Abstract
Escherichia coli nucleoside diphosphate kinase (eNDK) is an XTP:XDP phosphotransferase that plays an important role in the regulation of cellular nucleoside triphosphate concentrations. It is also one of several recently discovered DNases belonging to the NM23/NDK family. E. coli cells disrupted in the ndk gene display a spontaneous mutator phenotype, which has been attributed to the mutagenic effects of imbalanced nucleotide pools and errors made by replicative DNA polymerases. Another explanation for the increased mutation rates is that endk- cells lack the nuclease activity of the NDK protein that is essential for a DNA repair pathway. Here, we show that purified, cloned endk is a DNA repair nuclease whose substrate is uracil misincorporated into DNA. We have identified three new catalytic activities in eNDK that act sequentially to repair the uracil lesion: (i) uracil-DNA glycosylase that excises uracil from single-stranded and from U/A and U/G mispairs in double-stranded DNA; (ii) apyrimidinic endonuclease that cleaves double-stranded DNA as a lyase by forming a covalent enzyme-DNA intermediate complex with the apyrimidinic site created by the glycosylase; and (iii) DNA repair phosphodiesterase that removes 3′-blocking residues from the ends of duplex DNA. All three of these activities, as well as the nucleoside-diphosphate kinase, reside in the same protein. Based on these findings, we propose an editing function for eNDK as a mechanism by which the enzyme prevents mutations in DNA.
Nucleoside diphosphate kinases (NDKs) (EC 2.7.4.6) have been recognized for decades as a large and conserved family of enzymes that synthesize nucleoside triphosphates from nucleoside diphosphates and ATP. Given their broad substrate specificities and the nucleotide pool perturbations caused by the absence of NDK in Escherichia coli, it has been suggested that their function in vivo is to maintain the necessary nucleoside triphosphate concentrations (1, 2). The x-ray crystal structures of many NDKs, with and without nucleotide substrates, are well known (3). During the last decade, additional DNA metabolic activities have been demonstrated for NDKs, including sequence-dependent DNA binding and transcription (4-8) and site-specific cleavage of DNA (8-14). Moreover, the DNase activity of human NM23-H2/NDK (h2NDK) has been implicated in base excision repair (BER) on the basis of its DNA cleavage mechanism, which uses the ε-amino group of a conserved lysine as the active site nucleophile forming a covalent Schiff base intermediate with DNA, the hallmark mechanism of DNA glycosylase/apyrimidinic (AP) lyase BER enzymes (10, 14, 15). Thus far, however, h2NDK has been associated only with the cleavage of unusually structured but otherwise undamaged DNA (9-11, 14), and not with the repair of specific DNA lesions.
Mammalian NDKs are also known as NM23 (nonmetastatic) on the basis of their involvement in tumorigenesis and tumor progression (16, 17). The Drosophila NDK is AWD, a developmental protein encoded by the altered wing disk gene (18). In E. coli, deletion of the ndk gene results in increased spontaneous mutation rates and elevations in the CTP, dCTP, and dGTP pool sizes, suggesting that the mutations are caused by pool perturbations and the incorporation of inappropriate nucleotides into DNA (2, 19). However, the majority of mutations in ndk- cells were reported to be AT→TA changes by R. M. Schaaper (unpublished results cited in ref. 2), which could not have been caused directly by the above nucleotide pool imbalances. While also observing AT→TA transversions in their ndk- strains, the majority of the mutations sequenced by Miller et al. (20) were of the AT→GC transition type, which could have arisen as a consequence of the composition of the pool expansions reported above (2, 19).
Another explanation for the increased mutation rates is that E. coli ndk- cells lack the nuclease activity of the NDK protein that is essential for a DNA repair pathway (12, 14), particularly because a mutator phenotype is often the consequence of BER defects (21, 22). This hypothesis is supported by the observation of Miller et al. (20), that deletion of the ndk gene in a mutS- mismatch repair strain dramatically increases the frequency of AT→GC transitions, suggesting that the products of the mutS and ndk genes act synergistically in a common DNA repair pathway. Because Miller et al. (20) detected no base mismatchspecific glycosylase activity in purified E. coli NDK (eNDK), they concluded that the high frequency of mutations in the absence of NDK were caused by a combination of polymerase errors and nucleotide pool imbalances.
In this work, we sought to identify the DNA repair function of eNDK that we have previously proposed is associated with its DNase activity (10, 12, 14), with the expectation that this might also provide insight into the mutator effect of ndk null cells. We carried out tests on a variety of damaged DNA substrates that eNDK might act upon, and identified the base uracil, which, when mispaired with adenine or guanine, is a specific target for excision repair. We show here that eNDK is a multifunctional BER nuclease that acts sequentially, first as a uracil-DNA glycosylase (UDG), then as an endonuclease that cleaves the DNA backbone at the uracil-less site, and then as a 3′ repair phosphodiesterase that removes 3′ terminal products from DNA. All three of these activities seem to be associated with the eNDK polypeptide.
