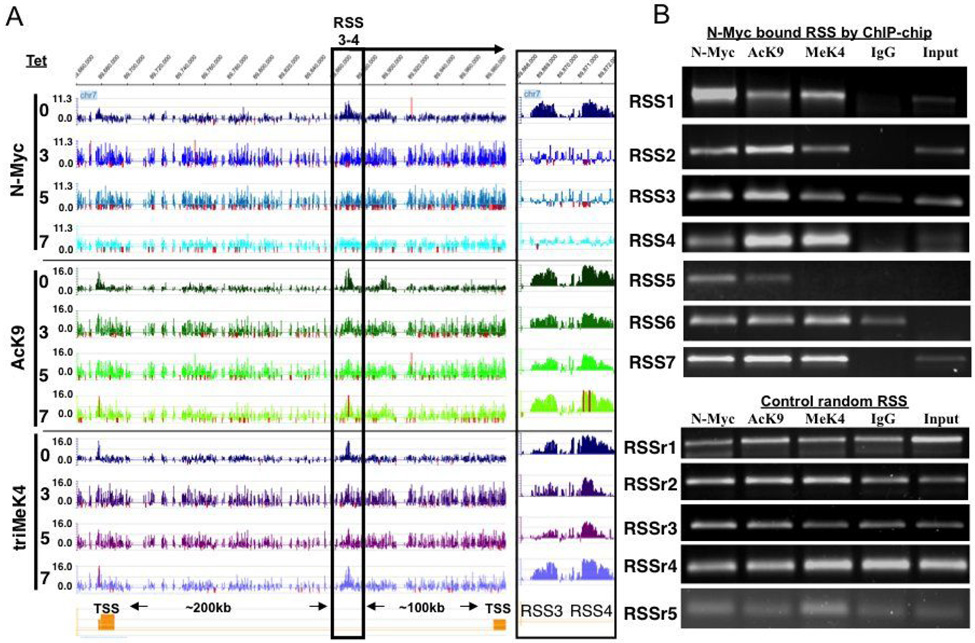
An example of two representative N-Myc bound RSS (RSS 3–4) is shown in Fig. 4A. N-Myc binding at these RSS is specific as it is ablated with Tet treatment. RSS 3–4 are 100–200kb away from the nearest TSS, yet AcK9 and triMeK4 in RSS 3–4 are strongly reduced at days 3–5 and re-elevated at day 7 (Fig. 4A, right box). As a control we performed straight ChIP assays on 7 N-Myc bound RSS and found all 7 confirmed, while 0/5 ChIPs confirmed enrichment for N-Myc binding or modified histones at randomly selected RSS (RSSr1–5; Fig. 4B). The functional consequences of N-Myc action at RSS remain unknown and we have not found evidence of regulation of nearest neighbor genes (for example expression microarray data for genes nearest to RSS3-4; Fig. S3) through an enhancer-like function, but this cannot be ruled out given the unpredictable distribution of the targets of enhancers and the fact that enhancers can be > 1Mb from their targets. The fact that at some RSS, N-Myc regulates both AcK9 and triMeK4 (Fig. 4), where at others (Fig. S4) it only regulates AcK9 suggests functional heterogeneity amongst RSS.
Figure 4. N-Myc bound and regulated RSS.
(A) Snapshot of Signalmap data of remote from transcriptional start site domains (RSS) that are bound and regulated by N-Myc. Inset at right of (A) is a blow up of boxed region to more clearly show peaks of RSS3 and RSS4. (B) ChIP confirmation attempts for 7 RSS bound by N-Myc. Enriched N-Myc binding with links to either AcK9 or triMeK4 were observed at 7/7 RSS tested, while 0/5 random RSS (RSSr1–5) exhibited such traits.
