Abstract
Prereplication complexes are assembled at eukaryotic origins of DNA replication in the G1 phase of the cell cycle, and they are activated in S phase by cyclin-dependent kinase (Cdk)2/cyclin E and Cdk2/cyclin A. Previous experiments using Xenopus nuclear assembly egg extracts suggested that Cdk1/cyclin A, which is normally active in early mitosis, can replace the function of Cdk2 in driving DNA replication, whereas Cdk1/cyclin B, which functions later in mitosis, cannot. Here, we use a completely soluble replication system derived from Xenopus egg extracts to show that Cdk1/cyclin B also can support DNA replication. The ability of mitotic Cdks to drive DNA replication raises the question of whether DNA replication is possible in mitosis. To address this question, chromatin containing prereplication complexes was driven into mitosis with Cdk1/cyclin B. Strikingly, upon addition of a replication extract, the chromatin underwent a complete round of DNA replication. Replicating mitotic chromosomes became visibly decondensed, and, after DNA replication was complete, they recondensed. Our results indicate that there is extensive overlap in the substrate specificity of the major metazoan Cdk/cyclin complexes and that mitosis is not fundamentally incompatible with DNA replication. The results suggest that origins that fail to initiate DNA replication in S phase might still be able to do so in mitosis.
During the eukaryotic cell cycle, DNA replication initiates in a two-step process (1). In G1, when cyclin-dependent kinase (Cdk) activity is low, the sequential binding to origins of the origin recognition complex, Cdc6, Cdt1, and the minichromosome maintenance complex, MCM2-7, leads to the formation of a prereplication complex (preRC). At the G1/S transition, the S-Cdks lead to activation of the preRC. This involves the loading of the initiation factor Cdc45, origin unwinding, and recruitment of DNA polymerases. Endoreduplication is inhibited because preRCs are disassembled during replication initiation and Cdks prevent de novo assembly of preRCs in S, G2, and M phases. Metazoans contain an additional inhibitor of preRC assembly called geminin (2), which inhibits Cdt1 function (3-5).
In yeast, DNA replication is stimulated by the S-phase cyclins Clb5/6 (Saccharomyces cerevisiae) and Cig2 (Schizosaccharomyces pombe), but, when these are deleted, the mitotic cyclins Clb2 (S. cerevisiae) and Cdc13 (S. pombe) can also drive S phase (6-8). In mammalian cells, antibody injection experiments and overexpression of dominant-negative Cdks indicate that entry into S phase requires Cdk2/cyclin E and Cdk2/cyclin A, the two major Cdks expressed at the beginning of S phase (9-12). The same studies found that there is no detectable effect on DNA replication when the M-Cdks, Cdk1/cyclin B and Cdk1/cyclin A, are inhibited. In Xenopus egg extracts, Cdk2/cyclin E is essential for DNA replication (13, 14). When Cdk2/cyclin E is removed from these extracts, Cdk2/cyclin A and Cdk1/cyclin A are able to support DNA replication (15, 16). These results indicate that a mitotic Cdk can drive S phase when S-Cdks are absent. Interestingly, Cdk1/cyclin B was reported not to promote S phase (14, 16).
Xenopus egg extracts have been used extensively to study DNA replication. Sperm chromatin added to interphase egg cytoplasm is assembled into nuclei that undergo DNA replication (”nuclear assembly extracts”) (17, 18). Any perturbation of the nuclear envelope in this system abolishes DNA replication (17, 19), which led to the hypothesis that higher-order nuclear structures are required for DNA replication. More recently, we developed a soluble, nucleus-independent assay (”nucleus-free system”) (see Fig. 1 A and ref. 20). In this system, DNA is first exposed to membrane-free Xenopus egg cytoplasm, which leads to the formation of preRCs. Subsequently, a concentrated nucleoplasmic extract (NPE) is added. NPE initiates DNA replication from existing preRCs while also preventing de novo preRC formation, resulting in a single round of DNA replication. The ability of NPE to stimulate DNA replication in the absence of nuclear structures is likely due to the fact that it contains high concentrations of one or more key replication factors. For example, we previously showed that the high concentration of Cdc7 present in NPE is important to achieve maximal rates of DNA replication (21).