Materials and Methods
Enzymes, Inhibitors, and Antibodies. eNDK was prepared by using the expression vector ndkec (a gift of M. Konrad, Max Planck Institute, Goettingen, Germany) in BL21(DE3) E. coli cells and purified by ammonium sulfate fractionation (60%), DEAE ion-exchange chromatography by gradient elution (≈150 mM NaCl), and by hydroxyapatite chromatography (flow through) as described (10-12). Control enzymes from E. coli were uracil DNA glycosylase (eUDG, ung), endonuclease III (EndoIII), endonuclease IV (EndoIV), endonuclease VIII (EndoVIII), exonuclease III (ExoIII), and human APE (all from Trevigen). Affinity-purified antibodies raised against eNDK were a gift of I. Lascu (University of Bordeaux, Bordeaux, France). The UDG inhibitor Ugi was from NEB.
DNA Substrates. Oligonucleotide substrates (top strands) WT (5′-CCTGCCCTGTGCAGCTGTGGG-3′), uracil containing (5′-CCTGCCCTGUGCAGCTGTGGG-3′), and their complementary strands were purchased from IDT. The AP (5′-CCTGCCCTGAPGCAGCTGTGGG-3′), and phosphoglycolate (PG) (5′-CCTGCCCTGTGCAGCTGTGGG-PG-3′) oligonucleotides and their complementary strands were obtained from Trevigen. The 5′ end of each top strand was 32P-end-labeled by using [γ-32P]ATP and T4 polynucleotide kinase. The labeled oligonucleotides were purified from unincorporated 32P by ethanol precipitation and annealed to form duplexes with 1.5-fold molar excess unlabeled complementary strands by heating for 10 min at 85°C and slow cooling. Each duplex oligonucleotide was subsequently gel purified. Sequencing ladders were prepared by using the WT oligonucleotide and a kit based on the Maxam and Gilbert procedure (Sigma).
Enzyme Assays. Uracil processing (removal of uracil from DNA and concomitant cleavage of the AP site) by eNDK was assayed in a 5-μl reaction buffer (10 mM Hepes-KOH, pH.7.4/100 mM KCl), with 1 unit of NDK and 0.1 pmol of 32P-labeled DNA. The reaction mixtures were incubated for 30 min at 37°C and then stopped by the addition of 2.5 μl of 95% formamide/20 mM EDTA. The products were analyzed on 20% TBE-urea containing polyacrylamide gels. Before the assays shown here, the reaction conditions were optimized with regard to time, temperature, pH, protein, and metal ion concentrations. One unit of NDK is defined as the amount of enzyme required to cleave 0.1 pmol of duplex DNA oligonucleotide in 30 min at 37°C under the assay conditions. AP site cleavage and 3′ phosphodiesterase activities were assayed in reaction buffer as above. UDG activity was assayed in reaction buffer containing 5 mM EDTA. Control enzymes were assayed by using the above protocols for NDK with units adjusted as recommended by the vendor. NDK activity was measured as described (12).
Results
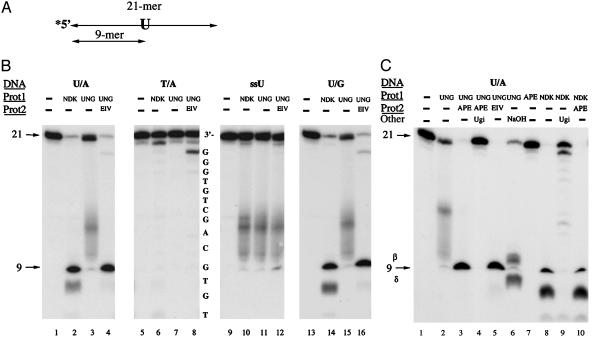
Uracil-Processing Activity of eNDK. eNDK uracil-repair activity was detected by using a 21-bp 32P-end-labeled duplex oligonucleotide, containing a centrally located uracil (10th base from the 5′ end; Fig. 1A) paired with adenine (U/A mispair) or with guanine (U/G mispair). To map the cleavage sites, we used 20% denaturing urea-acrylamide gels and Maxam and Gilbert sequencing ladders. With no addition of other enzymes, eNDK cleaved both the U/A and U/G oligonucleotides at the uracil incorporation site, producing a ≈9-nucleotide-long (9 nt) 32P-end-labeled DNA fragment and an additional product that was shorter by ≈1 nt or less (Fig. 1B, lanes 2 and 14). The specificity with which eNDK cleaves uracil-containing substrates suggests that both a uracil removing and an endonucleolytic activity are involved in the generation of the resulting fragments. Apparently, eNDK recognizes uracil in DNA, then cleaves the 5′ glycosylic bond of the uridine according to a UDG reaction (15, 21, 22), and then acts as an AP endonuclease upon this AP intermediate.
Fig. 1.
Uracil processing by eNDK. (A) Schematic view of the oligonucleotide substrate used in this experiment. Asterisk indicates the radioactively labeled terminus. (B) DNA cleavage analysis of U/A, T/A, ssU (single-stranded), and U/G oligonucleotides. Sequencing ladders prepared from the T/A substrate (partial sequence shown here) were run alongside samples. Enzymes and substrates used are indicated above lanes. The smeared appearance of UNG-treated DNA is because of instability of AP DNA and to the collapse of the helix from loss of base-stacking interactions. Reaction mixtures were separated on 20% sequencing gels, and the gels were subjected to autoradiography. (C) DNA cleavage analysis using the UDG inhibitor protein Ugi. See text for details.