N-Myc bound and regulated genic chromatin domains weakly influence transcription
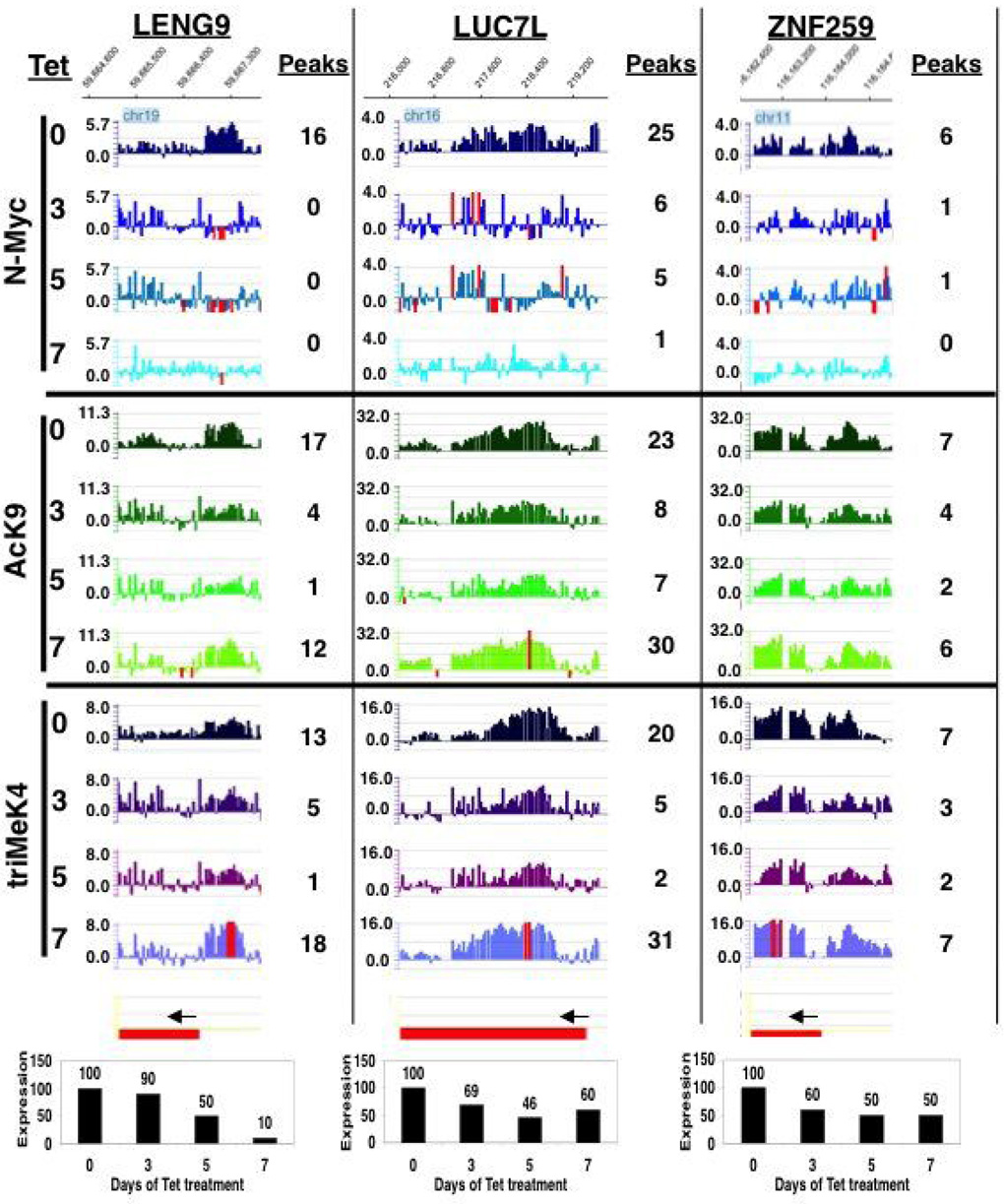
Parallel gene expression microarray and ChIP-chip data were compared (Table S2). Of the 66 N-Myc bound annotated genes for which a clear result was evident on the array, 63 (96%) had either AcK9 or triMeK4 present with 53 (80%) having both marks. Of the 63 genes with either or both marks, 38 (58%) lost the marks upon N-Myc depletion with 6/38 being restored at least 2 fold at day 7. Overall changes in expression of N-Myc bound genes were modest, most often in the range of 2 fold or less despite pronounced chromatin modification changes (Fig. 5). Further, the relative degree of loss of AcK9 and triMeK4 (e.g. as determined by peak counting represented by data to the right of the Signalmap histograms) at individual genes with depletion of N-Myc did not clearly correlate with the relative decrease in transcription at the 5 genes examined (the three in Fig. 5 and 2 additional N-Myc bound and regulated genes, DKC1 and CEP250). This suggests that even minor decreases in N-Myc mediated euchromatin are sufficient to drive the approximately 2-fold decrease in transcription that we typically see as a maximum decrease in Myc target genes upon Myc loss. Further, Myc-independent transcriptional machinery is in place that mediates a certain level of transcription.
Figure 5. N-Myc is a weak transcription factor even at genes it directly binds and at which it regulates AcK9 and triMeK4.
Snapshots of Signalmap data of three representative genes meeting the following criteria as putative N-Myc targets: N-Myc bound, N-Myc dependent histone modifications, and reduced levels of gene expression at days 3–5. Data at the bottom for each gene are mean expression levels from biological replicate expression microarray experiments. Data to the right of each Signalmap histogram represents manually counted peaks per cluster.