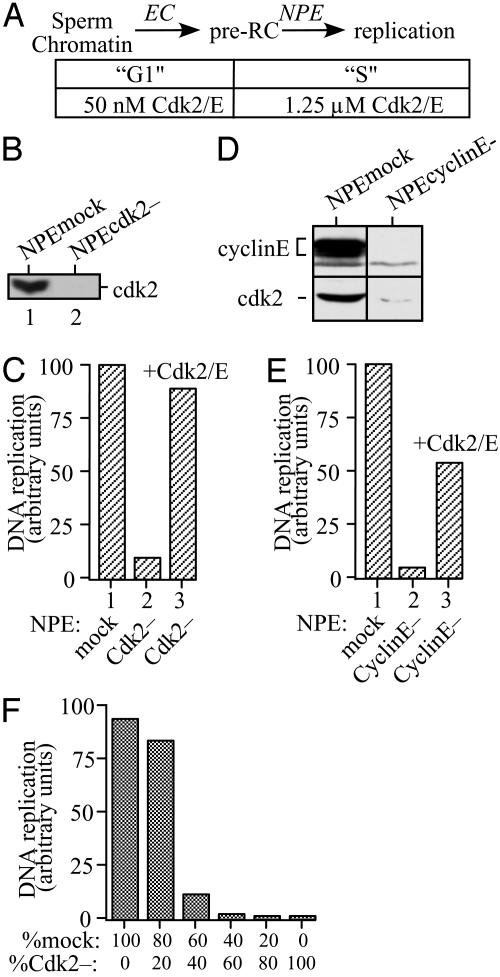
Fig. 1.
NPE stimulates DNA replication by supplying a high concentration of Cdk2/cyclin E. (A) Schematic representation of the nucleus-free DNA replication system. (B) A total of 0.5 μl of NPEmock (lane 1) or NPEcdk2- (lane 2) was assayed by Western blotting with anti-Cdk2 peptide antibody. (C) Sperm chromatin was incubated with untreated egg cytosol for 30 min and then supplemented with 2 volumes of NPEmock (reaction 1), NPECdk2- (reaction 2), or NPECdk2- supplemented with recombinant Cdk2/cyclin E (reaction 3). Reactions were stopped after 60 min. (D) A total of 1 μl of NPEmock (left lane) or NPEcyclinE- (right lane) was blotted with cyclin E or Cdk2 antibody. (E) Sperm chromatin incubated with untreated egg cytosol was supplemented with NPEmock (reaction 1), NPEcyclinE- (reaction 2), or NPEcyclinE- supplemented with recombinant Cdk2/cyclin E (reaction 3). Replication was measured after 30 min. (F) Sperm chromatin was incubated with untreated egg cytosol for 30 min and then supplemented with 2 volumes total of NPEmock and NPECdk2- mixed in different ratios, and replication was measured.
There were two plausible explanations for the inability of Cdk1/cyclin B to support DNA replication in nuclear assembly extracts seen previously. First, this mitotic protein kinase may not recognize key S-phase substrates. Alternatively, because Cdk1/cyclin B causes nuclear envelope breakdown, its DNA replication promoting activity may have been masked in this nucleus-dependent system. To distinguish between these possibilities, we have revisited the question of which Cdks can promote S phase using the nucleus-free system. When Cdk2/cyclin E is depleted from NPE, DNA replication is inactivated, and it can be restored by the addition of Cdk2/cyclin A or Cdk1/cyclin A. We now also demonstrate that Cdk1/cyclin B has full DNA replication promoting activity in the nucleus-free system. Our results raised the question of whether Cdk1/cyclin A or Cdk1/cyclin B may sometimes stimulate replication initiation in mitosis from origins that failed to fire in S phase. To test whether this is possible, we assembled preRCs on chromatin, drove them into mitosis, and then added NPE. Strikingly, replication initiated and a complete round of DNA synthesis occurred. Therefore, mitosis is not fundamentally incompatible with DNA replication. Our results argue that there is extensive overlap in the specificity of metazoan Cdks that are expressed during S, G2, and M phases, and they raise the possibility that some DNA replication could take place in mitosis.
Materials and Methods
Extract preparation and DNA replication were carried out as described (20). Briefly, Xenopus demembranated sperm chromatin was incubated for 20-30 min with egg cytosol at a concentration of 20,000 sperm per μl of extract, except in experiments shown in Fig. 4 (10,000 sperm per μl). Reactions were supplemented with two volumes of NPE containing [α-32P]dATP and stopped after 60 min, unless stated otherwise. Reaction products were separated by agarose gel electrophoresis, and DNA replication was measured by using a PhosphorImager (Molecular Dynamics) (22). To estimate the percentage of DNA replicated in Fig. 4C, the endogenous concentration of dATP in replication reactions was determined to be 100 μM, and this information was used to calculate how many nanograms of DNA had been synthesized (18).
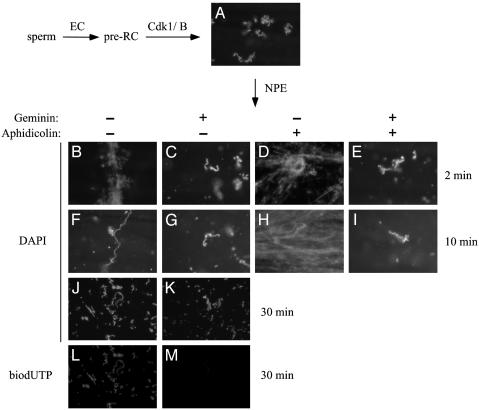
Fig. 4.