When the same oligonucleotide is treated with E. coli UDG (UNG), which is a simple monofunctional DNA glycosylase without an associated endonuclease activity, the uracil is removed by UNG creating an AP site that is quite labile, but UNG does not cleave the sugar phosphate backbone (Fig. 1B, lanes 3 and 15). The DNA chain at the AP site created by UNG can then be cleaved by an endonuclease, e.g., E. coli EndoIV, which specifically recognizes AP lesions but does not have an associated glycosylase activity. Clearly, the combined action of UNG and EndoIV generate the same 9-nt-long fragment, as does eNDK alone (lanes 4 and 16).
When uracil is replaced with thymine in the oligonucleotide (T/A), eNDK, like UNG, does not recognize this WT substrate (Fig. 1B, lanes 5-8), suggesting high specificity for the uracil base. It is worth noting here that, in the absence of a specific lesion, eNDK lacks significant nonspecific endonuclease activity. Although the single-stranded U-oligonucleotide substrate is deglycosylated by eNDK, as by UNG, the phosphodiester bond does not get cleaved (Fig. 1B, lanes 9-12), indicating that AP-site cleavage by eNDK is specific for double-stranded DNA, as is the case with EndoIV (lane 16).
The comigration of the main eNDK product of both the U/A and U/G cleaved substrates with that of the EndoIV cleaved DNA suggests that eNDK cleaves the backbone leaving 3′ hydroxyl ends. This was also suggested in parallel experiments in which the NDK reaction mixture was further treated with EndoIV or with human AP endonuclease, both of which cleave hydrolytically 5′ to the AP site producing 3′ terminal hydroxyls (21-23). No further products were obtained after these treatments (Fig. 1C, lane 10), suggesting that the 3′ end of the 9-mer is a hydroxyl group. Similar conclusions were reached in the case of human h2NDK cleaved DNA by using a 3′ end-labeling procedure (9).
eNDK, like all other UDG enzymes, is inhibited by Ugi, the highly structure-specific peptide inhibitor protein encoded by the uracil containing genome of bacteriophage PBS-2 (15, 21, 22; Fig. 1C, lanes 4 and 9). This suggests that the uracil recognition pocket targeted by Ugi is the same for eNDK as it is for other UDGs and that these protein families might be structurally related. An additional activity of eNDK that shortens the 3′ end of the 21-mer oligonucleotide by ≈1 nucleotide or less, on the other hand, is stimulated by Ugi (lane 9). This implies that the 3′ end processing activity of eNDK is separate from the UDG.
UDG Activity of eNDK. We next asked whether eNDK acts as a uracil DNA glycosylase independently of the cleavage activity. Although the glycosylase and AP lyase activities are mechanistically coupled in bifunctional BER enzymes, it is possible to separate them because glycosylases are resistant to EDTA treatment (21, 22), whereas many AP endonucleases, including that of NDK, are inhibited. Thus, to generate uracil-less AP sites, we carried out the uracil-processing experiments in the presence of 5 mM EDTA, thereby leaving the phosphodiester bonds intact. We next added AP site-cleaving enzymes that are EDTA resistant, e.g., EndoIII, which cleaves by β-elimination of the 3′ phosphate without additional processing (Fig. 2, lanes 3 and 8), and EndoVIII, which cleaves by successive β- and δ-eliminations (lanes 4 and 9). Spermidine (lanes 5 and 10) and hot alkali (Fig. 2, lanes 6 and 11) cleave AP sites by β or by successive β, δ eliminations (21, 22). Clearly, eNDK acts like a uracil glycosylase capable of generating AP sites that are susceptible to cleavage by these reagents, which, alone, do not cleave the uracil substrate (lanes 12-15). We can therefore conclude that eNDK exhibits glycosylase activity independent of AP site cleavage.
Fig. 2.
UDG activity of eNDK. Oligonucleotide-cleavage assay was carried out with the U/A oligonucleotide substrate in the presence of EDTA to prevent cleavage of the backbone by eNDK. Enzymes and substrates used are indicated above lanes. Reaction mixtures were separated on 20% sequencing gels, and the gels were subjected to autoradiography. See text for further details.
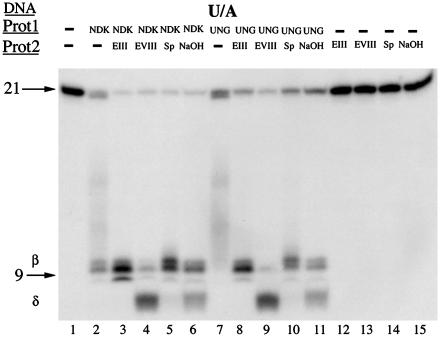
AP Endonuclease Activity of eNDK. Because all known glycosylase/lyases are able to recognize AP sites in DNA as substrates for cleavage (21-23), we expected that eNDK would also cleave AP sites independently of the glycosylase activity. We used a 32P-5′ end-labeled substrate that contains a centrally located AP site (opposite adenine) generated commercially by a glycosylase action (Fig. 3A). We found that such AP substrates were cleaved by eNDK at the expected position ≈9 nucleotides from the 5′ end (Fig. 3, lane 2), confirming that eNDK exhibits AP endonuclease activity independently of the glycosylase reaction. As was the case with the uracil-containing substrates, eNDK produced an additional fragment that migrated faster than the 9-nt product and ahead of the δ-elimination products of EndoVIII and NaOH (lanes 2-8). To a lesser degree, UNG plus EndoIV also generated the shorter fragment (lane 3).