Thirty out of the sixty six N-Myc bound genes exhibited day 0 expression at least 10% above day 3 expression levels, but their mean expression at day 0 was just 1.5 fold above day 3 levels for a mean 50% higher expression. Twenty-four N-Myc bound genes exhibited lower expression at day 0 versus day 3, but only by a mean of 28%. Thus there is a moderately strong link between our ChIP-chip and gene expression microarray studies. Out of the 66 N-Myc bound genes, 25 (38%) have been previously identified as Myc target genes (12) suggesting our ChIP-chip assay efficiently identified targets. At days 3 and 5, 165 and 306 genes were downregulated two-fold or more respectively, suggesting a progressive increase in transcriptional silencing with prolonged depletion of N-Myc (Table S3). The transcriptional effects of loss of N-Myc are not robust as only 50 and 75 genes are downregulated 3 fold or more.
Discussion
Myc is linked to widespread histone acetylation (16, 17) as well as overall active histone modifications in tumors and epidermal stem cells (41, 42). Beyond acetylation, Myc is required for the global maintenance of triMeK4 (16) possibly mediated by interaction with the histone demethylase LID, which targets triMeK4 for demethylation (32). Myc targets in ES cells also have a strong, nearly 100% association with triMeK4 (18). Here we for the first time use functional genomics to globally map Myc’s chromatin function finding a widespread role for Myc in the human genome including both genic and intergenic regions.
Myc requires a preexisting active chromatin state to bind and in turn influence chromatin itself (17). By combining a loss of function model with functional genomics, we have focused on defining Myc function after it binds. It is of interest, however, that the requirement of Myc for preexisting domains of AcK9 and triMeK4 to bind chromatin so clearly matches with Myc’s function to regulate AcK9 and triMeK4 after binding. This suggests possible positive autoregulation of its own chromatin function by Myc. Furthermore, the fact that essentially 100% of AcK9 and triMeK4 marks at day 7 in our studies when N-Myc is restored exist at the same locations where they existed at day 0 in the first place, suggests a specific epigenetic memory exists for Myc binding and function, most likely consisting of low levels of residual AcK9, triMeK4, or some combination of the two. Together our data and the requirement of Myc for preexisting AcK9 and triMeK4 suggests N-Myc predominantly maintains or enhances euchromatin and less often establishes it de novo.
Open chromatin and open questions
While there is growing evidence that Myc has a much more global role in regulation of transcription and chromatin than previously anticipated, this is the first functional genomics study that examines Myc chromatin activity using a loss of function model and that includes intergenic regions distant from classical promoters. We find strong evidence that Myc is required to maintain euchromatin in a widespread manner including at intergenic sites. While the functional meaning of such activity remains to be determined, two main possibilities exist. First, Myc maintains widespread intergenic euchromatin together with genic euchromatin as part of a global role in sustaining active overall nuclear chromatin. Second, Myc mediates euchromatin at remote sites as part of a previously uncharacterized enhancer function to regulate genes at a distance. At this point, data does not clearly support one model over the other. Nonetheless it is intriguing that intergenic binding sites for N-Myc are not enriched for E-boxes. While E-box independent binding has been reported and may be fairly widespread (17), here we provide the first evidence that such binding may be of particular importance for Myc intergenic function.
In the model system we have used in this study, the increase in N-Myc at day 7 gives us a unique tool to study the effects of reintroduction of N-Myc and the partial reversal of loss of euchromatic marks at that time suggests that N-Myc regulates a dynamic euchromatic program. One puzzle is why by ChIP-chip we do not observe an increase in N-Myc binding at that later time point coincident with the chromatin rescue. The simplest explanation is that N-Myc is acting at these domains to restore active chromatin, but that its binding is of an affinity too low to detect by ChIP-chip or by direct ChIP (Fig. S7). It is also possible going along with this notion that the loss of N-Myc sensitizes chromatin to reelevation of N-Myc such that a relatively weak amount of N-Myc activity has a more potent effect. A second explanation is that a significant part of the rescue at day 7 is indirect, perhaps due to N-Myc induces a target gene such as GCN5.
Another surprising observation in our studies was that a large fraction of euchromatic marks regulated by N-Myc were not at locations of detectable N-Myc binding. While it is formally possible that N-Myc is binding these regions in a manner undetectable by ChIP-chip, we believe it is may be more likely that one of three other models or some combination of the three are at work: (1) Myc regulates the expression of histone modifying enzymes (e.g. GCN5; Fig. S5) which are themselves Myc target genes and are responsible for the Myc-dependent chromatin effects at unbound regions, (2) Myc regulates the activity of histone modifying enzymes (e.g. LID/RPB2), and (3) Myc regulates chromatin at a distance such that Myc binding at one location can influence chromatin at another throw higher order chromatin structure. The fact that partially restored N-Myc at day 7 appears to rescue a substantial fraction of AcK9 and triMeK4 marks that were lost upon its depletion but that N-Myc binding is not detectable at that time similarly suggests either an indirect function for Myc or a type of binding that is not detectable by ChIP-chip but is nonetheless functional.