Mitotic chromatin decondenses temporarily during DNA replication. Sperm chromatin was incubated with egg cytosol containing (C, G, E, I, K, and M) or lacking (B, F, D, H, J, and L) geminin for 20 min. Subsequently, Cdk1/cyclin B was added to render the chromatin mitotic. After 40 min, reactions were supplemented with 2 volumes of NPE containing (D, E, H, and I) or lacking (B, C, F, G, and J--M)50 μg/ml aphidicolin. Some reactions contained bio-dUTP (J-M). Samples were withdrawn before (A), 2 min after (B-E), 10 min after (F-I), or 30 min after (J-M) NPE addition. Samples were stained with Hoechst (A-K) or with fluorescently labeled streptavidin (L and M) and visualized.
Recombinant human Cdk2/6-His-cyclin E, human Cdk2/6-His-cyclin A, and human 6-His-cyclin A were produced and purified as described (23). The cyclin/Cdk concentration was typically 0.2-0.4 mg/ml. The minimum concentration of each Cdk/cyclin required to achieve at least 50% DNA replication in Cdk2-depleted egg extracts was used. The minimum concentrations were ≈30 ng/μl for Cdk2/cyclin E, ≈3 ng/μl for cyclin A, and ≈3 ng/μl for Cdk2/cyclin A. Human Cdk1/cyclin B was produced in insect cells as described (24) and was used at ≈35 ng/μl to induce mitosis.
H1 kinase assays were performed in kinase buffer (250 mM sucrose, 10 mM MgCl2, 50 mM KCl, 10 mM Hepes, pH 7.7) supplemented with 40 μM cold ATP, ≈2 μCi of [γ-32]ATP, 0.2 mg/ml histone H1 (Sigma), and 50 mM β-glycerophosphate. Ten microliters of the above mixture was supplemented with 0.01 μl of the extract to be assayed and incubated at room temperature for 5 min, and the reaction was stopped by the addition of 11 μl of 2× SDS protein gel loading buffer. Reaction products were separated by PAGE, and the amount of incorporated radioactivity was measured by using a PhosphorImager.
Bio-dUTP immunofluorescence was performed essentially as described [31], except that chromatin was cross-linked with 4% formaldehyde in 10 volumes of buffer XBE2 (25), and chromatin was pelleted at 7,000 rpm in an HB-6 swinging bucket rotor.
Rabbit polyclonal antibodies against Xenopus Cdk2 protein were produced and affinity-purified by Bethyl Laboratories (Montgomery, TX) by using C-terminal peptide PFFRDVSRPTPHLI. For immunodepletions, these antibodies were bound to recombinant protein A-Sepharose Fast Flow (PAS, Amersham Pharmacia Biosciences) at concentrations of 15-20 mg/ml resin. One volume of extract was incubated three consecutive times for 1 h with 0.2 volumes of PAS/antibody beads. To deplete cyclin E, 20 μl of NPE was incubated for 3 h with PAS bound to 5 μg of affinity-purified cyclin E1 antibody (20). Western blotting was performed by using the above antibodies and anti-MCM7 (26). Anti-XCAP-E (Smc2) antibodies (27) were a kind gift from Dr. Tatsuya Hirano (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
Results
To determine the range of Cdks that can support DNA replication in the nucleus-free system, it was first necessary to remove endogenous Cdk2/cyclin E. Xenopus egg cytosol contains ≈50 nM Cdk2/cyclin E (28, 29), whereas the concentration of this kinase in NPE is ≈1.25 μM (20). We asked whether removal of Cdk2 from NPE is sufficient to abolish DNA replication. Sperm chromatin was incubated with undepleted egg cytosol to form preRCs and then supplemented with mock-depleted NPE (NPEmock) or NPE from which Cdk2 had been quantitatively removed (NPECdk2-; Fig. 1B). Compared with NPEmock, NPECdk2- was inactive for DNA replication (Fig. 1C, compare columns 1 and 2), and readdition of recombinant Cdk2/cyclin E restored DNA replication (Fig. 1C, column 3). Similarly, depletion of cyclin E from NPE, a procedure that codepletes >95% of Cdk2 (Fig. 1D), was sufficient to abolish DNA replication, and the effect was largely reversed by readdition of Cdk2/cyclin E (Fig. 1E). We conclude that cytosolic Cdk2/cyclin E is insufficient to support DNA replication, and that this protein kinase must be supplied by NPE. We next measured DNA replication when NPEmock and NPECdk2- were mixed in different ratios and added to preRCs formed in undepleted egg cytosol. As seen in Fig. 1F, the efficiency of DNA replication was strongly reduced if the amount of Cdk2 present in NPE was <80% the normal level, ≈1 μM. These results argue that a high concentration of Cdk2/cyclin E is critical for initiation of DNA replication. Similarly, in nuclear assembly egg extracts, it was recently reported that the intranuclear concentration of Cdk2/cyclin E required for DNA replication is also very high, at least 3 μM (30). Recombinant Cdk2/cyclin E was not sufficient to replace the function of NPE in initiating DNA replication (data not shown), as expected given the requirement for high concentrations of nuclear Cdc7 (21) and MCM10 (31).