Fig. 3.
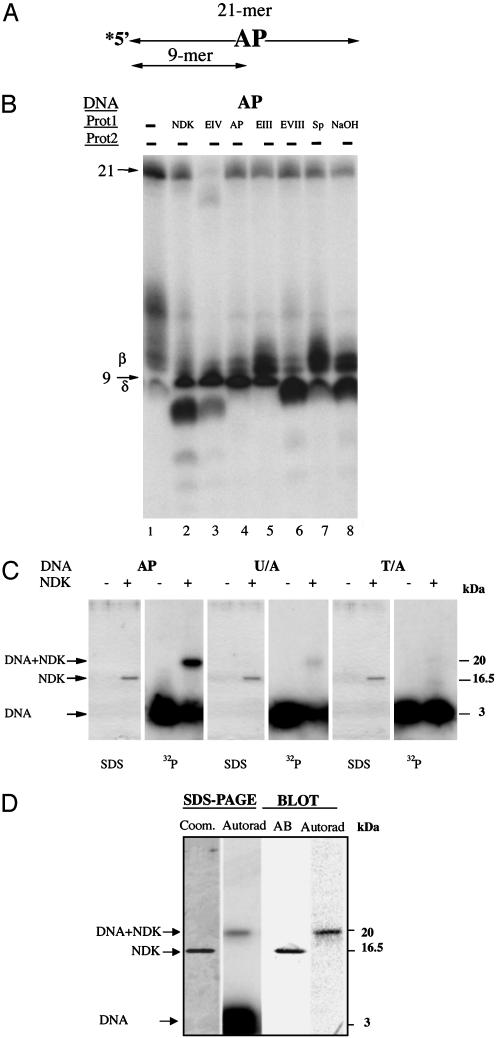
AP endonuclease activity of eNDK. (A) Schematic view of the oligonucleotide substrate used in this experiment, with the complementary strand having A opposite the AP site. Asterisk indicates the radioactively labeled terminus. (B) DNA cleavage analysis of AP-site DNA. Enzymes and substrates used are indicated above lanes. The smeared appearance of AP DNA by itself is because of its instability caused by the collapse of the helix (C). eNDK binds AP DNA covalently. Enzymes and substrates used are indicated above lanes. After reactions were stopped, samples were boiled for 10 min and then separated on SDS/15% PAGE gels followed by Coomassie blue staining and autoradiography. (D) Same as C, except only the AP substrate was used, and a duplicate gel was transferred to a PDF membrane without staining; after development, the membrane was also autoradiographed. Control lanes with eNDK alone in the absence of DNA did not produce radioactive signals (data not shown). See text for further details.
Whether the eNDK AP-site endonuclease activity is hydrolytic or whether it proceeds by β-elimination can be assessed via covalent bond formation with the AP site DNA. The presence of a covalent complex would support the idea that Schiff base is formed as an intermediate in the reaction pathway. If eNDK were a lyase and catalyzed β-elimination, it should form a covalent complex with the aldehyde form of the deoxyribose substrate, as deoxyribose residues at sites of base loss exist in an equilibrium between the open (aldehyde) form and the closed (furanose) form. In the aldehyde form, 3′ phosphodiester bonds are readily hydrolyzed by β-elimination reaction (21-23). As shown in Fig. 3C, eNDK does form an SDS and heat-resistant covalent complex with the AP substrate. This complex has an apparent mass of ≈20 kDa, which is the expected mass of the 16.5-kDa eNDK polypeptide bound to a 21-mer oligonucleotide migrating with an apparent mass of ≈3 kDa in SDS/15% PAGE gels. It was further confirmed by Western Blot analysis that the protein in the shifted band is eNDK covalently bound to DNA (Fig. 3D). This covalent complex formed between the AP site and eNDK strongly suggests β-elimination activity by a lyase mechanism. A weak complex is also formed with the uracil-containing substrate (U/A, Fig. 3C), but not with the WT T/A, which is not cleaved by eNDK (Fig. 1). The difficulty of capturing covalent complexes while the uracil base is still in place is probably because of a high turnover rate of the glycosylation reaction vs. the slower, rate-limiting AP-site cleavage; it is also easier to form a covalent bond with the ring opened up in the aldehyde form (21-23). The association of the cleavage with a uracil-glycosylase activity also strongly argues in favor of a lyase reaction and is consistent with previous findings indicating glycosylase/lyase activity in human h2NDK (10). Whereas eNDK retains the catalytic lysine of human h2NDK, with which we have demonstrated an NaBH4 reduced complex (10), we were unable to show such a complex with eNDK. This may be because of a mechanistic property of eNDK such as that proposed for the MutY glycosylase/lyase, which, although lyase capable, does not readily form an NaBH4-reduced complex (23).