While Myc is most famous for its role in cancer, normal Myc functions in stem cells are likely of great important. Myc’s role in the formation of iPS cells is intriguing (43), but the mechanisms by which it enhances iPS cell formation remain unknown. One theory is that Myc contributes to their induced self-renewal and pluripotency through global chromatin reprogramming (13) that may in essence set the overall chromatin table for the subsequent activity of other stem cell related TFs, even if it does not directly interact with those factors (18), a notion consistent with our findings in this study. However, other mechanisms are possible. The function of Myc in iPS cells is likely quite similar to its role in ESC. A specific global hyperactive chromatin state in ESC encompassing expansive intergenic regions, apparently key to maintenance of an ESC-state (44), could be regulated by Myc in ESC and locked in place during tumorigenesis such as neuroblastoma genesis. Ongoing functional genomics analyses of Myc and histone modifications in hESC and iPS cells should resolve many of these open questions.
Supplementary Material
Acknowledgements
We thank members of the Knoepfler lab for reading the manuscript. We want to thank Charles Nicolet and the UC Davis Expression Analysis Core for their excellent work on the expression microarray studies. This work was supported by K01 CA114400 and an award from the Brain Tumor Society (PSK) as well as R01 CA045240 and 1U54-HG004558 (PJF).
References
- 1.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA-binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 2.Amin C, Wagner AJ, Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol. Cell. Biol. 1993;13:383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amati B, Dalton S, Brooks MW, et al. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 4.Peukert K, Staller P, Schneider A, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandori C, Cowley SM, James LP, Eisenman RN. The MYC/MAX/MAD network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 6.Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD. Amplification of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Research. 1990;50:2347–2350. [PubMed] [Google Scholar]
- 7.Kohl NE, Kanda N, Schreck RR, et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35:359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 8.Schwab M, Alitalo K, Klempnauer KH, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 9.Su X, Gopalakrishnan V, Stearns D, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008 doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastoma correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 12.Dang CV, O'Donnell KA, Zeller KI, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Welstead GG, Schorderet P, Boyer LA. The reprogramming language of pluripotency. Curr Opin Genet Dev. 2008 doi: 10.1016/j.gde.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Cartwright P, McLean C, Sheppard A, et al. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 16.Knoepfler PS, Zhang XY, Cheng PF, et al. Myc influences global chromatin structure. Embo J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guccione E, Martinato F, Finocchiaro G, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong DJ, Liu H, Ridky TW, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank SR, Parisi T, Taubert S, et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced actylation of histone H4 and gene activation. Genes Dev. 2001;15:2069. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orian A, Grewal SS, Knoepfler PS, et al. Genomic binding and transcriptional regulation by the Drosophila myc and mnt transcription factors. Cold Spring Harb Symp Quant Biol. 2005;70:1–10. doi: 10.1101/sqb.2005.70.019. [DOI] [PubMed] [Google Scholar]
- 26.Orian A, van Steensel B, Delrow J, et al. Genomic binding by the Drosophila Myc, Max, Mad.Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cawley S, Bekiranov S, Ng HH, Kapranov P, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 28.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoepfler PS. Myc goes global: new tricks for an old oncogene. Cancer Res. 2007;67:5061–5063. doi: 10.1158/0008-5472.CAN-07-0426. [DOI] [PubMed] [Google Scholar]
- 30.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 32.Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krig SR, Jin VX, Bieda MC, et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ralser M, Querfurth R, Warnatz HJ, et al. An efficient and economic enhancer mix for PCR. Biochem Biophys Res Commun. 2006;347:747–751. doi: 10.1016/j.bbrc.2006.06.151. [DOI] [PubMed] [Google Scholar]
- 35.Harrow J, Denoeud F, Frankish A, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7 Suppl 1: S4:1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin VX, O'Geen H, Iyengar S, Green R, Farnham PJ. Identification of an OCT4 and SRY regulatory module using integrated computational and experimental genomics approaches. Genome Res. 2007;17:807–817. doi: 10.1101/gr.6006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey TL, Gribskov M. Score distributions for simultaneous matching to multiple motifs. J Comput Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- 39.Lutz W, Stohr M, Schurmann J, et al. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene. 1996;13:803–812. [PubMed] [Google Scholar]
- 40.Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 41.Frye M, Fisher AG, Watt FM. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS ONE. 2007;2:e763. doi: 10.1371/journal.pone.0000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu CH, van Riggelen J, Yetil A, et al. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Efroni S, Duttagupta R, Cheng J, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.