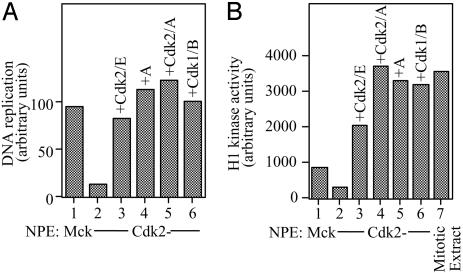
We next investigated which Cdk complexes can replace the function of Cdk2/cyclin E in DNA replication initiation. DNA replication in Cdk2-depleted NPE could be restored with recombinant Cdk2/cyclin E, Cdk2/cyclinA, or cyclin A alone (Fig. 2A, columns 3-5). In the latter case, we infer that cyclin A formed an active complex with endogenous Cdk1, because cyclin A did not restore DNA replication when Cdk1 was also immunodepleted with an antipeptide antibody (data not shown). Importantly, we found that recombinant Cdk1/cyclin B also fully restored DNA replication in Cdk2-depleted NPE (Fig. 2 A, column 6). We assayed histone H1 kinase activity of the various recombinant Cdks after addition to NPECdk2- at the same concentrations that were used for replication rescue experiments in Fig. 2 A. The total H1 kinase activity in NPECdk2- was ≈3-fold lower than in NPEMock (Fig. 2B, compare 1 and 2), indicating that the majority of H1 kinase activity in total extract is Cdk2-dependent. Addition of Cdk2/cyclin E, Cdk2/cyclin A, cyclin A alone, or Cdk1/cyclin B to NPECdk2- each resulted in similar levels of H1 kinase activity that were comparable to the level seen in mitotically arrested extracts (Fig. 2B, columns 3-7). Therefore, the concentration of Cdk1/cyclin B that normally exists in mitosis can support DNA replication in the context of our nucleus-free egg extracts.
Fig. 2.
Cdk1/cyclin B can support DNA replication. (A) Sperm chromatin was incubated with untreated egg cytosol and then supplemented with NPEmock (reaction 1) or NPECdk2- (reactions 2-6). NPE in reactions 3-6 was supplemented with recombinant Cdk2/cyclin E, cyclin A, Cdk2/cyclin A, and Cdk1/cyclin B, respectively. (B) Recombinant Cdk2/cyclin E, Cdk2/cyclin A, cyclin A, or Cdk1/cyclin B was added to NPECdk2- as in A, and the equivalent of 0.01 μl of each NPE/Cdk/cyclin mixture (reactions 3-6), as well as NPEmock (reaction 1), NPECdk2- alone (reaction2), or crude egg extract arrested in metaphase (reaction 7), was assayed for total H1 kinase activity.
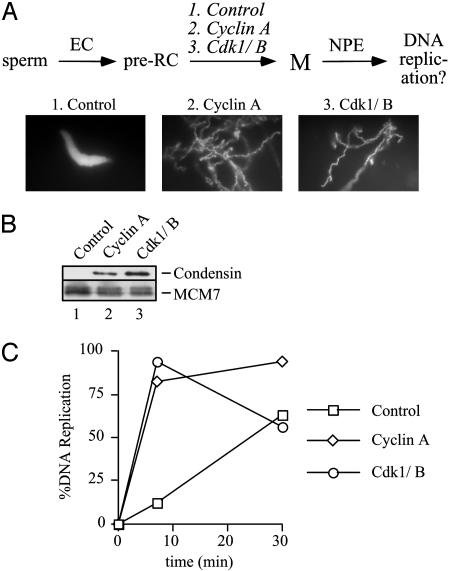
The observation that Cdk1/cyclin A and Cdk1/cyclin B can support DNA replication raises the question of whether DNA replication can take place in mitosis, the time when these kinases are most active. To begin to address this issue, we asked whether mitotic chromatin structure might represent a barrier to DNA replication; therefore, we carried out the experiment outlined in Fig. 3A. Sperm chromatin was incubated with interphase egg cytosol to form preRCs. Subsequently, we added cyclin A or Cdk1/cyclin B to induce mitosis, or buffer as a control. Within 20 min, the chromatin was condensed (Fig. 3A) and the condensin SMC2/XCAP-E was bound to the sperm (Fig. 3B and ref. 27). Importantly, preRCs persisted on the mitotic chromatin, as evidenced by the continued presence of MCM7 (Fig. 3B). At this stage, DNA replication had not begun because NPE was lacking. When NPE was added, both control chromatin and mitotic chromatin underwent DNA replication (Fig. 3C). Unexpectedly, in the presence of mitotic Cdks, DNA replication was completed after only 7 min, whereas in the control, it lasted at least 30 min, as is customary in this system (20). It is presently unclear whether the increased rate of DNA synthesis was due to more initiation events or faster replication fork progression. In the experiment shown in Fig. 3C, we carefully measured the amount of input DNA replicated (see Materials and Methods), and in the presence of M-Cdks it approached 100%. The same result was obtained if we used Cdk2-depleted NPE supplemented with Cdk1/cyclin B (data not shown), indicating that Cdk1/cyclin B can initiate DNA replication on precondensed chromatin. Fresh sperm chromatin added after the first round of DNA replication underwent rapid and complete condensation, demonstrating that the Cdk1/cyclin B added at the start of DNA replication was still active (data not shown). Therefore, preRCs bound to chromatin that is functionally in mitosis are able to undergo replication initiation, and replication forks are able to move efficiently through condensed chromatin in the continued presence of high Cdk1/cyclin B activity.