eNDK Exhibits 3′ Repair Phosphodiesterase Activity. As observed above, eNDK cleavage produced a 9-nt-long fragment with a 3′ OH terminus and an additional fragment of as yet unknown composition that migrated faster than the main product by ≈1 nt or less. This shorter fragment was obtained with all substrates used here (U/A, U/G, and AP; Figs. 1 and 3) and also with several other uracil-containing substrates that are unrelated in DNA sequence and nearest neighbor nucleotides to the 21-mer used here, suggesting that its formation is specific for the uracil-processing mechanism of eNDK. To a lesser degree, a similar species is also generated by eEndoIV, an AP endonuclease with 3′ phosphodiesterase, 3′ phosphatase, and, under low salt conditions, 3′-5′ exonuclease activities (24-27). Therefore, one possible source of these ≤8-mer products is a further processing of the 3′-OH end by a 3′ phosphodiesterase or 3′-5′ exonuclease activity. Another possibility that would also be consistent with the migration is the removal of an additional base by the UDG reaction.
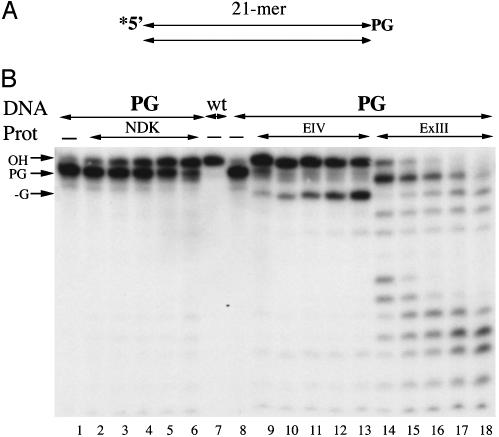
Were eNDK to cleave AP sites in DNA as a lyase generating 3′-α,β-aldehydes, it would also have to possess a 3′ repair phosphodiesterase activity to hydrolyze the sugarphosphate residues left by the α,β-eliminations to end up with a free 3′ hydroxyl. Here, we have identified such a 3′ repair phosphodiesterase activity in eNDK using a substrate with a 3′ PG attached to the 3′ end of the labeled oligonucleotide strand (Fig. 4A). Enzymatic removal of the 3′ PG by a phosphodiesterase would result in a 3′ OH and free phosphoglycolic acid, a reaction that can be monitored in a 20% sequencing gel because the 3′ OH oligonucleotide has a lower electrophoretic mobility than the 3′ PG substrate with the extra phosphoryl group (Fig. 4B). In a time course experiment, eNDK (lanes 2-6) removed 3′ PG efficiently, as judged by a gradual shift up in the mobility of the oligonucleotides. The control enzyme eEndoIV (lanes 9-13) also removed the 3′ PG ends and, in addition, one extra nucleotide catalyzed by its 3′-5′ exonuclease activity (24-28). E. coli ExoIII can also repair 3′ PG residues and sequentially remove nucleotides from the 3′ end (lanes 14-18). We conclude that eNDK possesses a 3′ repair phosphodiesterase activity capable of removing 3′ blocking lesions (e.g., 3′ deoxyribose-5-phosphate or 3′ phosphomonoester) specifically from duplex DNA that may occur following, and related to, the lyase incision reaction (15, 21-23). None of the three enzymes removed 3′ PG from single-stranded DNA (data not shown).
Fig. 4.
eNDK has 3′ repair phosphodiesterase activity. (A) Schematic view of oligonucleotide substrate used in this experiment. Asterisk indicates the radioactively labeled terminus. (B) Time course of phosphoglycolate removal from the 3′ end of the oligonucleotide by eNDK and EndoIV (0-20 min) and ExoIII (0-5 min). Enzymes and substrates used are indicated above lanes. Products are displayed on a 20% urea/PAGE gel. See text for further details.