Fig. 3.
Mitotic chromatin can replicate. Sperm chromatin was incubated with untreated egg cytosol to form preRCs, then buffer, cyclin A, or Cdk1/cyclin B was added. After an additional 20 min, reactions were supplemented with 2 volumes of NPE treated with buffer or the appropriate Cdk. (A) Chromatin samples were withdrawn immediately before NPE addition. After staining with Hoechst, the chromatin was examined under UV illumination. (B) Chromatin was isolated immediately before NPE addition, and chromatinbound proteins were analyzed by Western blotting by using anti-XCAP-E and MCM7 antibodies. (C) DNA replication was measured 7 and 30 min after NPE addition. The percentage of input DNA replicated was measured (see Materials and Methods) and graphed.
It is important to note that it was critical to establish preRCs on chromatin before it was rendered mitotic. Thus, if cyclin A or Cdk1/cyclin B was added to egg cytosol before sperm chromatin, no DNA replication was observed upon addition of NPE (data not shown), as expected because high concentrations of Cdk activity are inhibitory for de novo preRC assembly (32, 33). Therefore, as for DNA replication in S phase, DNA replication in mitotic extracts is totally dependent on preRCs that were previously assembled in a G1-like environment that lacks high Cdk activity.
We investigated how the passage of a DNA replication fork might affect the structure of highly compacted mitotic chromatin. As in Fig. 3, sperm was first incubated in egg cytosol to form preRCs, Cdk1/cyclin B was then added to induce mitosis, and, finally, NPE was added to initiate DNA replication. We found that, 2 min after addition of NPE, the chromatin decondensed visibly (Fig. 4, compare A and B), and the effect depended on the initiation of DNA replication because it was blocked when geminin was present during the initial incubation of the sperm with egg cytosol (Fig. 4C). Although some chromatin looks decondensed in Fig. 4C, it is merely out of focus (data not shown). By 10 min after the addition of NPE, when incorporation of radioactive dATP had plateaued (Fig. 3C), the chromatin resumed its condensed state (Fig. 4F). Recondensation at the 10-min time point depended on completion of DNA replication because when NPE contained aphidicolin to block DNA synthesis, chromatin decondensed and never recondensed (Fig. 4 D and H). The highly decondensed state observed after a 10-min incubation with aphidicolin was not a nonspecific effect of aphidicolin because chromatin remained fully condensed when the reaction also contained geminin (Fig. 4 E and I). Also, failure to recondense in aphidicolin was not due to checkpoint-mediated down-regulation of Cdk1 activity, because decondensation was observed when aphidicolin was added together with 5 mM caffeine (K.M. and J.C.W., unpublished results), which inhibits ATM and ATR checkpoint kinases (34). Together, these experiments show that DNA replication leads to a significant decondensation of chromatin. The fact that all of the chromatin on the slide undergoes decondensation (Fig. 4 B, D, and H) indicates that all of the DNA underwent replication initiation. Finally, the data argue that replication-mediated decondensation is reversible, because the chromatin recondenses when DNA replication is complete (Fig. 4F). To further prove that chromatin recondenses after DNA replication, we performed DNA replication as in Fig. 4 B, C, F, and G, but we included biotinylated dUTP (bio-dUTP). In this case, the condensed chromatin that reappears after incubation with NPE was labeled with bio-dUTP (Fig. 4L), but not when DNA replication was blocked with geminin (Fig. 4M). Thus, mitotic chromatin decondenses during DNA replication, after which it quickly recondenses.