Discussion
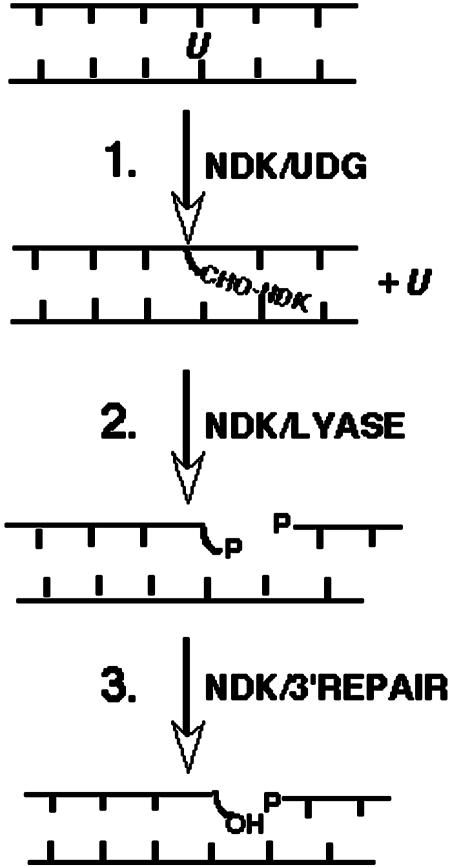
We have previously identified DNA-cleaving activities in several proteins of the NM23/NDK family, including human h2NDK (9-11), h1NDK (8, 9), and eNDK (12). Until now, the most extensively studied mechanism has been that of h2NDK, where a combination of biochemical, mutational, and structural analyses have identified it as a multifunctional BER-like nuclease with combined glcosylase/AP lyase activities, although no specific DNA damage or lesion has yet been identified as its substrate for repair. Because NM23/NDK proteins are highly conserved structurally and functionally as NDKs, and the important catalytic residues of the h2NDK nuclease (Lys-12, Arg-88, and Arg-105) have also been conserved between species, we anticipated that other members of the NM23/NDK family might also share the important features, if not the details, of this DNA repair mechanism (10-12, 14). In this paper, we have identified a DNA repair activity in eNDK whose substrate is uracil misincorporated into DNA. We show that eNDK is a multifunctional DNA glycosylase/AP lyase BER enzyme that acts sequentially by first releasing free uracil base from DNA, then cleaving the DNA backbone, and then further processing the cleaved DNA ends. These steps are outlined schematically in Fig. 5.
Fig. 5.
Schematic of the proposed steps in eNDK-mediated uracil repair. Each step is detailed in the text.
The first step in uracil processing is catalyzed by UDG activity that excises uracil from both U/A and U/G mispairs in DNA. Although E. coli already encodes a UDG enzyme, termed UNG, this enzyme is a simple monofunctional glycosylase with no associated AP lyase activity. Indeed, UNG was the first DNA glycosylase to be discovered (29) and is among the most well characterized biochemically (15, 21-23). eNDK/UDG, like UNG/UDG (30, 31), removes uracil from both double- and single-stranded DNA. The glycosylase activity of eNDK is inhibited by the bacteriophage-encoded Ugi protein, as is the activity of all other known UDG enzymes (32). This indicates that UDGs and eNDK are structurally related with respect to a uracil-specific binding pocket. This pocket is probably different from the nucleotide-binding site used by the NDK reaction because the latter exhibits broad specificity with respect to all nucleosides (1, 3). Sequence and three-dimensional structure alignments of UNG and eNDK have not suggested significant similarities; however, they did show that eNDK and UNG proteins have similar folds, both composed of compactly folded, four-stranded β-sheets surrounded by α-helixes (3, 15), but with the β-strands arranged in different orientations.
The second step of the repair reaction is DNA cleavage. The initial UDG activity suggests that the enzyme first cleaves the 5′ glycosylic bond of the deoxyuridine, then goes on to act as an endonuclease upon this intermediate by making incisions at the AP site created by the removal of uracil. The endonuclease associated with the UDG activity is specific for duplex DNA. NDK also incises intact AP sites in DNA independently of the glycosylase, demonstrating that it is an AP site repair endonuclease.
Whether eNDK processes BER, after removal of uracil, as a class II AP endonuclease through hydrolysis of the phosphodiester bond 5′ to the AP site, or as a class I AP lyase that cleaves the C—O bond 3′ to the AP site by β-elimination, will require further analysis. Whereas pure AP endonucleases, like EndoIV and ExoIII, do not form covalent complexes with their DNA substrates and do not remove a base, AP lyases, like EndoIII and EndoVIII, are bifunctional, acting both as glycosylases and AP lyases and form covalent enzyme-DNA complexes (15, 21-23).
Our current data suggest that the incision activity of eNDK is an AP lyase, both because it is associated with a glycosylase activity and because it forms a covalent enzyme-DNA complex with the AP site DNA. After cleavage by β, or β- and δ-eliminations by a lyase reaction, it seems that the enzyme further processes the cleaved DNA ends by a 3′ repair activity, such as a3′ phosphodiesterase or 3′-5′ exonuclease. The major cleavage product of eNDK seems to have free 3′ OH ends, both because it comigrates with the products of E. coli EndoIV and human AP endonuclease in 20% sequencing gels and because no further processing takes place upon additional incubations with these enzymes whose final products are 3′ hydroxyls. Our data also support the presence of a 3′ repair phosphodiesterase activity in eNDK, which removes 3′ phosphoglycolate residues from double-stranded DNA ends. Under some circumstances, eNDK also shows a weak 3′-5′ exonuclease activity on duplex DNA, e.g., at high protein concentrations, or when the glycosylase activity is not required or is inhibited, e.g., with the AP site substrate, and in the presence of the Ugi inhibitor. Either of these two types of potential exonucleolytic activities of eNDK would be able to remove the 3′ blocking residues generated by a lyase reaction and to provide the 3′ OH ends necessary for repair polymerization and ligation in vivo. The somewhat faster migrating ≈8-nt fragment generated by eNDK may be a product of further processing of the lyase cleaved backbone by the 3′ phosphodiesterase or 3′-5′ exonuclease activity. Another possibility is that eNDK removes a 3′ OH group or a second base next to the uracil; both would be consistent with the migration of these fragments.