Discussion
In budding and fission yeast, mitotic B-type Cdks have S phase-promoting activity (6-8). By contrast, in metazoans, only Cdk2/cyclin E, Cdk2/cyclin A, and Cdk1/cyclin A have been shown to support DNA replication (15, 16), whereas Cdk1/cyclin B was reported not to possess this activity (14, 16). Using a nucleus-free DNA replication system derived from Xenopus egg extracts, we show here that Cdk1/cyclin B also can support DNA replication. Therefore, in metazoans as in yeast, Cdks that act late in the cell cycle are able to catalyze events that normally occur earlier. The converse does not appear to be true, because high concentrations of Cdk2/cyclin E do not induce mitosis (16). This hierarchy is likely designed to prevent premature mitosis during S phase. Our findings suggest that the previous failure to uncover the DNA replication activity of Cdk1/cyclin B was most likely due to Cdk1/cyclin B-mediated nuclear envelope breakdown, a key requirement for DNA replication in nuclear assembly egg extracts. Indeed, after completion of our studies, a report using nuclear assembly extracts showed that intermediate levels of Cdk1/cyclin B that do not lead to nuclear envelope breakdown can drive DNA replication (35). Interestingly, it was recently shown that certain cancer cells are able to proliferate in the absence of Cdk2 activity (36). Our results suggest that DNA replication in these Cdk2-deficient cells may be stimulated by Cdk1/cyclin A and/or Cdk1/cyclin B.
Cdk1/cyclin A and Cdk1/cyclin B drive the dramatic changes in cellular structure and function observed in mitosis by adding stimulatory and inhibitory phosphates to numerous proteins. For example, phosphorylation by Cdk1/cyclin B leads to activation of the condensin complex (37). Phosphorylated condensin binds to chromatin, causing chromosome compaction by means of a mechanism that may involve positive supercoiling of DNA (27, 38). In contrast, phosphorylation by Cdk1/cyclin B inhibits the function of the nuclear lamins, leading to nuclear envelope breakdown (39). Notably, the high concentrations of Cdk1/cyclin B present in mitosis are inhibitory for transcription, insuring that certain types of metabolism are not allowed in mitosis (ref. 40 and references therein). In contrast, we found that when chromatin containing preRCs is driven into a mitotic state with Cdk1/cyclin B or cyclin A, the preRCs are subsequently able to undergo initiation, and a complete round of chromosomal DNA replication ensues in the continuous presence of high mitotic Cdk activity. Therefore, once a preRC has been established, neither the initiation step nor the elongation step of DNA replication appears to be inhibited by condensed chromatin structure. Importantly, Cdk1/cyclin B leads to hyperphosphorylation of many DNA replication factors in egg extracts, including MCM4 and RPA (ref. 32 and our unpublished results), but these phosphorylation events do not appear to be inhibitory for DNA replication. Therefore, unlike transcription, the mitotic state is not inhibitory for DNA replication. It is important to emphasize that DNA replication in mitotic extracts depends on already established preRCs, indicating that it obeys cell cycle control mechanisms that limit DNA replication to a single round per cell cycle.
The question arises of whether DNA replication can take place in mitosis under physiological conditions and, if so, whether this ever happens in cells. In budding yeast arrested in mitosis with nocodozole, transient lowering of Cdk levels leads to reassembly of preRCs, origin firing, and a complete round of DNA replication (41). In Xenopus egg extracts, the initiation and elongation steps of DNA replication require a transport-competent nuclear envelope (17, 19, 20, 42, 43). Therefore, once the nuclear envelope has broken down in mitosis, DNA replication is not expected to occur. In our experiments, we bypassed the requirement for a nuclear envelope through the addition of NPE. However, in somatic cells, the requirement for a nuclear envelope may be much less stringent. Johnson and Rao (44) showed that, when a cell in mitosis was fused with an S-phase cell, the S-phase nucleus underwent rapid envelope breakdown and premature chromosome condensation. Importantly, the condensed chromatin underwent significant levels of DNA replication. This observation shows that in somatic cells, a pseudomitotic state is compatible with DNA replication and that the nuclear envelope is not strictly required.
Assuming that DNA replication is possible in mitosis, does this normally occur? Inhibition of Cdk1 activity in somatic cells has no visible effect on DNA replication (11, 12), consistent with the observation that the vast majority of DNA replication takes place in S phase. However, execution of anaphase with any unreplicated DNA leads to chromosome breakage (mitotic catastrophe); therefore, we find appealing the possibility that if any origins fail to initiate replication in S phase, they could still do so in mitosis through the action of Cdk1/cyclin A and/or Cdk1/cyclin B. Similarly, if a replication fork does not complete duplication of its replicon in S phase, it could still do so in mitosis. The time between nuclear envelope breakdown and anaphase is highly variable, but on average it comprises ≈40 min (45), allowing a significant period for DNA replication. A priori, replication of small amounts of DNA in mitosis is not expected to disrupt chromosome segregation, as long as normal mitotic chromatin structure is reestablished before anaphase. We found that DNA replication caused significant decondensation of mitotic chromatin, as might be expected if proteins that compact DNA are displaced by the replication fork. However, the chromatin rapidly recondensed when DNA replication was complete, indicating that proper chromatin structure was restored. A potential complication of our model is that unreplicated DNA is thought to down-regulate Cdk1 activity (22). However, this effect has so far been observed only in the presence of DNA replication inhibitors and may not apply to small amounts of unreplicated DNA in an otherwise normal cell. Indeed, when the rate of DNA replication is reduced because of limited preRC assembly, yeast cells undergo anaphase with unreplicated DNA, leading to chromosome fragmentation (46). We envision that mitotic DNA replication probably occurs infrequently and involves small amounts of DNA at random locations. As such, detecting mitotic DNA replication with available techniques is expected to be difficult. However, if a protein inhibitor of DNA replication that acts after preRC assembly is identified in the future, one could ask whether its expression in mitosis increases the incidence of mitotic catastrophes and genomic instability.
Conclusion
We have shown that Cdk1/cyclin B is just as active in promoting DNA replication as are Cdk2/cyclin E, Cdk2/cyclin A, and Cdk1/cyclin A, indicating that there is extensive overlap in the substrate specificity of these various Cdk/cyclin complexes. In addition, we show that biochemical changes brought about by M-Cdks, including chromosome condensation, pose no barrier to initiation events that occur after preRC formation or movement of the replication fork during elongation. Our results raise the possibility that the permissive window for DNA replication is longer than previously envisioned, extending from the G1/S transition until the moment cells undergo anaphase.
Acknowledgments
We thank T. Hirano for the condensin antibody; Randy King for Cdk1/cyclin B; Tatsuro Takahashi and James Wohlschlegel for help with preparation of Cdk2/cyclin E; Anindya Dutta, Tatsuro Takahashi, Elaine Elion, and Christin Cvetic for comments on the manuscript; and Julian Blow and the members of our laboratory for helpful discussions. This work was supported by National Institutes of Health Grant GM62267 and a Burroughs Wellcome Career Award (to J.C.W.).
Abbreviations: Cdk, cyclin-dependent kinase; NPE, nucleoplasmic extract; preRC, prereplication complex; MCM, minichromosome maintenance complex.
References
- 1.Bell, S. P. & Dutta, A. (2002) Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- 2.McGarry, T. J. & Kirschner, M. W. (1998) Cell 93, 1043-1053. [DOI] [PubMed] [Google Scholar]
- 3.Mihaylov, I. S., Kondo, T., Jones, L., Ryzhikov, S., Tanaka, J., Zheng, J., Higa, L. A., Minamino, N., Cooley, L. & Zhang, H. (2002) Mol. Cell. Biol. 22, 1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tada, S., Li, A., Maiorano, D., Mechali, M. & Blow, J. J. (2001) Nat. Cell Biol. 3, 107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wohlschlegel, J. A., Dwyer, B. T., Dhar, S. K., Cvetic, C., Walter, J. C. & Dutta, A. (2000) Science 290, 2309-2312. [DOI] [PubMed] [Google Scholar]
- 6.Haase, S. B. & Reed, S. I. (1999) Nature 401, 394-397. [DOI] [PubMed] [Google Scholar]
- 7.Fisher, D. L. & Nurse, P. (1996) EMBO J. 15, 850-860. [PMC free article] [PubMed] [Google Scholar]
- 8.Amon, A., Irniger, S. & Nasmyth, K. (1994) Cell 77, 1037-1050. [DOI] [PubMed] [Google Scholar]