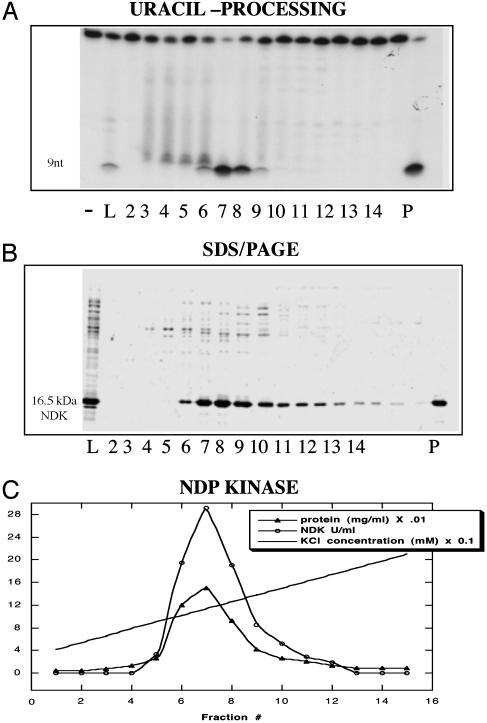
Copurification of the cleavage activity assayed on native EMSA gels with the NDK activity and the NDK protein has suggested previously that these activities reside in the same protein (12). We obtained similar results here on the copurification of the combined uracil-glycosylase/lyase activity with both the NDK protein and the NDK catalytic activity (Fig. 6). It is therefore unlikely that the uracil-processing activity we have described in this paper is because of a contamination by the concerted action of several independently acting trace amounts of nucleases. Moreover, EndoIV and ExoIII, the only known AP endonucleases of E. coli, show dissimilar cleavage specificities from eNDK, have no associated glycosylase activity, are both hydrolytically acting, and do not employ covalent chemistry. Additionally, EndoIV is not present in high copy numbers in E. coli unless induced (22). Nor could the cleavage activity in our eNDK preparations be because of other known lyases in E. coli, because the chemistry dictates that lyases must first act as glycosylases by attacking the same N—O bond. To our knowledge, no glycosylase/lyases exist in E. coli that can recognize uracil. UNG contamination is also unlikely, as UNG has no associated AP lyase activity; a glycosylase must always be associated with a lyase to open up the ring for covalent bond formation. A mutational analysis should further substantiate these newly described eNDK properties.
Fig. 6.
Uracil processing (A), eNDK protein (B), and NDK activity (C) coelute from a DEAE column. Numbers shown under each panel represent protein-containing fractions. L, DEAE load; P, purified eNDK product. Substrate oligonucleotide used in the cleavage assay (A) was U/A.
Additional questions remain. For instance, what is the significance of the association of these repair activities in a single tetrameric protein with a 66-kDa mass? Before this study, a combined UDG glycosylase/AP lyase activity has never been identified in E. coli, or in any other organism. Is the function of eNDK simply to provide redundancy for uracil processing in DNA, or is it nobler? Thus far, only UNG/UDG has been assigned the task of uracil removal in E. coli. The observation that the UDG activity of eNDK is capable of excising uracil when U is paired with either A or G suggests that in vivo eNDK counteracts the consequences of both dUMP misincorporation opposite adenine during DNA replication and of cytosine deamination. A U/G mispair is known to be mutagenic and the mutator phenotype of mutants lacking the monofunctional UNG/UDG has been attributed to failure of deaminated cytosine repair (15, 21-23, 33). However, little is known about the physiological effects of U/A mispairs. The robust U/A and U/G repair activities in eNDK provoke the speculation that eNDK plays the role of a proofreader for errors produced by DNA polymerases during replication and replication-associated DNA repair, which may explain the apparent colocalization of NDK with DNA replicative complexes (2). It is possible, therefore, that the mutator phenotype of eNDK is because of a failure of this proofreading activity and to the lack of a functional DNA repair pathway, rather than to dNTP pool imbalances as has been proposed earlier (2, 19, 20). Future experiments should focus on genetic analyses to assess the functional significance of the NDK-catalyzed DNA repair pathway and of the NDK-catalyzed dNTP phosphate transfer in this process, as well as on the elucidation of its crystal structure complexed with uracil-based substrates. Whether the human enzymes can also release uracil or other bases will be important to determine, given their known involvement in gene regulation and cancer. The observations reported in this paper have offered new insights into the behavior of NM23/NDK proteins in living cells and a potential DNA repair pathway guarding the integrity of the genome.
Acknowledgments
We thank M. Konrad for the ndkec plasmid, I. Lascu for antibody, J. Stock for discussions, and Y. Xu for structural information. This work was supported by National Institutes of Health/National Cancer Institute Grant CA76496 (to E.H.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NDK, nucleoside diphosphate kinase; eNDK, Escherichia coli NDK; BER, base excision repair; UDG, uracil-DNA glycosylase; AP, apyrimidinic; PG, phosphoglycolate.
References
- 1.Lascu, I. & Gonin, P. (2000) J. Bioenerg. Biomembr. 32, 237-246. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, M. A., Ray, N. B., Olcott, M. C., Hendricks, S. P. & Mathews, C. K. (2000) J. Bioenerg. Biomembr. 32, 259-267. [DOI] [PubMed] [Google Scholar]
- 3.Janin, J., Dumas, C., Morera, S., Xu, Y., Meyer, P., Chiadmi, M. & Cherfils, J. (2000) J. Bioenerg. Biomembr. 32, 215-225. [DOI] [PubMed] [Google Scholar]
- 4.Postel, E. H., Berberich, S. J., Flint, S. J. & Ferrone, C. A. (1993) Science 261, 478-480. [DOI] [PubMed] [Google Scholar]
- 5.Berberich, S. J. & Postel, E. H. (1995) Oncogene 10, 2343-2347. [PubMed] [Google Scholar]
- 6.Ji, L., Arcinas, M. & Boxer, L. M. (1995) J. Biol. Chem. 270, 13392-13398. [DOI] [PubMed] [Google Scholar]
- 7.Postel, E. H., Berberich, S. J., Rooney, J. W. & Kaetzel, D. M. (2000) J. Bioenerg. Biomembr. 32, 277-284. [DOI] [PubMed] [Google Scholar]
- 8.Ma, D., Xing, Z., Liu, B., Pedigo, N. G., Zimmer, S. G., Bai, Z., Postel, E. H. & Kaetzel, D. M. (2002) J. Biol. Chem. 277, 1560-1567. [DOI] [PubMed] [Google Scholar]
- 9.Postel, E. H. (1999) J. Biol. Chem. 274, 22821-22829. [DOI] [PubMed] [Google Scholar]
- 10.Postel, E. H., Abramczyk, B. M., Levit, M. N. & Kyin, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postel, E. H., Abramczyk, B. A., Gursky, S. K. & Xu, Y. (2002) Biochemistry 41, 6330-6337. [DOI] [PubMed] [Google Scholar]
- 12.Levit, M. N., Abramczyk, B. M., Stock, J. B. & Postel, E. H. (2002) J. Biol. Chem. 277, 5163-5167. [DOI] [PubMed] [Google Scholar]
- 13.Fan, Z., Beresford, P. J., Oh, D. Y., Zhang, D. & Lieberman, J. (2003) Cell 117, 659-672. [DOI] [PubMed] [Google Scholar]
- 14.Postel, E. H. (2003) J. Bioenerg. Biomembr. 35, 31-40. [DOI] [PubMed] [Google Scholar]
- 15.Krokan, H. E., Standal, R. & Slupphaug, G. (1997) Biochem. J. 325, 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartsough, M. & Steeg, P. S. (2000) J. Bioenerg. Biomembr. 32, 301-308. [DOI] [PubMed] [Google Scholar]
- 17.Lacombe, M.-L., Milon, L., Munier, A., Mehus, J. G. & Lambeth, D. O. (2000) J. Bioenerg. Biomembr. 32, 247-258. [DOI] [PubMed] [Google Scholar]
- 18.Timmons, L. & Shearn, A. (2000) J. Bioenerg. Biomembr. 32, 293-300. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Q., Zhang, X., Almaula, N., Mathews, C. & Inouye, M. (1995) J. Mol. Biol. 254, 337-341. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H., Funchain, P., Clendenin, W., Huang, T., Nguyen, A., Wolff, E., Yeung, A., Chiang, J. H., Garibyan, L., Slupska, M. M. & Yang, H. (2002) Genetics 162, 5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 22.Nickoloff, J. A. & Hoekstra, M. F. (1998) DNA Damage and Repair, Vol. 1: DNA Repair in Prokaryotes and Lower Eukaryotes (Humana, Totowa, NJ).
- 23.McCullough, A. K., Dodson, M. L. & Lloyd, R. S. (1999) Annu. Rev. Biochem. 68, 255-285. [DOI] [PubMed] [Google Scholar]
- 24.Levin, J. D., Johnson, A. W. & Demple, B. (1988) J. Biol. Chem. 263, 8066-8071. [PubMed] [Google Scholar]
- 25.Siwek, B., Bricteux-Gregoire, S., Bailly, V. & Verly, W. G. (1988) Nucleic Acids Res. 16, 5031-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerins, S. M., Collins, R. & McCarthy, T. V. (2003) J. Biol. Chem. 278, 3048-3054. [DOI] [PubMed] [Google Scholar]
- 27.Demple, B., Johnson, A. & Fung, D. (1986) Proc. Natl. Acad. Sci. USA 83, 7731-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mol, C. D., Kuo, C.-F., Thayer, M. M., Cunningham, R. P. & Tainer, J. A. (1995) Nature 374, 381-386. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl, T. (1974) Proc. Natl. Acad. Sci. USA 71, 3649-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosfield, D. J., Daniels, D. S., Mol, C. D., Putnam, C. D., Parikh, S. S. & Tainer, J. A. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 315-347. [DOI] [PubMed] [Google Scholar]
- 31.Parikh, S. S., Putnam, C. D. & Tainer, J. A. (2000) Mutat. Res. 460, 183-199. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Z. & Mosbaugh, D. W. (1989) J. Biol. Chem. 264, 1163-1171. [PubMed] [Google Scholar]
- 33.Duncan, B. K. & Weiss, B. (1982) J. Bacteriol. 151, 750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]