- 9.Ohtsubo, M., Theodoras, A. M., Schumacher, J., Roberts, J. M. & Pagano, M. (1995) Mol. Cell. Biol. 15, 2612-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagano, M., Pepperkok, R., Verde, F., Ansorge, W. & Draetta, G. (1992) EMBO J. 11, 961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Heuvel, S. & Harlow, E. (1993) Science 262, 2050-2054. [DOI] [PubMed] [Google Scholar]
- 12.Girard, F., Strausfeld, U., Fernandez, A. & Lamb, N. J. (1991) Cell 67, 1169-1179. [DOI] [PubMed] [Google Scholar]
- 13.Fang, F. & Newport, J. W. (1991) Cell 66, 731-742. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, P. K., Chevalier, S., Philippe, M. & Kirschner, M. W. (1995) J. Cell Biol. 130, 755-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevalier, S., Tassan, J. P., Cox, R., Philippe, M. & Ford, C. (1995) J. Cell Sci. 108, 1831-1841. [DOI] [PubMed] [Google Scholar]
- 16.Strausfeld, U. P., Howell, M., Descombes, P., Chevalier, S., Rempel, R. E., Adamczewski, J., Maller, J. L., Hunt, T. & Blow, J. J. (1996) J. Cell Sci. 109, 1555-1563. [DOI] [PubMed] [Google Scholar]
- 17.Newport, J. (1987) Cell 48, 205-217. [DOI] [PubMed] [Google Scholar]
- 18.Blow, J. J. & Laskey, R. A. (1986) Cell 47, 577-587. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan, M. A., Mills, A. D., Sleeman, A. M., Laskey, R. A. & Blow, J. J. (1988) J. Cell Biol. 106, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter, J., Sun, L. & Newport, J. (1998) Mol. Cell 1, 519-529. [DOI] [PubMed] [Google Scholar]
- 21.Walter, J. C. (2000) J. Biol. Chem. 275, 39773-39778. [DOI] [PubMed] [Google Scholar]
- 22.Dasso, M. & Newport, J. W. (1990) Cell 61, 811-823. [DOI] [PubMed] [Google Scholar]
- 23.Wohlschlegel, J. A., Dwyer, B. T., Takeda, D. Y. & Dutta, A. (2001) Mol. Cell. Biol. 21, 4868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai, D., Wessling, H. C., Fisher, R. P. & Morgan, D. O. (1995) Mol. Cell. Biol. 15, 345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacCallum, D. E., Losada, A., Kobayashi, R. & Hirano, T. (2002) Mol. Biol. Cell 13, 25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter, J. & Newport, J. (2000) Mol. Cell 5, 617-627. [DOI] [PubMed] [Google Scholar]
- 27.Hirano, T. & Mitchison, T. J. (1994) Cell 79, 449-458. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi, H., Golsteyn, R., Poon, R., Stewart, E., Gannon, J., Minshull, J., Smith, R. & Hunt, T. (1991) Cold Spring Harbor Symp. Quant. Biol. 56, 437-447. [DOI] [PubMed] [Google Scholar]
- 29.Rempel, R. E., Sleight, S. B. & Maller, J. L. (1995) J. Biol. Chem. 270, 6843-6855. [DOI] [PubMed] [Google Scholar]
- 30.Moore, J. D., Kornbluth, S. & Hunt, T. (2002) Mol. Biol. Cell 13, 4388-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wohlschlegel, J. A., Dhar, S. K., Prokhorova, T. A., Dutta, A. & Walter, J. C. (2002) Mol. Cell 9, 233-240. [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson, M., Madine, M., Dalton, S. & Gautier, J. (1996) Proc. Natl. Acad. Sci. USA 93, 12223-12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua, X. H. & Newport, J. (1998) J. Cell Biol. 140, 271-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M. & Abraham, R. T. (1999) Cancer Res. 59, 4375-4382. [PubMed] [Google Scholar]
- 35.Moore, J. D., Kirk, J. A. & Hunt, T. (2003) Science 300, 987-990. [DOI] [PubMed] [Google Scholar]
- 36.Tetsu, O. & McCormick, F. (2003) Cancer Cell 3, 233-245. [DOI] [PubMed] [Google Scholar]
- 37.Kimura, K., Hirano, M., Kobayashi, R. & Hirano, T. (1998) Science 282, 487-490. [DOI] [PubMed] [Google Scholar]
- 38.Kimura, K., Rybenkov, V. V., Crisona, N. J., Hirano, T. & Cozzarelli, N. R. (1999) Cell 98, 239-248. [DOI] [PubMed] [Google Scholar]
- 39.Heald, R. & McKeon, F. (1990) Cell 61, 579-589. [DOI] [PubMed] [Google Scholar]
- 40.Hartl, P., Gottesfeld, J. & Forbes, D. J. (1993) J. Cell Biol. 120, 613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahmann, C., Diffley, J. F. & Nasmyth, K. A. (1995) Curr. Biol. 5, 1257-1269. [DOI] [PubMed] [Google Scholar]
- 42.Cox, L. S. (1992) J. Cell Sci. 101, 43-53. [DOI] [PubMed] [Google Scholar]
- 43.Hughes, M., Zhang, C., Avis, J. M., Hutchison, C. J. & Clarke, P. R. (1998) J. Cell Sci. 111, 3017-3026. [DOI] [PubMed] [Google Scholar]
- 44.Johnson, R. T. & Rao, P. N. (1970) Nature 226, 717-722. [DOI] [PubMed] [Google Scholar]
- 45.Rieder, C. L., Schultz, A., Cole, R. & Sluder, G. (1994) J. Cell Biol. 127, 1301-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lengronne, A. & Schwob, E. (2002) Mol. Cell 9, 1067-1078. [DOI] [PubMed] [Google Scholar]